Abstract
The early initiation of appropriate antimicrobial therapy is critical for improving the prognosis of patients with septicemic melioidosis. Thus, the use of a rapid molecular diagnosis may affect the outcome of this disease, which has a high mortality rate. We report the development of two TaqMan real-time PCR assays (designated 8653 and 9438) that detect the presence of two novel genes unique to Burkolderia pseudomallei. The analytical sensitivity and specificity of the assays were assessed with 91 different B. pseudomallei isolates, along with 96 isolates and strains representing 28 other bacterial species, including the closely related Burkholderia/Ralstonia. The two assays performed equally well with both purified DNA and crude cell lysates, with 100% analytical specificity for the detection of B. pseudomallei. The limit of detection was 50 fg of DNA (equivalent to six bacterial genomes) per PCR for both assay 8563 and 9438. We also evaluated these assays with DNA extracted from blood specimens taken from 45 patients with culture-confirmed septicemic melioidosis or other septicemias. Of the 28 melioidosis blood specimens, assays 8653 and 9438 gave sensitivities of 71% (20/28) and 54% (15/28), respectively. Effectively, all fatal cases of septicemic melioidosis were detected by 8653. For the 17 non-melioidosis blood specimens, specificities of 82% (14/17) and 88% (15/17) were obtained for assays 8653 and 9438, respectively. The real-time PCR assays developed in this study provide alternative, rapid molecular tools for the specific detection of B. pseudomallei, and this may be of particular use in the early diagnosis and treatment of septicemic melioidosis.
Burkholderia pseudomallei is a flagellated gram-negative saprophyte commonly found in the soil and water of regions of endemicity such as Southeast Asia and northern Australia. It causes melioidosis, an infection with protean clinical manifestations including acute septicemia, pulmonary infection, and chronic visceral and soft tissue abscesses (32). Infection usually occurs when compromised skin surfaces come into contact with soil or water containing the bacterium (6, 14). A high incidence of severe melioidosis such as acute pneumonia and septicemia can occur through inhalation, and this has been the situation observed particularly during bad weather events and during the heavy rainfall in regions of endemicity (3, 5, 15). There is no licensed vaccine against B. pseudomallei infection. The successful management of patients with severe melioidosis depends on the early diagnosis and initiation of appropriate antimicrobial therapy. It has been reported that 50% of patients with septicemic melioidosis die within 48 h after the onset of symptoms (2, 28), indicating that early diagnosis improves the likelihood of survival.
Currently, the definitive diagnosis of melioidosis relies on bacterial culture. Although this method remains the “gold standard” for pathogen identification, isolation of B. pseudomallei in clinical specimens requires the use of special selective media, and this may require up to 5 days of incubation for a positive result. If enrichment media are used instead, the presence of B. pseudomallei in nonsterile specimens such as sputum and throat swabs may be overlooked due to the speedier growth of other commensal organisms. As a result of these factors, many patients who are clinically suspected of having septicemic melioidosis remain without a culture-confirmed diagnosis, and this increases their likelihood of receiving inappropriate antimicrobial therapy (11, 26). Serological tests to detect B. pseudomallei-specific antibodies such as the indirect hemagglutination assay and enzyme-linked immunosorbent assay have been used clinically, but they are not reliable and useful for the diagnosis of active infections in melioidosis-endemic regions such as Thailand where seroconversion is common (2, 4, 23). B. pseudomallei-specific antigen detection methods using monoclonal antibodies such as the latex agglutination assay and direct immunofluorescence appear to be useful for rapid identification of B. pseudomallei in Thailand, but these tests are not commercially available (21, 24, 33). Commercially available biochemical tests such as API 20NE have been reported to misidentify B. pseudomallei as a number of other bacterial species (12, 16).
The importance of B. pseudomallei both as a lethal pathogen and as a potential biothreat agent (20) has led to the development of various PCR-based detection methods, in both conventional end-point and real-time formats, for specific and rapid identification of the bacterium for earlier diagnosis of melioidosis (7, 8, 9, 13, 17, 18, 27, 29, 30, 34). These PCR assays have targeted 16S rRNA genes, 23S rRNA genes, the flagellin C gene (fliC), the ribosomal protein subunit S21 gene (rspU), a single-nucleotide polymorphism in a putative antibiotic-resistant gene (termed P27), and the (named orf2, orf11, orf13, and SCU2) type III secretion system genes of B. pseudomallei. These reports showed that the real-time PCR assays (17, 18, 27, 29, 30, 34) demonstrated greater analytical sensitivity, rapidity, and ease of use than conventional PCR. However, with the exception of a real-time PCR assay reported more recently that targets orf2 of the TTS1 system, which was evaluated with clinical specimens from patients in Australia (17, 18), the other reported real-time PCR assays were evaluated only with pure bacterial DNA cultures or spiked human blood samples.
In this paper, we report the development of two real-time PCR assays that target novel loci that are both specific only to B. pseudomallei and the evaluation of both assays using DNA extracted from blood specimens collected from 45 patients diagnosed with septicemia caused by B. pseudomallei or other organisms.
MATERIALS AND METHODS
Collection of bacteria.
A total of 187 bacterial isolates and strains were used in this study (Table 1). They included 91 B. pseudomallei isolates, 37 isolates of seven closely related Burkholderia/Ralstonia species (9 B. mallei, 20 B. thailandensis, and 3 B. cepacia isolates and 5 isolates of four other species), as well as 57 isolates of 18 clinically relevant pathogens (including three biothreat agents), and two plant pathogens. Of the 91 B. pseudomallei isolates, 69 were obtained from clinical samples over a period from 1988 to 2004 from various hospitals in Singapore. Thirty-two isolates were obtained during the outbreak of melioidosis in Singapore between January and April in 2004 (15). Other B. pseudomallei isolates included 13 clinical isolates from Thailand, 4 animal isolates (2 each from Singapore and Malaysia), 3 soil isolates from Singapore, and 2 type strains from the American Type Culture Collection (ATCC). The identities of the B. pseudomallei isolates were confirmed by bacterial culture using Ashdown agar medium plates (Bloxwich Pte. Ltd., Singapore) and by biochemical characteristics using API20NE (BioMerieux, Marcy, France).
TABLE 1.
Bacterial strains used in this study to determine the specificity of the real-time PCR assays 8653 and 9438
| Organism (total no.) | Source | Year(s) of collectiona | No. of isolates or strains tested (sample type) | No. of positive PCR results for:
|
|
|---|---|---|---|---|---|
| Assay 8653 | Assay 9438 | ||||
| Burkholderia pseudomallei (n = 91) | Clinical isolates from an outbreak of melioidosis in Singapore | 2004 | 32 (cell lysate) | 32 | 32 |
| Clinical isolates from Singapore | 1993-2002 | 30 (cell lysate) | 30 | 30 | |
| Clinical isolates from Singapore | 1988-1989 | 7 (purified DNA) | 7 | 7 | |
| Animal isolates from Singapore | 1986-1987 | 2 (purified DNA) | 2 | 2 | |
| Soil isolates from Singapore | 1996-2001 | 3 (purified DNA) | 3 | 3 | |
| Animal isolates from Malaysia | 1996 | 2 (purified DNA) | 2 | 2 | |
| Clinical isolate K96243 from Khon Kaen, Thailand | 1996 | 1 (purified DNA) | 1 | 1 | |
| Clinical isolates from Khon Kaen, Thailand | 2006 | 12 (cell lysate) | 12 | 12 | |
| ATCC strains 23343 and 15682 | 1994 | 2 (purified DNA) | 2 | 2 | |
| Burkholderia/Ralstonia (n = 37) | |||||
| Burkholderia mallei | ATCC and NCTC | 1920-1961 | 9 (purified DNA) | 0 | 0 |
| Burkholderia thailandensis | Clinical isolates from Khon Kaen, Thailand | 2006 | 4 (cell lysate) | 0 | 0 |
| Clinical isolate from central and northeast Thailand | 2000 | 1 (purified DNA) | 0 | 0 | |
| Soil isolates from central and northeast Thailand | 2004 | 9 (cell lysate) | 0 | 0 | |
| Soil isolates | 2000 | 5 (purified DNA) | 0 | 0 | |
| ATCC 700388 | 1994 | 1 (purified DNA) | 0 | 0 | |
| Burkholderia cepacia | Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 |
| ATCC strains | 1999 | 2 (purified DNA) | 0 | 0 | |
| Burkholderia gladioli | NCTC12378 | 1999 | 1 (purified DNA) | 0 | 0 |
| Burkholderia caryophylli | ATCC 25418 | 1999 | 1 (purified DNA) | 0 | 0 |
| Ralstonia pickettii | ATCC 27511, NCTC11149 | 1999 | 2 (purified DNA) | 0 | 0 |
| Ralstonia solanacearum | ATCC 33193 | 1997 | 1 (purified DNA) | 0 | 0 |
| Biothreat agents (n = 3) | |||||
| Bacillus anthracis | NCTC8234 | 2000 | 1 (purified DNA) | 0 | 0 |
| Francisella tularensis | NCTC10857 | 2000 | 1 (purified DNA) | 0 | 0 |
| Yersinia pestis | NCTC5923 | 2000 | 1 (purified DNA) | 0 | 0 |
| Clinically relevant bacteria (n = 54) | |||||
| Acinetobacter baumannii | Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 |
| Aeromonas sobria | Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 |
| Campylobacter jejuni | ATCC 29428 | 1999 | 1 (purified DNA) | 0 | 0 |
| Citrobacter (C. freundii and C. koseri) | Clinical isolates from Khon Kaen, Thailand | 2006 | 3 (cell lysate) | 0 | 0 |
| Clostridium difficile | ATCC 9689 | 1999 | 1 (purified DNA) | 0 | 0 |
| Escherichia coli | Clinical isolates from Khon Kaen, Thailand | 2006 | 5 (cell lysate) | 0 | 0 |
| Enterobacter | Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 |
| Klebsiella pneumoniae | Clinical isolates from Khon Kaen, Thailand | 2006 | 9 (cell lysate) | 0 | 0 |
| Pseudomonas aeruginosa | ATCC 27853 | 1997 | 1 (purified DNA) | 0 | 0 |
| Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 | |
| Proteus mirabilis | Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 |
| Salmonella enterica subsp. | Clinical isolates from Khon Kaen, Thailand | 2006 | 19 (cell lysate) | 0 | 0 |
| Staphylococcus aureus | ATCC 25923 | 1997 | 1 (purified DNA) | 0 | 0 |
| Clinical isolates from Khon Kaen, Thailand | 2006 | 6 (cell lysate) | 0 | 0 | |
| Staphylococcus epidermidis | Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 |
| Group B Streptococcus | |||||
| Yersinia enterocolitica | Clinical isolate from Khon Kaen, Thailand | 2006 | 1 (cell lysate) | 0 | 0 |
| ATCC 9610 | 2000 | 1 (purified DNA) | 0 | 0 | |
| Plant pathogen (n = 2) | |||||
| Erwinia amylovora | ATCC 19382 | 1994 | 1 (purified DNA) | 0 | 0 |
| Xanthomonas axonopodis | ATCC 43911 | 1994 | 1 (purified DNA) | 0 | 0 |
The year of collection is not necessarily the same as the actual year in which the isolates or strains were obtained from the original sources; the year given for some strains represents the point when our laboratory received them.
The 96 non-B. pseudomallei bacterial isolates or strains were collected from environmental samples and patients in Thailand (B. thailandensis and clinically relevant bacterial strains) and type strains from ATCC and the National Collection of Type Cultures (NCTC).
Collection of blood specimens from patients.
Blood specimens were collected from 45 hospitalized patients with septicemia in the northeastern region of Thailand in 1999 (see Table 3). These samples were a part of those studied by Suputtamongkol et al. (25). Of the 45 patients, 28 had melioidosis as the final diagnosis, while the other 17 had septicemia caused by six different bacterial species, namely, B. cepacia, Escherichia coli, Enterobacter sp., Klebsiella pneumoniae, Staphylococcus aureus, and Streptococcus sp. Of the specimens obtained from 28 patients with melioidosis, 21 were positive by blood culture assays, and 7 were blood culture negative but had positive cultures from other sites (see Table 3).
TABLE 3.
Evaluation of the real-time PCR assays with DNA extracted from blood specimensa
| Clinical diagnosis and blood specimen | Patient information
|
Mean CT value (no. of CT values used) for:
|
|||
|---|---|---|---|---|---|
| Clinical outcome | Type of positive culture | Assay 8653 | Assay 9438 | Assay for orf11 | |
| Septicemia by Burkholderia pseudomallei | |||||
| BP1 | Survived | B | 32.23 (5) | 32.56 (2) | 30.21 (2) |
| BP2 | Survived | B | 32.68 (5) | 34.28 (7) | 32.13 (2) |
| BP3 | Survived | B/P/TS | 33.80 (5) | 33.54 (6) | 31.28 (2) |
| BP4 | Died | B/TS/U | 30.87 (5) | 33.73 (4) | 33.53 (2) |
| BP5 | Died | B/S/TS | 36.03 (6) | 37.48 (4) | 37.35 (2) |
| BP6 | Survived | B/P | 32.92 (3) | 34.16 (7) | NA |
| BP7 | Died | B/P/S/TS/U | 25.32 (5) | 24.35 (6) | 22.71 (2) |
| BP8 | Survived | P | 34.60 (4) | 38.74 (4) | Neg |
| BP9 | Survived | P | 34.77 (3) | 36.77 (3) | 38.23 (1) |
| BP10 | Died | B/P/S/U | 32.50 (5) | 34.06 (4) | NA |
| BP11 | Survived | B/P/S/U | 33.49 (4) | 37.48 (4) | Neg |
| BP12 | Survived | B/P | 34.64 (5) | 36.90 (7) | 37.07 (2) |
| BP13 | Survived | P/TS | 37.80 (5) | 36.63 (2) | 39.12 (2) |
| BP14 | Died | B | 32.30 (9) | 33.67 (6) | 35.10 (2) |
| BP15 | Survived | B | 35.59 (6) | 34.61 (2) | 42.27 (1) |
| BP16 | Survived | B | 33.64 (2) | Neg | NA |
| BP17 | Survived | B | 33.66 (3) | Neg | 32.96 (2) |
| BP18 | Survived | P | 35.41 (3) | Neg | Neg |
| BP19 | Died | B/S | 34.62 (9) | Neg | 37.53 (2) |
| BP20 | Died | B/TS | 33.02 (2) | Neg | Neg |
| BP21 | Survived | B | Neg | Neg | Neg |
| BP22 | Survived | B/TS | Neg | Neg | Neg |
| BP23 | Survived | P | Neg | Neg | Neg |
| BP24 | Survived | S/TS | Neg | Neg | Neg |
| BP25 | Survived | B/P | Neg | Neg | Neg |
| BP26 | Survived | B | Neg | Neg | Neg |
| BP27 | Survived | P | Neg | Neg | Neg |
| BP28 | Survived | B/P/U | Neg | Neg | Neg |
| Septicemia by Burkholderia cepacia | |||||
| CI1 | Died | B/S | Neg | Neg | Neg |
| Septicemia by Escherichia coli | |||||
| CI2 | Survived | B | Neg | Neg | Neg |
| CI3 | Survived | B | Neg | Neg | Neg |
| CI4 | Survived | B/U | Neg | Neg | Neg |
| CI5 | Survived | B | 33.56 (6) | 33.83 (5) | 39.31 (2) |
| Septicemia by Enterobacter sp. | |||||
| CI6 | Survived | B | Neg | Neg | Neg |
| Septicemia by Klebsiella pneumoniae | |||||
| CI7 | Survived | B | 29.92 (4) | 30.03 (6) | 33.3 (2) |
| CI8 | Died | B/S | Neg | Neg | Neg |
| CI9 | Survived | B/S/U | Neg | Neg | Neg |
| CI10 | Survived | B | Neg | Neg | Neg |
| CI11 | Survived | B | Neg | Neg | Neg |
| Septicemia by Staphylococcus aureus | |||||
| CI12 | Died | B/P/S | Neg | Neg | Neg |
| CI13 | Survived | B | 36.27 (5) | Neg | Neg |
| CI14 | Survived | B/U | Neg | Neg | Neg |
| Septicemia by Streptococcus sp. | |||||
| CI15 | Died | B | Neg | Neg | Neg |
| CI16 | Survived | B/CSF | Neg | Neg | Neg |
| CI17 | Died | B | Neg | Neg | Neg |
Evaluation of the real-time PCR assays was performed with DNA extracted from blood specimens collected from 45 septicemic patients admitted to various hospitals in the northeastern region, Thailand. Abbreviations: B, blood; P, pus; S, sputum; TS, throat swab; U, urine culture; CSF, cerebrospinal fluid culture; Neg, negative amplifications; NA, not enough DNA samples were left for performing these real-time PCRs.
Six milliliters of heparinized blood samples was collected from each patient within 36 h of admission. Buffy coat was obtained by centrifugation at 3,000 rpm for 20 min and stored at −20°C until use.
Preparation of bacterial DNA.
Bacterial isolates were cultured on Luria-Bertani (LB) agar medium at 37°C for 24 to 48 h. A single colony was then inoculated into 10 ml of LB culture broth and incubated at 37°C overnight. Total chromosomal DNA of each isolate was extracted and purified using a QIAGEN Genomic DNA kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The concentration and purity of the DNA preparations were determined by measuring the optical density at 260 and 280 nm, using a spectrophotometer (Ulitrospec 2000; Pharmacia Biotech).
For the preparation of crude bacterial cell lysates, about five bacterial colonies were suspended in 200 μl of distilled water or 1× phosphate-buffered saline and then heat inactivated at 95°C for 15 min. The boiled bacterial suspension was briefly vortexed and spun down at 16,000 × g, and 1 μl of this preparation was used directly for a real-time PCR.
For blood specimens, total DNA was extracted from buffy coat samples using a QIAGEN Mini DNA kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The DNA sample was eluted in 200 μl of elution buffer, and one 5-μl aliquot was used per real-time PCR.
Real-time PCR assays.
The real-time PCR assays, designated assay 8653 and assay 9438, were developed based on previously reported B. pseudomallei-unique sequences (19). The DNA sequences of the primers and probes and the genomic location of the two genes are shown in Table 2. The primers and probes were selected using Primer Express (Applied Biosystem) software, and their genomic locations were analyzed against two complete B. pseudomallei genomes available from the public database (K96243 chromosome 2 [GenBank accession no. NC_006351] and 1710b chromosome II [GenBank accession no. NC_007435]). Each of the probes was labeled with a reporter dye, 6-carboxyfluorescein (FAM), and a quencher dye, 6-carboxytetramethylrhodamine (TAMRA). The expected size of the amplified product for both assays was 81 base pairs.
TABLE 2.
Description of the primers and probes and their genomic locations for the real-time PCR assays developed in this study
| Assay | Sequence of primer and probe (5′-3′) | Locus (or loci) targeted in the complete genome of strains K96243/1710b |
|---|---|---|
| 8653 | Forward ATCGAATCAGGGCGTTCAAG | BPSS1187/BURPS1710b_A0179, encoding a hypothetical protein |
| Reverse CATTCGGTGACGACACGACC | ||
| FAM-CGCCGCAAGACGCCATCGTTCAT-TAMRA | ||
| 9438 | Forward CGATCTCGTCAAGGTGTCGG | BPSS2089/BURPS1710b_A1189, encoding a hypothetical protein |
| Reverse TTGACCTGGATGGCAAAGAAG | ||
| FAM-TTGCCTCAGTCACGCGCACGT-TAMRA |
The 8653 and 9438 real-time PCR assays with purified genomic DNA and crude cell lysates were performed in a Roche LightCycler instrument (Roche Diagnostics, Germany) in the DSO laboratory and in a Rotor Gene 3000 instrument (Corbett Life Science, Australia) in the Centre for Research and Development of Medical Diagnostic Laboratories (CMDL), Khon Kaen University, whereas the two real-time PCR assays with DNA extracted from clinical buffy coat samples were performed only in the Rotor Gene 3000 instrument.
Specificities of the 8653 and 9438 assays were determined with purified genomic DNA or crude cell lysates from the 91 B. pseudomallei isolates and 96 isolates or type strains of 28 other bacterial species. The sensitivity of the real-time PCR assays was determined by using triplicates of 10-fold serial dilutions of the purified K96243 genomic DNA on both a LightCycler and a Rotor Gene 3000 instrument; the limit of detection was obtained by triplicates of twofold serial dilutions of the purified genomic DNA from 100 fg to 12.5 fg per PCR.
A final volume of a 20-μl reaction mixture was performed on the LightCycler. The reaction mixture contained 1× PCR buffer, 3 mM of MgCl2, a 200 μM concentration of each dNTP, 500 nM of forward and reverse primers, 250 nM of probe, 10 μg of bovine serum albumin, and 1 unit of PlatinumTaq DNA polymerase (Invitrogen Life Technology), in addition to 1 μl of DNA template. The reaction mixture for assay 9438 was denatured initially at 95°C for 2 min, followed by 45 cycles of 95°C for 5 s, 51°C for 10 s, and 72°C for 10 s and a final step at 40°C for 20 s. The cycling parameters for assay 8653 were the same, except for the 56°C annealing temperature. The entire cycle for either of the two assays was about 30 min. A final volume of a 25-μl reaction mixture was used on the Rotor Gene 3000. The parameters were initial denaturation at 95°C for 3 min, followed by 45 cycles of 95°C for 30 s, 51°C (for assay 9438) or 56°C (for assay 8653) for 30 s, and 72°C for 30 s. The entire cycle lasted for about 2 hours. All of the real-time PCRs were carried out in duplicates or triplicates, and at least two separate experiments were performed.
Real-time PCR assay targeting orf11 of TTS1.
Performance of the real-time PCR 8653 and 9438 assays was also compared to that described by Thibault et al. (27). The current study was also performed with DNA samples from clinical blood specimens and bacterial cell lysates in a Rotor Gene 3000 instrument. This assay amplifies a 110-base-pair fragment of the orf11 gene of the TTS1 gene cluster. The DNA sequences of the forward and reverse primers and the hydrolysis probe used in this study were as described previously (27), but the PCR was modified as follows: a final volume of a 25-μl reaction mixture contained 2.5 μl of 1× PCR buffer, 3.5 mM of MgCl2, a 200 μM concentration of each dNTP, 12.5 μM of forward and reverse primers, 6.5 μM of probe, and 1 unit of PlatinumTaq DNA polymerase (Invitrogen Life Technology), in addition to 1 μl of DNA template. The reaction mixture was denatured initially at 95°C for 5 min, followed by 45 cycles of 95°C for 10 s and 60°C for 45 s.
Data analysis.
The DNA sequences of the individual primers and probes were compared in silico by using BLAST software against the nucleotide database, and the respective amplified product sequences were analyzed against the Bacterial Genome Database in National Center for Biotechnology Information (NCBI) at the website http://www.ncbi.nlm.nih.gov/BLAST/.
Baseline adjustments and cycle threshold (CT) values of each real-time PCR assay were automatically calculated by the respective LightCycler and Rotor Gene 3000 programs, Roche Molecular Biochemical version 3.5 and Rotor-Gene version 6.0.27. A mean CT value was calculated from the total individual CT values obtained from separate real-time PCRs on that sample.
RESULTS
B. pseudomallei-specific DNA targets.
The individual primers and probes were analyzed using BLAST against the NCBI nucleotide database in order to assess the specificity of the assays in silico. The results showed that the DNA sequences of all of the primers and probes were highly unique to B. pseudomallei, with no significant similarities to other sequences.
Further BLAST analysis against the Bacterial Genome Database also indicated that both 81-bp DNA targets were highly conserved among all of the reported B. pseudomallei genomes, including two completed genomes (strains K96243 and 1710b) and eight unfinished genomes, except for one unfinished B. pseudomallei strain 1655 genome which had no match with the 81-bp target for the 8653 assay. No significant similarities were found between the 81-bp DNA targets and any other closely related Burkholderia/Ralstonia genomes available from the database including nine B. mallei, one B. thailandensis, and five Ralstonia spp. isolates or the existing genomes of other bacterial species.
Analytical specificity.
Assays 8653 and 9438 were 100% specific for the detection of B. pseudomallei; no cross-reactivity occurred with isolates of any of the other 28 bacterial species included as controls in this study. Both assays could also directly detect B. pseudomallei in crude cell lysates, and the quality of PCR amplification was comparable to those obtained with the purified genomic DNA (Table 1). The assays were also tested with DNA extracted from blood samples from two healthy volunteers and showed that the assays had no cross-reactivity with human DNA. Repetition of the two real-time PCR assays confirmed the results were reproducible.
Analytical sensitivity.
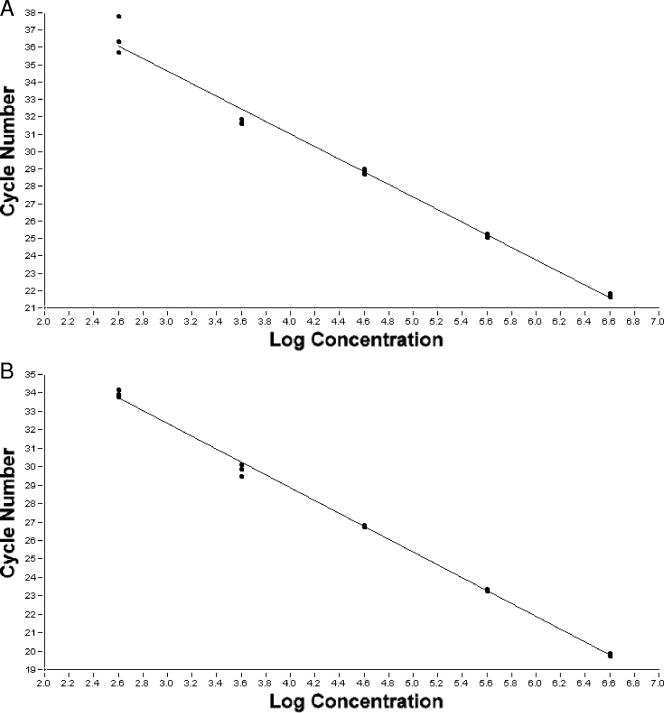
A linear dynamic range of 106 to 102 fg per PCR was observed constantly for both the 8563 and the 9438 assay with both the LightCycler (Fig. 1) and the Rotor Gene 3000 (data not shown), and a detection limit of 50 fg (equivalent to 6 genomes) per PCR was obtained for both assays with both real-time PCR instruments.
FIG. 1.
Linear dynamic range of the real-time PCR assays 8653 (A) and 9438 (B). The assays were performed with triplicates of 10-fold dilutions of the purified genomic DNA extracted from B. pseudomallei strain K96243. The regression line representing the linear range of 106 to 102 fg per PCR was obtained using LightCycler software (Roche Molecular Biochemical, version 3.5) for both assays (A and B), respectively.
Evaluation of assays with clinical blood samples.
The usefulness of the two real-time PCR assays for the diagnosis of melioidosis was evaluated with DNA samples extracted from blood specimens taken from 28 melioidosis cases and 17 non-melioidosis septicemia cases (Table 3). Of the 28 melioidosis blood specimens, 15 were real-time PCR positive with both the 8653 and the 9438 assay, with another 5 blood specimens positive only with the 8653 assay, giving an overall sensitivity of 71% (20/28) and 54% (15/28) for the 8653 and 9438 assays, respectively. Higher PCR sensitivities of 76% (16/21) and 57% (12/21) for both of the real-time PCR assays were observed when the analysis was confined to the 21 culture-positive blood specimens. Seven samples from cases with fatal outcomes all tested positive (100%) on assay 8653, while 5/7 (71%) were positive on 9438. This was observed especially for the blood specimen BP7, with which both real-time PCR assays consistently produced strong amplification and CT values of 25.32 (8653) and 24.35 (9438), which were lower than the CT values produced with all other positive specimens. These were about eight and ten cycles earlier than the average cycle number of 33.54 and 34.60, calculated from the individual mean CT values obtained with assay 8653 and 9438, respectively.
For the 17 blood specimens from non-melioidosis patients, 14 and 15 samples were PCR negative by assay 8653 and 9438, representing an overall specificity of 82% and 88% for the two assays, respectively. The three PCR-positive blood specimens (CI5 and CI7, positive by both 8653 and 9438, and CI13, positive only by 8653) were from respective patients with septicemia caused by one of the three organisms E. coli, K. pneumoniae, and S. aureus (Table 3).
We also compared the performance of our two assays with that of a previously reported real-time PCR assay which detects the B. pseudomallei-specific orf11 of TTS1 (27) with the 45 blood specimens described above. The results are shown in Table 3, and the results obtained with the orf11 assay were similar to those obtained with the 8653 and 9438 assays. This comparison also indicates that assay 8653 is more sensitive than the 9438 and orf11 assays when used with blood specimens. Furthermore, of the three non-melioidosis blood specimens that had false-positive PCR results with 9438 and/or 8653, two specimens (CI5 and CI7) were also positive with the orf11 assay. This suggests that the false-positive results may be due to cross-contamination of these specimens with genomic DNA from B. pseudomallei.
DISCUSSION
Melioidosis is associated with a significant incidence of fatal outcomes in regions of Southeast Asia and northern Australia. More than half of the mortality occurs within the first 48 h of admission and up to 90% for septicemic patients without immediate treatment (1, 2, 10, 28). In regions in which melioidosis is not endemic, B. pseudomallei is also a concern because of its potential release during a bioterror event. Therefore, a rapid and reliable method for specific identification of B. pseudomallei would be useful for early diagnosis and initiation of appropriate antimicrobial therapy.
In this study, two TaqMan real-time PCR assays targeting novel genetic DNA markers that are unique to B. pseudomallei have been developed in parallel for rapid and specific identification of the organism. Our results indicate both assays performed with 100% analytical specificity with purified genomic DNA and crude bacterial lysates of B. pseudomallei, with no cross-reaction observed when samples were tested against a panel of other related bacterial species. Both assays are sensitive analytically, able to detect as few as six genome equivalents constantly in a single reaction when tested with purified genomic DNA. These results indicate that either of the two real-time PCR assays can be used for the rapid and specific verification of B. pseudomallei in crude bacterial cultures or even with materials suspected to contain or contaminated with B. pseudomallei. However, as these two assays target two different B. pseudomallei-specific genes, the two assays could also be used in combination to cross-validate the specificity and hence increase the confidence level for the detection of B. pseudomallei in suspected bacterial cultures and contaminants.
The performance of these assays has also been evaluated with blood specimens from 28 septicemic melioidosis patients admitted to hospitals in a northeastern part of Thailand where B. pseudomallei is a common etiology of septicemia (1). We have shown that a similar set of blood specimens were PCR positive with both assays, but assay 8653 is more sensitive than 9438. The sensitivity of the assays correlates with the clinical information from the patients (clinical outcome and their bacterial culture results). All seven culture-positive blood specimens (7/7) from fatal cases were PCR positive with assay 8653 and 71% (5/7) with assay 9438. Higher sensitivities of 76% and 57% were obtained among the 21 culture-positive blood samples than the overall values of 71% and 54% obtained with 8653 and 9438, respectively. The sensitivity of either 8653 or 9438 is comparable with that of the reported assay targeting orf2 of the TTS1 system, which was evaluated with blood specimens from 33 patients in Australia, and this assay detected 74% or 17% of culture-positive blood specimens with or without septic shock, respectively (17).
The false-negative PCR results for the melioidosis blood specimens could be due to a lower concentration of B. pseudomallei in the blood of the melioidosis patients. It has been reported that the concentration of B. pseudomallei in the blood samples from patients with septicemic melioidosis ranges from 1 to 1,000 CFU/ml (22, 28), and a separate report showed that 45% of septicemic melioidosis patients have less than 1 CFU/ml bacterium in the blood (31). This low level of bacterium in the blood presents a challenge, even to PCR. However, as suggested by our results, even though PCR may not be able to identify all patients with septicemic melioidosis, the assays were positive for all or most of the cases that eventually succumbed to infection. Rapid diagnosis may thus make a difference in these cases.
The assays also appear to have identified three blood specimens as B. pseudomallei positive, although the blood cultures isolated other bacterial species. These false-positive results, however, may be due to cross-contamination of the samples with genomic DNA from B. pseudomallei. Comparison of the results of our two assays with those obtained using the previously reported assay of orf11 of the TTS1 system suggests that of the three false-positive results, at least two are likely to be due to cross-contamination of the specimens with genomic DNA of B. pseudomallei. Alternatively, though unlikely, coinfection may be present in these cases. This comparison involved both the examination of the real-time PCR results and gel electrophoresis analysis of the PCR products which showed DNA fragments of expected sizes amplified from these blood specimens (data not shown).
In conclusion, the use of real-time PCR assays for the rapid and specific detection of B. pseudomallei appears to be promising for clinical use. Large-scale clinical studies of the use of these real-time PCR assays in clinical settings would also be critical to determine the cost effectiveness and degree of improvement to clinical outcome, if any, of such an approach compared to traditional methods in the management of septicemic melioidosis.
Acknowledgments
This work was funded by the Defense Research Directorate, Defense Science and Technology Agency, Singapore, and by the Centre for Research and Development of Medical Diagnostic Laboratories, Khon Kaen University, Thailand. C.L. was a TRF research scholar funded by the Thailand Research Fund, Thailand; G.L. was supported by the Wellcome Trust Research Development Award in Tropical Medicine, United Kingdom (reference number 069426).
We thank the infectious disease physicians and the clinical microbiologists in Singapore and Thailand for providing bacterial isolates and clinical specimens. The clinical data and samples were from a Thailand Research Fund (TRF) project.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Chaowagul, W., N. J. White, D. A. Dance, Y. Wattanagoon, P. Naigowit, T. M. Davis, S. Looareesuwan, and N. Pitakwatchara. 1989. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J. Infect. Dis. 159:890-899. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, A. C., S. P. Jacups, D. Gal, M. Mayo, and B. J. Currie. 2006. Extreme weather events and environmental contamination are associated with case-clusters of melioidosis in the Northern Territory of Australia. Int. J. Epidemiol. 35:323-329. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, A. C., M. O'Brien, K. Freeman, G. Lum, and B. J. Currie. 2006. Indirect hemagglutination assay in patients with melioidosis in northern Australia. Am. J. Trop. Med. Hyg. 74:330-334. [PubMed] [Google Scholar]
- 5.Currie, B. J., and S. P. Jacups. 2003. Intensity of rainfall and severity of melioidosis, Australia. Emerg. Infect. Dis. 9:1538-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dance, D. A. B. 1991. Melioidosis: the tip of the iceberg? Clin. Microbiol. Rev. 4:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dharakul, T., S. Songsivilai, S. Viriyachitra, V. Luangwedchakarn, B. Tassaneetritap, and W. Chaowagul. 1996. Detection of Burkholderia pseudomallei DNA in patients with septicemic melioidosis. J. Clin. Microbiol. 34:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gal, D., M. Mayo, E. Spencer, A. C. Cheng, and B. J. Currie. 2005. Short report: application of a polymerase chain reaction to detect Burkholderia pseudomallei in clinical specimens from patients with suspected melioidosis. Am. J. Trop. Med. Hyg. 73:1162-1164. [PubMed] [Google Scholar]
- 9.Haase, A., M. Brennan, S. Barrett, Y. Wood, S. Huffam, D. O'Brien, and B. Currie. 1998. Evaluation of PCR for diagnosis of melioidosis. J. Clin. Microbiol. 36:1039-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heng, B. H., K. T. Goh, E. H. Yap, H. Loh, and M. Yeo. 1998. Epidemiological surveillance of melioidosis in Singapore. Ann. Acad. Med. Singapore 27:478-484. [PubMed] [Google Scholar]
- 11.Inglis, T. J., D. B. Rolim, and J. L. Rodriguez. 2006. Clinical guideline for diagnosis and management of melioidosis. Rev. Inst. Med. Trop. 48:1-4. [DOI] [PubMed] [Google Scholar]
- 12.Inglis, T. J., D. Chiang, G. S. Lee, and L. Chor-Kiang. 1998. Potential misidentification of Burkholderia pseudomallei by API 20NE. Pathology 30:62-64. [DOI] [PubMed] [Google Scholar]
- 13.Kunakorn, M., K. Raksakait, C. Sethaudom, R. W. Sermswan, and T. Dharakul. 2000. Comparison of three PCR primer sets for diagnosis of septicemic melioidosis. Acta Trop. 74:247-251. [DOI] [PubMed] [Google Scholar]
- 14.Leelarasamee, A., and S. Bovornkitti. 1989. Melioidosis: review and update. Rev. Infect. Dis. 11:413-425. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., J. P. Loh, L. T. Aw, E. P. Yap, M. A. Lee, and E. E. Ooi. 2006. Rapid molecular typing of Burkholderia pseudomallei, isolated in an outbreak of melioidosis in Singapore in 2004, based on variable-number tandem repeats. Trans. R. Soc. Trop. Med. Hyg. 100:687-692. [DOI] [PubMed] [Google Scholar]
- 16.Lowe, P., C. Enger, and R. Norton. 2002. Comparison of automated and nonautomated systems for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 40:4625-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meumann, E. M., D. Gal, R. T. Novak, M. E. Kaestli, M. Mayo, J. P. Hanson, E. Spencer, M. B. Glass, J. E. Gee, P. P. Wilkins, and B. J. Currie. 2006. Clinical evaluation of a type III secretion system real-time PCR assay for diagnosing melioidosis. J. Clin. Microbiol. 44:3028-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novak, R. T., M. B. Glass, J. E. Gee, D. Gal, M. J. Mayo, B. J. Currie, and P. P. Wilkins. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J. Clin. Microbiol. 44:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong, C., C. H. Ooi, D. Wang, H. Chong, K. C. Ng, F. Rodrigues, M. A. Lee, N. Pitakwatchara, and P. Tan. 2004. Patterns of large-scale genomic variation in virulent and avirulent Burkholderia species. Genome Res. 14:2295-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Report summary. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samosornsok, N., A. Lulitanond, N. Saenla, N. Anuntagool, S. Wongratanacheewin, and S. Sirisinha. 1999. Sort report: evaluation of a monoclonal antibody-based latex agglutination test for rapid diagnosis of septicemic melioidosis. Am. J. Trop. Med. Hyg. 61:735-737. [DOI] [PubMed] [Google Scholar]
- 22.Simpson, A. J., P. A. Howe, V. Wuthiekanun, and N. J. White. 1999. A comparison of lysis centrifugation, pour plate, and conventional blood culture methods in the diagnosis of septicaemic melioidosis. J. Clin. Pathol. 52:616-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirisinha, S., N. Anuntagool, T. Dharakul, P. Ekpo, S. Wongratanacheewin, P. Naigowit, B. Petchclai, V. Thamlikitkul, and Y. Suputtamongkol. 2000. Recent developments in laboratory diagnosis of melioidosis. Acta Trop. 74:235-245. [DOI] [PubMed] [Google Scholar]
- 24.Steinmetz, I., A. Reganzerowski, B. Brenneke, S. Haussler, A. Simpson, and N. J. White. 1999. Rapid identification of Burkholderia pseudomallei by latex agglutination based on an exopolysaccharide-specific monoclonal antibody. J. Clin. Microbiol. 37:225-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suputtamongkol, Y., W. Chaowagul, P. Chetchotisakd, N. Lertpatanasuwun, S. Intaranongpai, T. Ruchutrakool, D. Budhsarawong, P. Mootsikapun, V. Wuthiekanun, N. Teerawatasook, and A. Lulitanond. 1999. Risk factors for melioidosis and bacteremic melioidosis. Clin. Infect. Dis. 29:408-413. [DOI] [PubMed] [Google Scholar]
- 26.Suputtamongkol, Y., A. Rajchanuwong, W. Chaowagul, D. A. B. Dance, M. D. Smith, V. Wuthiekanun, A. L. Walsh, S. Pukrittayakamee, and N. J. White. 1994. Ceftazidime versus amozicillin/clavulanate in the treatment of severe melioidosis. Clin. Infect. Dis. 19:846-853. [DOI] [PubMed] [Google Scholar]
- 27.Thibault, F. M., E. Valade, and D. R. Vidal. 2004. Identification and discrimination of Burkholderia pseudomallei, B. mallei, and B. thailandensis by real-time PCR targeting type III secretion system genes. J. Clin. Microbiol. 42:5871-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiangpitayakorn, C., S. Songsivilai, N. Piyasangthong, and T. Dharakul. 1997. Speed of detection of Burkholderia pseudomallei in blood cultures and its correlation with the clinical outcome. Am. J. Trop. Med. Hyg. 57:96-99. [DOI] [PubMed] [Google Scholar]
- 29.Tomaso, H., T. L. Pitt, O. Landt, A. S. Dahouk, H. C. Scholz, E. C. Reisinger, L. D. Sprague, I. Rathmann, and H. Neubauer. 2005. Rapid presumptive identification of Burkholderia pseudomallei with real-time PCR assays using fluorescent hybridization probes. Mol. Cell Probes 19:9-20. [DOI] [PubMed] [Google Scholar]
- 30.U'Ren, J. M., M. N. Van Ert, J. M. Schupp, W. R. Easterday, T. S. Simonson, R. T. Okinaka, T. Pearson, and P. Keim. 2005. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 43:5771-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh, A. L., M. D. Smith, V. Wuthiekanun, Y. Suputtamongkol, W. Chaowagul, D. A. Dance, B. Angus, and N. J. White. 1995. Prognostic significance of quantitative bacteremia in septicemic melioidosis. Clin. Infect. Dis. 21:1498-1500. [DOI] [PubMed] [Google Scholar]
- 32.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 33.Wuthiekanum, V., V. Desakorn, G. Wongsuvan, P. Amornchai, A. C. Cheng, B. Maharjan, D. Limmathurotsakul, W. Chierakul, N. J. White, N. P. Day, and S. J. Peacock. 2005. Rapid immunofluorescence microscopy for diagnosis of melioidosis. Clin. Diagn. Lab. Immunol. 12:555-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap, E. P. H., S. M. Ang, S. G. K. Seah, and S. M. Phang. 2002. Homogenous assays for the rapid PCR detection of Burkholderia pseudomallei 16S rRNA gene on a real-time fluorometric capillary thermocycler, p. 59-70. In U. Reischl, C. Wittwer, and F. Cockerill (ed.), Rapid cycle real-time PCR—methods and applications: microbiology and food analysis. Springer, New York, NY.