Abstract
Sole reliance on biochemical methods can limit the clinical microbiology laboratory's ability to identify bacterial pathogens. This study describes the incorporation of DNA pyrosequencing-based identification for routine pathogen identification of atypical clinical isolates in a large children's hospital. The assay capitalized on the highly conserved nature of 16S rRNA genes by positioning amplification and sequencing primers in conserved target sequences flanking the variable V1 and V3 regions. A total of 414 isolates of 312 pediatric patients were tested by DNA pyrosequencing during the time period from December 2003 to July 2006. Seventy-eight different genera were specified by DNA pyrosequencing, and isolates were derived from diverse specimen types. By integrating DNA sequencing of bacterial pathogens with conventional microbiologic methods, isolates that lacked a definitive identification by biochemical testing yielded genus- or species-level identifications in approximately 90% of cases by pyrosequencing. Improvements incorporated into the assay process during the period of clinical testing included software enhancements, improvements in sequencing reagents, and refinements in database search strategies. Coupled with isolation by bacteriologic culture and biochemical testing, DNA pyrosequencing-based bacterial identification was a valuable tool that markedly improved bacterial pathogen identification in a pediatric hospital setting.
Bacterial pathogens may not be identified in the clinical laboratory by routine morphological and biochemical methods. Difficult-to-identify pathogens may yield inconsistent or inconclusive results by biochemical testing with either manual or automated methods (5, 28). Molecular methods provide microbiologists with additional tools that may supplement biochemical testing for bacterial pathogen identification (29). In contrast to targeted molecular methods such as real-time PCR, global molecular approaches such as DNA sequencing offer attractive strategies for the identification of unknown pathogens from clinical specimens. Uncultured bacterial pathogens have been identified directly in human tissue by dideoxy DNA sequencing and have previously highlighted the importance of sequence-based identification (2, 25). Sanger or dideoxy DNA sequencing of all or part of the 16S rRNA gene has been commonly used for pathogen identification of cultured organisms in many studies (18). However, commercial kits for diagnostic sequencing have had limited impact on clinical laboratories. The implementation of DNA sequencing for routine pathogen identification has been limited by the technical demands and labor-intensive nature of dideoxy sequencing methods.
The implementation of DNA sequencing in diverse clinical laboratory settings requires the availability of user-friendly technologies that minimize labor and maximize cost-effectiveness. DNA pyrosequencing, or sequencing by synthesis, was first introduced in 1996 as a rapid and less expensive alternative to traditional Sanger DNA sequencing (27). Since its inception in the mid-1990s, DNA pyrosequencing assays have been developed for diverse applications, including genotyping, single nucleotide polymorphism detection, and microorganism identification (21). Pyrosequencing has been used to detect point mutations in antiviral or antimicrobial resistance genes as a strategy for molecular resistance testing (12, 20, 34). Pyrosequencing has been applied to organism identification by combining short-stretch DNA sequencing with signature matching in the well-characterized phylogenetic target, the 16S rRNA gene (14, 30), in addition to in a variety of target genes in bacteria (9, 13, 24, 32). Although pyrosequencing yields limited amounts of DNA sequence information, highly informative target sequences within the 16S rRNA gene facilitated the identification of pathogens, such as Helicobacter pylori (23) and Mycobacterium species (31). Pyrosequencing of the 16S rRNA gene was also used to develop a “molecular Gram stain” in order to rapidly classify bacteria as gram positive or gram negative by using molecular methods (16). A follow-up study (17) documented that molecular Gram stain results agreed with culture results in 85.7% of cases versus agreement in only 35.7% of cases with conventional Gram stains. Pyrosequencing also categorized pathogens associated with cases of neonatal sepsis based on group-specific signature sequences (15).
Microbial DNA sequencing applications often target regions within 16S rRNA genes for broad-range identification of different groups or individual species. Bacterial 16S rRNA genes consist of eight highly conserved and nine variable regions (33). V1 and V3 represent two distinct variable regions within the 16S rRNA gene, and these regions are the targets for the pyrosequencing-based identification assay presented in this study. The assay capitalizes on the highly conserved nature of 16S rRNA genes by positioning amplification and sequencing primers in the conserved regions flanking variable regions, specifically V1 and V3, thereby allowing primers to theoretically amplify most bacterial pathogens. The primers targeting the V1 and V3 regions were originally developed for pyrosequencing-based classification of bacteria using 10 nucleotides from each region (14), and the same primers were also utilized for the detection of bacterial contamination of water samples used in PCRs (10).
The molecular microbiology laboratory at Texas Children's Hospital implemented routine DNA pyrosequencing based on previously described parameters (14) in order to identify clinical isolates refractory to biochemical identification. DNA pyrosequencing-based identification coupled with culture findings and biochemical results provided a robust approach for more accurate identification of bacterial pathogens. The data presented here included a total of 414 patient isolates evaluated during a 31-month time period from December 2003 to July 2006. This polyphasic strategy facilitated a cost-effective approach for improved diagnostic bacteriology services by integrating DNA sequencing with conventional methods in the clinical laboratory.
MATERIALS AND METHODS
Selection of bacterial isolates.
Specimens were obtained from samples submitted to the diagnostic microbiology laboratory at Texas Children's Hospital (Houston, TX) for routine culture. The bacterial isolates were derived from a variety of specimen types and were submitted for DNA pyrosequencing if organisms were refractory to definitive biochemical identification by manual or automated methods. A total of 414 bacterial isolates were not identified by routine biochemical testing, including by automated (VITEK Legacy; bioMérieux, Inc., France) and manual methods (API 20 E and API 20 NE strips; bioMérieux). Samples were deemed refractory if a definitive identification could not be made (<90% probability-based identification) or if discrepant results were obtained by biochemical and morphological evaluation. Suspected Burkholderia cepacia isolates were automatically submitted for confirmation by pyrosequencing. Mycobacterial and fungal isolates were excluded from consideration in this study.
DNA extraction.
Pure bacterial cultures were submitted for DNA extraction on plated media, and suspected mixed cultures were purified or rejected. Bacterial DNA was extracted with the Mo Bio UltraClean microbial DNA kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. A 10-μl inoculating loop of bacteria served as starting material for the extraction, and chromosomal DNA was eluted in a final volume of 35 μl elution buffer. DNA quantitation was performed by absorbance spectrophotometry of purified DNA in the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
DNA amplification.
Separate PCRs were amplified for the V1 and V3 regions. Each 50-μl reaction mixture consisted of 0.8 mM deoxynucleoside triphosphates (Applied Biosystems, Foster City, CA), 2.5 mM MgCl2 (Applied Biosystems), GeneAmp 10× PCR Gold buffer (Applied Biosystems), 0.2 μM of each primer, 1.25 U AmpliTaq Gold DNA polymerase LD (Applied Biosystems), and 10 nanograms of bacterial DNA. Previously described primers were used to amplify the V1 and V3 regions of 16S rRNA genes (14). Nucleotide positions refer to positions in the Escherichia coli 16S rRNA gene. Bio-pBR5 (positions 6 to 27, 5′-biotin-GAAGAGTTTGATCATGGCTCAG-3′) and pBR-V1 (positions 120 to 101, 5′-TTACTCACCCGTCCGCCACT-3′) were used for V1 amplification, and Bio-B-V3 (positions 1047 to 1027, 5′-biotin-ACGACAGCCATGCAGCACCT-3′) and pJBS.V3 (positions 947 to 967, 5′-GCAACGCGAAGAACCTTACC-3′) were used for V3 amplification. PCR was performed in the GeneAmp PCR system 9700 (Applied Biosystems) under the following cycling parameters: 10 min at 95°C, 35 cycles of 95°C for 40 s, 55°C for 40 s, and 72°C for 60 s, followed by a single cycle of 72°C for 60 s.
DNA pyrosequencing.
The amplified products for V1 and V3 were prepared for pyrosequencing by using the recommended protocol for the vacuum prep tool (Biotage AB, Uppsala, Sweden). For each reaction, 40 μl of the biotinylated PCR product was used in the preparation. To prepare the sequencing plate, purified PCR products were resuspended in 40 μl of annealing buffer with 0.3 μM sequencing primer. Primers pBR-V1 and pJBS.V3 were used as DNA sequencing primers for the V1 and V3 regions, respectively. Pyrosequencing was originally performed on the PSQMA instrument (Biotage) using the PSQ SQA kit, but transition to the PSQ Gold SQA reagent kit was made on March 16, 2005.
Organism identification.
The resulting DNA pyrograms were automatically analyzed by the PSQMA software (version 2.1). Initially, all pyrograms were manually reviewed and the reviewer determined the number of bases that were used in the subsequent search. In December 2005, the process was revised so that the bases that were assigned “good quality” or “check quality” by the software were automatically used in the search, with a manual review of all bases utilized when no clear match was observed. The selected bases were used to search the Ribosomal Database Project (RDP), version 9.36 (http://rdp.cme.msu.edu/) (3). A minimum of 15 bases in at least one of the two regions (V1 and V3) was required to proceed in searching the RDP database. The sequence match tool of the RDP website was used with search preferences assigned to type strains, sequences from individual isolates, nearly full-length sequences (>1,200 bases), and “good quality” sequences. Organisms with a 100% DNA sequence match were compared with the microbiologic findings, such as morphological and biochemical features, and final bacterial identifications were established using this polyphasic strategy.
Sanger sequencing of the 16S rRNA gene.
A subset of clinical isolates evaluated by DNA pyrosequencing was also studied by conventional Sanger (dideoxy) sequencing. The oligonucleotide primers 16S 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 16S 1541R (5′-AAGGAGGTGATCCAGCCGCA-3′) were used for PCR amplification of a 1,533-bp amplicon. Following PCR amplification, the 16S 8F oligonucleotide primer was employed for DNA sequencing by Seqwright DNA Technology Services (Houston, TX). Resulting sequences were truncated for maximum quality and searched using the RDP and GenBank databases.
RESULTS
Summary of organisms identified by DNA pyrosequencing.
A total of 414 cultured bacterial isolates from 312 patients at Texas Children's Hospital were evaluated by DNA pyrosequencing between December 2003 and July 2006. During the initial phase of assay implementation, duplicate isolates from the same patient and same specimen were tested, and identical identifications were obtained. Of the 414 isolates tested, 372 isolates (approximately 90%) were definitively identified by DNA-pyrosequencing-based identification, with 364 of 372 isolates (98%) identified to the genus level and 189 of 372 isolates (51%) identified to the species level. All isolates identified by DNA pyrosequencing were considered potentially clinically significant and would have been reported by routine microbiological methods. The bacterial isolates represented 78 different genera and reflected the high degree of biological diversity in this atypical group (Table 1). By Gram stain morphology, bacilli (gram negative or gram positive) were predominant in number and indicated that rod-shaped organisms were often difficult to identify in the clinical laboratory. Of the 372 isolates definitively identified by DNA pyrosequencing, potential pathogens included 217 gram-negative bacilli (58%), 92 gram-positive bacilli (24%), 35 gram-positive cocci (10%), 14 gram-negative cocci (4%), and 14 miscellaneous organisms (4%). The miscellaneous group included gram-variable, coccobacillary, or other variable-morphology organisms. The 372 suspected pathogens identified by DNA pyrosequencing included isolates from 157 peripheral blood specimens (42%), 130 respiratory specimens (35%), 60 miscellaneous (wounds and superficial sites) specimens (16%), 17 gastrointestinal specimens (5%), and 8 cerebrospinal fluid specimens (2%). Potential pathogens from 157 peripheral blood specimens that yielded pyrosequencing-based identifications included 66 gram-positive bacilli (42%), 52 gram-negative bacilli (33%), 26 gram-positive cocci (16%), 9 miscellaneous organisms (6%), and 4 gram-negative cocci (3%). Notably, gram-positive bacteria, such as Microbacterium and Rothia spp., were identified multiple times by DNA pyrosequencing, while yielding ambiguous results by biochemical testing (Table 1). Organisms were identified from 130 different respiratory tract specimens, including 80 sputum samples (62%), 21 tracheal aspirates (16%), 17 bronchoalveolar lavages (13%), and 12 other respiratory specimens (9%). The organisms identified from respiratory tract specimens included 118 gram-negative bacilli (91%), 6 gram-negative cocci (4%), 3 gram-positive bacilli (2%), 2 gram-positive cocci (2%), and 1 miscellaneous organism (1%).
TABLE 1.
Summary of organisms identified by DNA pyrosequencing grouped by morphology and sorted by frequencya
| Morphology and organism | No. of isolates |
|---|---|
| Gram-negative cocci or coccobacilli | |
| Neisseria spp. not meningitidis/gonorrhoeae | 7 |
| Moraxella spp. | 6 |
| Gram-positive cocci | |
| Kocuria kristinae | 4 |
| Staphylococcus aureus | 4 |
| Leuconostoc spp. | 3 |
| Streptococcus intermedius | 3 |
| Coagulase-negative Staphylococcus | 2 |
| Rothia mucilaginosa | 2 |
| Staphylococcus spp., but not Staphylococcus aureus | 2 |
| Gram-positive bacilli | |
| Microbacterium spp. | 11 |
| Bacillus spp. but not Bacillus anthracis or Bacillus cereus | 9 |
| Microbacterium/Cellulomonas spp. | 8 |
| Actinomyces spp. | 7 |
| Corynebacterium spp. | 5 |
| Bacillus mycoides | 4 |
| Actinomyces odontolyticus | 3 |
| Cellulomonas spp. | 3 |
| Clostridium tertium | 3 |
| Propionibacterium spp. | 3 |
| Arthrobacter spp. | 2 |
| Bacillus cereus | 2 |
| Bacillus spp. | 3 |
| Brevibacterium casei | 2 |
| Curtobacterium spp. | 2 |
| Exiguobacterium acetyliticum | 2 |
| Paenibacillus spp. | 2 |
| Propionibacterium propionicum | 2 |
| Gram-negative bacilli or coccobacilli | |
| Pseudomonas aeruginosa | 33 |
| Pseudomonas spp. | 16 |
| Achromobacter xylosoxidans | 11 |
| Campylobacter coli | 10 |
| Stenotrophomonas maltophilia | 10 |
| Acinetobacter spp. | 9 |
| Burkholderia cepacia complex | 9 |
| Burkholderia spp. | 6 |
| E. coli | 6 |
| Achromobacter ruhlandii | 5 |
| Achromobacter spp. | 5 |
| Brevundimonas spp. | 4 |
| Klebsiella pneumoniae | 4 |
| Pseudomonas pseudoalcaligenes | 4 |
| Alcaligenes spp. | 3 |
| Burkholderia plantarii | 3 |
| Cupriavidus respiraculi | 3 |
| Klebsiella spp. | 3 |
| Neisseria elongata | 3 |
| Acinetobacter lwoffii | 2 |
| Actinobacillus actinomycetemcomitans | 2 |
| Burkholderia multivorans | 2 |
| Capnocytophaga spp. | 2 |
| Eikenella spp. | 2 |
| Enterobacter cloacae | 2 |
| Enterobacter spp. | 2 |
| Escherichia spp. | 2 |
| Flavobacterium spp. | 2 |
| Haemophilus influenzae | 2 |
| Kingella spp. | 2 |
| Neisseria weaveri | 2 |
| Pasteurella spp. | 2 |
| Pseudomonas spp., but not Pseudomonas aeruginosa | 2 |
| Ralstonia spp. | 2 |
| Shewanella spp. | 2 |
| Vibrio cholerae or Vibrio mimicus | 2 |
| Miscellaneous organisms | |
| Aeromonas spp. | 2 |
| Rhodococcus corynebacterioides | 2 |
| Rothia spp. | 2 |
Only organisms with two or more isolates have been included. Seventy-nine organisms yielded only single isolates. See the supplemental material for a complete list of the organisms identified.
Summary of isolates not identified by DNA pyrosequencing.
Of the 414 samples, 38 (9%) isolates could not be identified by DNA pyrosequencing. Isolates that were refractory to definitive molecular identification included organisms derived from 17 peripheral blood specimens (45%), 10 respiratory specimens (26%), 9 miscellaneous specimens (24%), and 2 gastrointestinal specimens (5%). Sixteen (42%) isolates belonging to this refractory group yielded fewer than 15 bases of adequate quality in the V1 and V3 regions of 16S rRNA genes. Gram stain morphologies of isolates in this group included 18 gram-negative bacilli (47%), 10 miscellaneous isolates (26%), 4 gram-positive bacilli (11%), 4 gram-positive cocci (11%), and 2 gram-negative cocci (5%).
All 38 isolates not identified by DNA pyrosequencing were submitted for Sanger (dideoxy) DNA sequencing. Eleven isolates either produced no sequence after multiple attempts or provided no significant matches. Eight isolates provided matches to two genera, 5 isolates were identified to the genus level, and 14 isolates were identified to the species level. Table 2 provides a list of organisms identified from this group of 38 isolates not identified by DNA pyrosequencing.
TABLE 2.
Organisms identified by Sanger sequencing of the 16S rRNA gene that could not be identified by DNA pyrosequencing
| Organism per Sanger sequencing | No. of isolates |
|---|---|
| Achromobacter xylosoxidans | 1 |
| Clostridium propionicum | 1 |
| Cupriavidus respiraculi | 1 |
| Enterobacter hormaechei | 2 |
| Enterobacter or Pantoea spp. | 3 |
| Escherichia or Shigella spp. | 2 |
| Kingella or Neisseria spp. | 2 |
| Klebsiella oxytoca | 1 |
| Klebsiella pneumoniae | 1 |
| Kocuria kristinae | 2 |
| Leucobacter komagatae | 1 |
| Microbacterium spp. | 1 |
| Neisseria meningitidis | 1 |
| Pseudomonas or Stenotrophomonas spp. | 1 |
| Pseudomonas spp. | 1 |
| Rhodococcus spp. | 1 |
| Schineria spp. | 1 |
| Serratia marcescens | 1 |
| Sporosarcina spp. | 1 |
| Williamsia serinedens | 1 |
| Xenophilus azovorans | 1 |
Only 4 of 414 isolates yielded discordant identifications. In one instance, pyrosequencing provided a match to only Neisseria spp., and this result was inconsistent with microbial morphological and biochemical data. The remaining three samples included three different specimens from one patient. The DNA sequence obtained for the three isolates matched the only known sequence of Mycobacterium gordonae, but the smear findings were not consistent with acid-fast bacilli. The isolate was identified as Rhodococcus equi by extensive phenotypic studies. DNA pyrosequencing was repeated on all three isolates, and resequencing confirmed that the organisms were isolates of Rhodococcus equi. The initial error was due to misinterpretation of homopolymeric length of a polycytosine residue tract. The sequencing software originally determined the peak to consist of three cytosine residues and resulted in an exact match with Mycobacterium gordonae. However, repeated pyrosequencing correctly determined the peak to consist of four cytosine residues and resulted in an exact match with Rhodococcus equi, the organism that correlated best with conventional microbiologic data.
Correlation of DNA pyrosequencing data with data obtained by Sanger DNA (dideoxy) sequencing.
A total of 27 isolates previously identified by DNA pyrosequencing were submitted for Sanger (dideoxy) DNA sequencing, but informative sequences were obtained for 26 of 27 isolates. For this group of isolates, the average number of bases obtained by Sanger sequencing was 601, and this number contrasts with the relatively low average of 63 bases (V1 plus V3) for the final 4-month period of this pyrosequencing study. Comparative DNA sequencing results obtained by pyrosequencing and Sanger sequencing are presented in Table 3. When DNA pyrosequencing yielded identifications and Sanger sequence was obtained (n = 26), the results matched the genera (26 of 26) obtained with Sanger sequencing in 100% of the cases examined (Table 3).
TABLE 3.
Comparisons of DNA pyrosequencing-based identifications and results obtained by Sanger sequencing of the 16S rRNA gene
| Organism per pyrosequencing | Organism per Sanger sequencing | % Agreement |
|---|---|---|
| Achromobacter spp. | Achromobacter xylosoxidans | 100 |
| Burkholderia cepacia complex | Burkholderia spp. | 100 |
| Burkholderia spp. | Burkholderia spp. | 100 |
| Burkholderia spp. | Burkholderia spp. | 100 |
| Campylobacter coli | Campylobacter jejuni/ Campylobacter coli | 100 |
| Corynebacterium spp. | Corynebacterium pseudodiphtheriticum/ Corynebacterium propinquum | 100 |
| Curtobacterium spp. | Curtobacterium spp. | 100 |
| Eggerthella lenta | Eggerthella lenta | 99 |
| Kocuria kristinae | Kocuria kristinae | 97 |
| Microbacterium spp. | Microbacterium hominis | 98 |
| Microbacterium spp. | Microbacterium oxydans/ Microbacterium maritypicum | 99 |
| Moraxella spp. | Moraxella nonliquefaciens | 99 |
| Neisseria elongata | Neisseria elongata | 98 |
| Neisseria weaveri | Neisseria weaveri | 99 |
| Ochrobactrum tritici | Ochrobactrum anthropi | 100 |
| Pseudomonas aeruginosa | Pseudomonas aeruginosa | 100 |
| Pseudomonas aeruginosa | Pseudomonas alcaliphila | 100 |
| Rhodococcus corynebacterioides | Rhodococcus corynebacterioides | 99 |
| Rhodococcus equi | Rhodococcus equi | 99 |
| Rothia mucilaginosa | Rothia mucilaginosa | 93 |
| Salmonella enterica serovar Typhi | Salmonella enterica serovar Typhi | 100 |
| Shewanella spp. | Shewanella spp. | 100 |
| Staphylococcus aureus | Staphylococcus aureus | 100 |
| Staphylococcus spp. not aureus | Staphylococcus sciuri | 100 |
| Streptococcus mitis group | Streptococcus mitis/Streptococcus infantis | 100 |
| Streptococcus thermophilus | Streptococcus salivarius/thermophilus | 99 |
Two bacterial isolates yielded different species identifications by Sanger sequencing versus DNA pyrosequencing, although the genus identifications were identical in both cases. One isolate obtained from peripheral blood was identified as Ochrobactrum anthropi by Sanger sequencing. Although O. anthropi was an exact match by pyrosequencing, the polyphasic strategy that included pyrosequencing, microbial morphology, and biochemical data supported the identification of Ochrobactrum tritici. Another isolate was identified as Pseudomonas aeruginosa by pyrosequencing but yielded the species designation Pseudomonas alcaliphila by dideoxy sequencing. The reference sequence of P. alcaliphila also presented an exact match with the pyrosequencing data, but the polyphasic strategy combining pyrosequencing and biochemical data yielded an identification of P. aeruginosa. This example highlights the challenge of identifying atypical P. aeruginosa isolates from patients with cystic fibrosis, as others have noted (7). Accurate species identification of P. aeruginosa in cystic fibrosis patients has important therapeutic implications, and 33 P. aeruginosa isolates were identified by DNA pyrosequencing in the absence of reliable biochemical identification (Table 1). As expected, most of the P. aeruginosa isolates submitted for DNA pyrosequencing in this study were cultured from respiratory tract specimens of children with cystic fibrosis.
Performance improvements with advances in pyrosequencing chemistry and informatics.
Several improvements to the pyrosequencing methodology have been incorporated since the assay implementation in December 2003. Improvements occurring during the time period of the study included the introduction of enhanced assay reagents (PSQ Gold; Biotage, Inc.), a key sequencing software upgrade (version 2.1 PSQ 96MA; Biotage, Inc.), and improvements in the search engine of the RDP. The average number of cumulative bases per isolate was 61 for the entire study (V1 plus V3 sequence data), and Fig. 1A depicts the changes in the mean numbers of cumulative bases per 4-month interval throughout the course of the study.
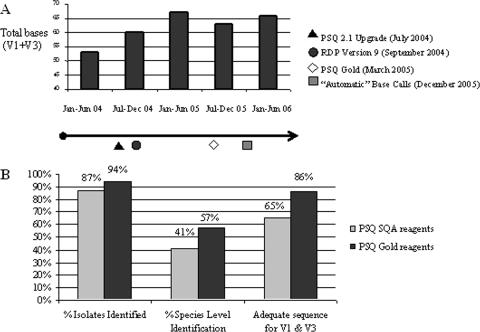
FIG. 1.
Enhancements in DNA pyrosequencing improved overall assay performance. (A) The mean numbers of interpretable bases obtained (when both the V1 and V3 regions were combined) showed a general increase during the course of the study, and specific refinements in DNA pyrosequencing chemistry and bioinformatics approaches are highlighted as discrete events in the timeline under the bar graph. (B) The implementation of PSQ Gold (Biotage) reagents included the addition of recombinant E. coli-derived, single-stranded DNA binding protein (Ssb), and the transition to PSQ Gold reagents resulted in increased percentages of bacterial isolates identified to the genus level (% Isolates Identified), increased percentages of species-level identifications (% Species Level Identification), and increased percentages of isolates with interpretable DNA sequence information in both variable regions, V1 and V3 (Adequate sequence for V1 & V3).
The introduction of PSQ Gold reagents in March 2005 included the addition of recombinant E. coli-derived, single-stranded binding protein (Ssb) and improved the overall quality of pyrosequencing data. Figure 1B depicts the performance improvements observed in several assay parameters at discrete time intervals during the study period. A total of 187 isolates were tested with the original PSQ reagents, and 227 isolates were tested with the PSQ Gold reagents. The PSQ Gold reagents improved the percentage of isolates yielding a definitive genus- or species-level identification from 87% to 94% of organisms. The percentage of isolates identified to the species level also improved from 41% to 57% of organisms submitted for pyrosequencing. The ability to obtain informative sequences in both V1 and V3 improved from 65% to 86% of isolates under evaluation, and the average read lengths increased from 34 bases to 36 bases and from 33 bases to 34 bases for V1 and V3, respectively, during the study period.
Two key developments during the study period improved the bioinformatics tools for analyses of DNA pyrosequencing data. The software for DNA pyrosequencing was upgraded from version 1.0 to version 2.1 in July 2004, and the new software version included improved algorithms for accurate base calling in homopolymeric regions. In September 2004, the RDP released version 9 (version 8 was used previously). A key development in version 9 was the incorporation of an updated, phylogenetically consistent, higher-order bacterial taxonomy as proposed by Garrity et al. (8). Other improved features in the RDP allowed users to select type isolates for particular bacterial species depending on established criteria, facilitating more focused refinement of the sequence matching process. DNA sequences from a total of 113 isolates were queried using RDP, version 8, and species-level identifications were made for 36% of the isolates. Of the 259 isolates sequenced and analyzed with RDP, version 9, species-level identifications were established for 57% of the isolates. Changes in sequencing chemistry as described were occurring in parallel and may be partly responsible for the improved accuracy of pathogen identification.
DISCUSSION
We have shown that DNA pyrosequencing significantly improved the ability of the diagnostic laboratory to determine definitive genus- or species-level identifications for diverse bacterial pathogens. Of the 414 bacterial isolates submitted for molecular studies due to the lack of reliable biochemical identification, 372 isolates (approximately 90%) were identified by DNA pyrosequencing. Most isolates submitted for pyrosequencing were either gram-negative or gram-positive bacilli (309 or 82% of all isolates). Gram-negative bacilli represented the single largest morphological category and comprised the vast majority (91%) of respiratory tract isolates. Interestingly, gram-positive bacilli represented the largest group of difficult-to-identify organisms from peripheral blood cultures. A total of 372 isolates have been correctly identified by DNA pyrosequencing, including 78 different genera representing 16 different specimen types. The diverse set of genera indicates the wide breadth of biological diversity represented by unusual clinical isolates in the clinical laboratory. A polyphasic strategy that included DNA pyrosequencing in the diagnostic laboratory improved bacterial pathogen identification in a children's hospital environment.
As shown in the current study, specific phenotypic groups, such as nonfermenting gram-negative bacilli and gram-positive bacilli may be difficult to identify despite successful culture and attempted biochemical testing. Nonfermenting gram-negative bacilli represent a group of pathogens that are difficult to identify by conventional biochemical methods and may require DNA sequencing for identification (1). In this study, gram-negative bacilli accounted for 91% of respiratory tract isolates and 58% of all isolates by Gram stain morphology. Prior studies have highlighted the identification challenges with unusual gram-negative bacilli. Compared to results of extensive phenotypic methods, partial 16S rRNA gene sequencing identified 97.2% and 89.2% of atypical, aerobic, gram-negative bacilli to the genus and species levels, respectively, in one study (29). DNA sequencing yielded unambiguous results with the highest concordance to established microorganism identifications relative to fatty acid profiling and carbon source utilization patterns. Even gram-negative bacilli that are usually considered straightforward, such as P. aeruginosa, may be challenging to identify by phenotypic methods. Pediatric patients with cystic fibrosis often yield P. aeruginosa isolates that are not identified by phenotypic methods (7), and this study included 33 such isolates that were identified unambiguously only by DNA pyrosequencing.
Gram-positive organisms, such as those of the genera Microbacterium and Rothia, pose challenges for biochemical approaches to bacterial identification. The genus Microbacterium represents a group of gram-positive bacilli that yielded multiple clinical isolates refractory to biochemical identification in this study. A prior study (19) documented the potential clinical significance of Microbacterium isolates obtained from the peripheral blood of patients with myeloid leukemia. This study (19) also highlighted the limitations of phenotypic methods for Microbacterium species identification. Interestingly, all Microbacterium isolates in this study were also obtained from peripheral blood specimens, emphasizing their potential importance in immunocompromised children. Routine identification of gram-positive cocci may benefit from DNA sequencing applications. The performance of DNA pyrosequencing was superior to that of the VITEK 2 biochemical testing panel for streptococcal speciation, and 75% of the group-level identifications were concordant between both methods (11). Rothia species represent a distinct group of gram-positive cocci that were detected in this set of isolates identified by DNA pyrosequencing. Specifically, Rothia mucilaginosa was identified from several pediatric patients, and its identification highlights the potential pathogenicity of this organism in immunocompromised patients. This species was reclassified from the genus Stomatococcus (4) and represents a challenging group of organisms that often are not classified or are misidentified by conventional approaches.
Advances in the chemistry of DNA pyrosequencing improved the performance of this methodology during the time of clinical testing covered in this study. The addition of recombinant E. coli single-stranded DNA binding protein to the pyrosequencing reagents (PSQ Gold) enhanced the performance of pyrosequencing in prior published studies (6, 26) and in the current study. The addition of Ssb represents a key modification in pyrosequencing chemistry that has been associated with improvements in read length, reduction in mispriming events, increased enzymatic efficiency, and greater accuracy of sequencing data (26). This study suggests that the addition of recombinant Ssb resulted in improved bacterial identification. Further refinements in DNA pyrosequencing may further increase read lengths and augment the ability to improve diagnostic accuracy. A recent study implicated the diminished efficiency of apyrase as a possible culprit of limited read lengths and provided evidence for superior performance of a three-enzyme system (minus apyrase) (22). This study provided evidence that replacement of apyrase with a carefully orchestrated washing step may improve read lengths to greater than 300 bases (22).
The application of bioinformatics strategies to pathogen identification presents several challenges. First, databases used for the identification of clinical isolates should utilize phylogenetically validated databases, such as the RDP (http://rdp.cme.msu.edu), instead of open sequence databases, such as GenBank or EMBL, that lack phylogenetic validation of microorganisms. Prior bacterial identification studies have usually queried generic databases, such as GenBank, with success. However, database standards should be carefully considered in the context of phylogeny, especially when considering DNA sequencing strategies for the identification of clinical isolates on a routine basis. Secondly, local databases may augment comprehensive internet-accessible sequence databases by enabling the storage of specific sequence information of local isolates or pathogenic clones. Patterns of sequence variation as demonstrated in the study examining alpha-hemolytic streptococci (11) may be examined temporally and in the context of disease and pathogen. The commercial IdentiFire database (Biotage) facilitates the storage of DNA sequences obtained from local clinical isolates. The software offers the ability to immediately search a user-created database using the pyrosequencing data. Such local databases facilitate comparisons of isolates circulating in particular geographical regions or individual hospitals. These local sequence databases also make faster searches possible with greater diagnostic accuracy since a restricted pathogen database can be easily queried and matched with recurrent phenotypic features of local difficult-to-identify pathogens. An initial evaluation of this approach has shown that the vast majority of identifications can be made using automated searches of local databases as opposed to a manual review of pyrosequencing data with open database queries.
As part of a comprehensive polyphasic strategy, pathogen identification could be enhanced in the diagnostic laboratory by the integration of DNA sequencing with biochemical testing and other phenotypic approaches. However, several limitations should be addressed before widespread implementation of such methodologies can occur in the clinical setting. Drawbacks to the assay include the inability to definitively identify approximately 10% of isolates submitted for sequencing in this study, primarily due to limited read lengths. Refinements in this technology resulting in increased amounts of sequencing data by real-time sequencing will strengthen the ability to accurately identify pathogens to the species level. Additional sequencing primers, which target different regions of the 16S rRNA gene, intergenic sequences, or other coding sequences, may facilitate improved diagnostic accuracy despite limited sequencing data yields. The inability to sequence lengthy homopolymeric tracts represents another hurdle for DNA pyrosequencing. Improvements in base calling by software-related algorithm enhancements, manual data interpretation, and refinements in the sequencing chemistry have generated more robust capabilities of handling 3- to 5-base homopolymers. Longer homopolymers should be avoided during the assay design process. Similar to other sequencing strategies, DNA pyrosequencing is limited by the need for pure bacterial isolates. Mixed cultures lead to poor sequence quality and inconclusive data and may limit diagnostic yields with clinical isolates if not effectively purified on plated medium prior to sequencing. Sources of exogenous bacterial DNA contamination that may confound DNA sequence-based identification include reagents used for assay performance. Initial development of DNA pyrosequencing applications in this study emphasized the importance of bacterial DNA contamination in sequencing reagents. One source of bacterial DNA contamination was determined to be commercial preparations of thermostable DNA polymerases, and 16S rRNA gene amplification of contaminating DNA precluded the generation of interpretable sequence data. Upon switching to a specialized formulation of a thermostable DNA polymerase that minimized the presence of bacterial DNA (e.g., AmpliTaq Gold LD [Applied Biosystems]), background DNA sequence signals were minimized and the reliability of DNA pyrosequencing improved markedly.
The implementation of routine DNA sequencing in the clinical laboratory necessitates multiple considerations, including workflow, process integration, laboratory reporting, and personnel competencies. Multiple advantages of DNA pyrosequencing over conventional dideoxy sequencing strategies include user friendliness, a streamlined labor component, and enhanced overall cost-effectiveness. In clinical laboratory settings today, the molecular diagnostics and microbiology laboratories are separate or molecular microbiology is a distinct section within a larger microbiology operation. Because cultured isolates are handled and tested initially in the microbiology laboratory, guidelines must be established for the submission of isolates for DNA sequencing. Cutoff values, such as probability-based biochemical identifications below 90%, provide discrete boundaries for the determination of decision-making points in the workup. Microbiologists must be intimately involved in the process in order to determine when an isolate requires molecular strategies for identification so that resources are managed effectively. The responsibility of selecting appropriate organisms for DNA pyrosequencing lies with the microbiologist at the medical technologist and managerial levels. Specimens that require further evaluation may necessitate the involvement of the technical or medical director. Submitted samples are sequenced in the molecular pathology laboratory, and resulting database queries are returned to the microbiologist(s) for the compilation of polyphasic (genotypic and phenotypic) data of each isolate prior to making a final determination. The collaborative nature of this process requires a laboratory structure and workflow that facilitates multiple interactions, including sample processing, data interpretation, and laboratory reporting. The ongoing, although belated, implementation of molecular methods for pathogen identification will foster enhanced collaborations in clinical laboratories and ultimately will improve the diagnostic accuracy of infectious diseases.
Supplementary Material
Acknowledgments
We acknowledge the work of Ebony Courtney, Kyle Menne, Janet Becker, Pamela Zapalac, and the entire Microbiology Section of the Department of Pathology at Texas Children's Hospital. In addition, we thank Julia Sponaugle, Amanda Hassler, and Amorette Kwan for their assistance during the initial development phase and Karla Sternberg and Tiffany Morgan for assistance with manuscript preparation. We are also grateful to Biotage, especially Rene Myers, for technical assistance and informatics guidance.
J.V. received financial support from the National Institutes of Health (VTEU N01 A1025465), the Defense Advanced Research Projects Agency, and the Department of Pathology at Texas Children's Hospital for the performance of these studies.
Footnotes
Published ahead of print on 25 July 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bosshard, P. P., R. Zbinden, S. Abels, B. Boddinghaus, M. Altwegg, and E. C. Bottger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 44:1359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buller, R. S., M. Arens, S. P. Hmiel, C. D. Paddock, J. W. Sumner, Y. Rikhisa, A. Unver, M. Gaudreault-Keener, F. A. Manian, A. M. Liddell, N. Schmulewitz, and G. A. Storch. 1999. Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N. Engl. J. Med. 341:148-155. [DOI] [PubMed] [Google Scholar]
- 3.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. D., R. A. Hutson, V. Baverud, and E. Falsen. 2000. Characterization of a Rothia-like organism from a mouse: description of Rothia nasimurium sp. nov. and reclassification of Stomatococcus mucilaginosus as Rothia mucilaginosa comb. nov. Int. J. Syst. Evol. Microbiol. 50:1247-1251. [DOI] [PubMed] [Google Scholar]
- 5.Downes, A., C. Thornhill, and R. H. Mallard. 1998. An evaluation of Vitek—an automated system for bacterial identification and susceptibility testing. Commun. Dis. Public Health 1:206-207. [PubMed] [Google Scholar]
- 6.Ehn, M., A. Ahmadian, P. Nilsson, J. Lundeberg, and S. Hober. 2002. Escherichia coli single-stranded DNA-binding protein, a molecular tool for improved sequence quality in pyrosequencing. Electrophoresis 23:3289-3299. [DOI] [PubMed] [Google Scholar]
- 7.Ferroni, A., I. Sermet-Gaudelus, E. Abachin, G. Quesne, G. Lenoir, P. Berche, and J. L. Gaillard. 2002. Use of 16S rRNA gene sequencing for identification of nonfermenting gram-negative bacilli recovered from patients attending a single cystic fibrosis center. J. Clin. Microbiol. 40:3793-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrity, G. M., J. A. Bell, and T. G. Lilburn. 2004. Taxonomic outline of the prokaryotes, p. xv-xxi. In G. M. Garrity, D. J. Brenner, N. R. Krieg, J. T. Staley, D. R. Boone, P. De Vos, M. Goodfellow, F. A. Rainey, and K.-H. Schleifer (ed.), Bergey's manual of systematic bacteriology, 2nd ed., Springer-Verlag, New York, NY.
- 9.Gharizadeh, B., M. Akhras, M. Unemo, B. Wretlind, P. Nyren, and N. Pourmand. 2005. Detection of gyrA mutations associated with ciprofloxacin resistance in Neisseria gonorrhoeae by rapid and reliable pre-programmed short DNA sequencing. Int. J. Antimicrob. Agents 26:486-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grahn, N., M. Olofsson, K. Ellnebo-Svedlund, H. J. Monstein, and J. Jonasson. 2003. Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol. Lett. 219:87-91. [DOI] [PubMed] [Google Scholar]
- 11.Haanperä, M., J. Jalava, P. Huovinen, O. Meurman, and K. Rantakokko-Jalava. 2007. Identification of alpha-hemolytic streptococci by pyrosequencing the 16S rRNA gene and by use of VITEK 2. J. Clin. Microbiol. 45:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins, K. L., C. Arnold, and E. J. Threlfall. 2007. Rapid detection of gyrA and parC mutations in quinolone-resistant Salmonella enterica using pyrosequencing technology. J. Microbiol. Methods 68:163-171. [DOI] [PubMed] [Google Scholar]
- 13.Innings, A., M. Krabbe, M. Ullberg, and B. Herrmann. 2005. Identification of 43 Streptococcus species by pyrosequencing analysis of the rnpB gene. J. Clin. Microbiol. 43:5983-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonasson, J., M. Olofsson, and H. J. Monstein. 2002. Classification, identification and subtyping of bacteria based on pyrosequencing and signature matching of 16S rDNA fragments. APMIS 110:263-272. [DOI] [PubMed] [Google Scholar]
- 15.Jordan, J. A., A. R. Butchko, and M. B. Durso. 2005. Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. J. Mol. Diagn. 7:105-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi, N., T. W. Bauer, D. Togawa, I. H. Lieberman, H. Sakai, T. Fujishiro, M. J. Tuohy, and G. W. Procop. 2005. A molecular Gram stain using broad range PCR and pyrosequencing technology: a potentially useful tool for diagnosing orthopaedic infections. Diagn. Mol. Pathol. 14:83-89. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, N., T. W. Bauer, M. J. Tuohy, I. H. Lieberman, V. Krebs, D. Togawa, T. Fujishiro, and G. W. Procop. 2006. The comparison of pyrosequencing molecular Gram stain, culture, and conventional Gram stain for diagnosing orthopaedic infections. J. Orthop. Res. 24:1641-1649. [DOI] [PubMed] [Google Scholar]
- 18.Kolbert, C. P., P. N. Rys, M. Hopkins, D. T. Lynch, J. J. Germer, C. E. O'Sullivan, A. Trampuz, R. Patel. 2004. 16S ribosomal DNA sequence analysis for identification of bacteria in a clinical microbiology laboratory, p. 361-377. In D. H. Persing, F. D. Tenover, J. Versalovic, Y.-W. Tang, E. R. Unger, D. A. Relman, and T. J. White (ed.), Molecular microbiology. Diagnostic principles and practice. ASM Press, Washington, DC.
- 19.Lau, S. K., P. C. Woo, G. K. Woo, and K. Y. Yuen. 2002. Catheter-related Microbacterium bacteremia identified by 16S rRNA gene sequencing. J. Clin. Microbiol. 40:2681-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindbäck, E., M. Unemo, M. Akhras, B. Gharizadeh, H. Fredlund, N. Pourmand, and B. Wretlind. 2006. Pyrosequencing of the DNA gyrase gene in Neisseria species: effective indicator of ciprofloxacin resistance in Neisseria gonorrhoeae. APMIS 114:837-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh, S. 2007. Pyrosequencing applications. Methods Mol. Biol. 373:15-24. [DOI] [PubMed] [Google Scholar]
- 22.Mashayekhi, F., and M. Ronaghi. 2007. Analysis of read length limiting factors in pyrosequencing chemistry. Anal. Biochem. 363:275-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monstein, H., S. Nikpour-Badr, and J. Jonasson. 2001. Rapid molecular identification and subtyping of Helicobacter pylori by pyrosequencing of the 16S rDNA variable V1 and V3 regions. FEMS Microbiol. Lett. 199:103-107. [DOI] [PubMed] [Google Scholar]
- 24.Pai, R., J. Limor, and B. Beall. 2005. Use of pyrosequencing to differentiate Streptococcus pneumoniae serotypes 6A and 6B. J. Clin. Microbiol. 43:4820-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Relman, D. A., S. Falkow, P. E. LeBoit, L. A. Perkocha, K. W. Min, D. F. Welch, and L. N. Slater. 1991. The organism causing bacillary angiomatosis, peliosis hepatis, and fever and bacteremia in immunocompromised patients. N. Engl. J. Med. 324:1514. [DOI] [PubMed] [Google Scholar]
- 26.Ronaghi, M. 2000. Improved performance of pyrosequencing using single-stranded DNA-binding protein. Anal. Biochem. 286:282-288. [DOI] [PubMed] [Google Scholar]
- 27.Ronaghi, M., S. Karamohamed, B. Pettersson, M. Uhlen, and P. Nyren. 1996. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242:84-89. [DOI] [PubMed] [Google Scholar]
- 28.Stager, C. E., and J. R. Davis. 1992. Automated systems for identification of microorganisms. Clin. Microbiol. Rev. 5:302-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tärnberg, M., T. Jakobsson, J. Jonasson, and U. Forsum. 2002. Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rDNA variable V1 and V3 regions. APMIS 110:802-810. [DOI] [PubMed] [Google Scholar]
- 31.Tuohy, M. J., G. S. Hall, M. Sholtis, and G. W. Procop. 2005. Pyrosequencing as a tool for the identification of common isolates of Mycobacterium sp. Diagn. Microbiol. Infect. Dis. 51:245-250. [DOI] [PubMed] [Google Scholar]
- 32.Wahab, T., S. Hjalmarsson, R. Wollin, and L. Engstrand. 2005. Pyrosequencing Bacillus anthracis. Emerg. Infect. Dis. 11:1527-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Z. J., M. Z. Tu, J. Liu, X. L. Wang, and H. Z. Jin. 2006. Comparison of amplicon-sequencing, pyrosequencing and real-time PCR for detection of YMDD mutants in patients with chronic hepatitis B. World J. Gastroenterol. 12:7192-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.