Abstract
Vibrio vulnificus is an opportunistic, highly invasive human pathogen with worldwide distribution. V. vulnificus strains are commonly divided into three biochemical groups (biotypes), most members of which are pathogenic. Simple sequence repeats (SSR) provide a source of high-level genomic polymorphism used in bacterial typing. Here, we describe the use of variations in mutable SSR loci for accurate and rapid genotyping of V. vulnificus. An in silico screen of the genomes of two V. vulnificus strains revealed thousands of SSR tracts. Twelve SSR with core motifs longer than 5 bp in a panel of 32 characterized and 56 other V. vulnificus isolates, including both clinical and environmental isolates from all three biotypes, were tested for polymorphism. All tested SSR were polymorphic, and diversity indices ranged from 0.17 to 0.90, allowing a high degree of discrimination among isolates (27 of 32 characterized isolates). Genetic analysis of the SSR data resulted in the clear distinction of isolates that belong to the highly virulent biotype 3 group. Despite the clonal nature of this new group, SSR analysis demonstrated high-level discriminatory power within the biotype 3 group, as opposed to other molecular methods that failed to differentiate these isolates. Thus, SSR are suitable for rapid typing and classification of V. vulnificus strains by high-throughput capillary electrophoresis methods. SSR (≥5 bp) by their nature enable the identification of variations occurring on a small scale and, therefore, may provide new insights into the newly emerged biotype 3 group of V. vulnificus and may be used as an efficient tool in epidemiological studies.
Vibrio vulnificus, a gram-negative aquatic bacterium, is a ubiquitous and opportunistic, highly invasive human pathogen. Infections in humans are acquired through skin wounds or the consumption of contaminated seafood, with the main clinical syndromes being primary septicemia, wound infection, and gastroenteritis (12, 41). Immunocompromised individuals or those with chronic liver disease are at higher risk for lethal infection (20). Small numbers of V. vulnificus infections are reported annually. However, human infections result in a high hospitalization rate and a fatality rate that can reach up to 50%, with at times a gap of only 24 h between hospitalization and death (20, 34, 37). The widely accepted methods for the identification and typing of V. vulnificus strains are mainly biochemical and require numerous tests. Other tests, such as pulsed-field gel electrophoresis, amplified fragment length polymorphism analysis, multilocus enzyme electrophoresis, and the random amplification of polymorphic DNA, have also been applied for the classification of V. vulnificus strains, providing comparable, yet sometimes different, results (e.g., see references 16 and 59). Strains of V. vulnificus are biochemically divided into three different biotypes that can be isolated from a variety of hosts, such as fish, eels, and oysters, and a variety of habitats, including water, sediment, and plankton (16, 21, 22, 60). Different biotypes can emerge due to geographic isolation or niche separation (16). Biotype 1, isolated mostly from shellfish in coastal estuarine areas, is the most common group worldwide and is responsible for numerous clinical cases of infection (12, 34). Biotype 2 was isolated from diseased eels for the first time in 1982 (51) and was later identified as an opportunistic human pathogen (1). Biotype 3, initially found in Israel in 1996, is associated mainly with fish and is the etiological agent of many incidents of severe infections in humans (3). Recent work claims that biotype 3 evolved by the hybridization of genomes of biotypes 1 and 2 (5).
Simple sequence repeats (SSR), or microsatellites, also termed variable-number tandem repeats (VNTR), are a class of short DNA sequence motifs that are repeated in a head-to-tail fashion at a specific locus. Slipped-strand mispairing and recombination are believed to be the causes of SSR variability (31, 39, 56). SSR were previously found to be highly polymorphic among strains of several bacterial species (15, 55) but are mostly stable at the strain level. The relatively high degree of variation at SSR loci, particularly those with core motifs of 2 to 9 bp, enables the efficient use of SSR for bacterial typing and various genetic analyses (e.g., see references 25, 26, 32, and 55). As a result, in the last few years, multiple-locus SSR analysis, also termed multiple-locus VNTR analysis, has been increasingly recognized as the method of choice for genotyping a number of pathogens, such as Bacillus anthracis (26), Borrelia species (11), Salmonella enterica (33), Enterococcus faecium and Enterococcus faecalis (52, 53), Escherichia coli (28, 56), Staphylococcus aureus (14), and Vibrio cholerae (9).
This study presents a multiple-locus SSR analysis (or multiple-locus VNTR analysis) of V. vulnificus with emphasis on the newly emerged, highly virulent biotype 3 group. The genomes of two V. vulnificus strains (8, 29) were screened for the presence of SSR tracts. The selected SSR sites were used for the establishment and assessment of a method based on length variation at SSR loci for the accurate and rapid typing of V. vulnificus isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The study included a set of 32 strains of V. vulnificus (Table 1): 10 isolates of biotype 1, 6 isolates of biotype 2, 15 isolates of biotype 3, and 1 isolate with an undetermined biotype. Twenty of the strains were isolated from clinical or environmental sources in Israel between 1996 and 2003. An additional 56 environmental strains were isolated from fish in Israel during September and October 2004 (Table 1). The final species validation was done by the detection of the vvh gene as described below. All bacterial strains were cultured on both thiosulfate-citrate-bile salts-sucrose agar plates and Luria agar plates with a final concentration of 1.5% NaCl.
TABLE 1.
V. vulnificus isolates in this study
| Isolate identification | Subidentification(s) | Biotype | Sourcea | Type (origin)b |
|---|---|---|---|---|
| 9028A/95 | v201 | 1 | CDC | Clinical |
| 9030/95 | v202 | 1 | CDC | Clinical |
| 9032/95 | v203 | 1 | CDC | Clinical |
| 9020/96 | v204 | 1 | CDC | Clinical |
| 9021/96 | v205 | 1 | CDC | Clinical |
| 9022/96 | v206 | 1 | CDC | Clinical |
| 03:96.8.6 | v207 | 2 | L. Hoi, Denmark | Env (diseased eel) |
| 03:96.8.7 | v208 | 2 | L. Hoi, Denmark | Env (diseased eel) |
| 03/04:95.8.161 | v209 | 2 | L. Hoi, Denmark | Env (diseased eel) |
| 03/04:95.8.162 | v210 | 2 | L. Hoi, Denmark | Env (diseased eel) |
| 04:96.7.137 | v211 | 2 | L. Hoi, Denmark | Env (diseased eel) |
| 04:96.7.138 | v212 | 2 | L. Hoi, Denmark | Env (diseased eel) |
| 145/96 | v213 | 3 | IMH, Israel | Clinical |
| 58/97 | v214 | 3 | IMH, Israel | Clinical |
| 11028/97 | v215 | 3 | IMH, Israel | Clinical |
| 162/97 | v216 | 3 | IMH, Israel | Clinical |
| 1033/97 | v217 | 3 | IMH, Israel | Clinical |
| 1/98 | v218 | 3 | IMH, Israel | Clinical |
| 1/99 | v219 | 3 | IMH, Israel | Clinical |
| 5/00 | v220 | 3 | IMH, Israel | Clinical |
| 8/00 | v221 | 3 | IMH, Israel | Clinical |
| 1/01 | v222 | 3 | IMH, Israel | Clinical |
| 10/02 | v223 | 3 | IMH, Israel | Clinical |
| 7/03 | v224 | 3 | IMH, Israel | Clinical |
| 5/1 | v225 | 1 | IMH, Israel | Env (water) |
| 2322 | v226 | 1 | IMH, Israel | Env (fish) |
| 101/4 | v227 | NDc | IMH, Israel | Env (fish) |
| 2321 | v228 | 1 | IMH, Israel | Env (fish) |
| 105 | v229 | 3 | IMH, Israel | Env (fish) |
| 6a | v230 | 1 | IMH, Israel | Env (fish) |
| 12 | v231 | 3 | IMH, Israel | Env (fish) |
| 8/03e | v232 | 3 | IMH, Israel | Env (fish) |
| New environmental isolatesd | VVyb1 | 3 | Israel | Env (fish) |
| VVyb2 to VVyb10 | 1 | Israel | Env (fish) | |
| VVyb11 to VVyb58e | ND | Israel | Env (fish) |
All isolates were part of the Israeli Ministry of Health (IMH) collection. CDC, Centers for Disease Control and Prevention.
Env, environmental.
ND, not determined biochemically.
Set of 56 strains isolated in September and October 2004.
Isolates VVyb38 and VVyb45 were missing.
DNA preparation.
High-quality DNA extraction from pure cultures was performed (15, 49). Briefly, a loopful of V. vulnificus colonies was washed with SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 8.0]). The cell suspension was incubated overnight at 50°C with proteinase K solution (20 mg/ml in Tris-EDTA [TE]). Sodium dodecyl sulfate was added to a final concentration of 1%, and the mixture was incubated at 55°C for 1 h, followed by phenol-chloroform extraction and ethanol precipitation. The DNA was stored at −20°C.
Crude and fast DNA extraction methods were also carried out. Briefly, a loopful of scraped V. vulnificus colonies was suspended in 0.5 ml of TE buffer and the mixture was heated for 10 min at 80°C and centrifuged for 10 min at 16,060 × g. The pellet was resuspended in 100 μl of TE buffer, and the mixture was heated for 5 min at 100°C. The solution was centrifuged for 2 min at 16,060 × g, and the supernatant was stored at −20°C.
Computerized analysis of SSR distribution in genomes.
The complete genomic sequences of two V. vulnificus strains, YJ016 (8) and CMCP6 (29), were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). The two chromosomes of strain YJ016 were designated VV and VVA, and those of strain CMCP6 were designated VV1 and VV2. Both chromosomes (3.3 Mb and 1.8 Mb) were screened for perfect SSR (i.e., exact-repeat motifs) with a minimum of more than two repeats by using the SSR computer program (15; http://www.technion.ac.il/∼anne/choice3.html).
Locus and primer selection.
The 12 loci with the longest SSR (those having both the longest core motifs and the highest numbers of repeats) found among both chromosomes of the two strains by in silico analysis were selected for this study. We did not include sites that showed similarity to phage or prophage sequences (based on the NCBI BLAST tool analysis). These 12 sites included SSR loci with core motifs ranging from 6 to 8 bp. Unique primers flanking the SSR sequences were designed to generate PCR products of 100 to 350 bp by using the Gene Runner software (version 3.05; Hastings Software Inc.). The assigned locus names included the chromosomal designation and the downstream gene number (based on the NCBI plus-strand orientation) (see Table SA1 in the supplemental material). Each locus was tested for uniqueness in the V. vulnificus genome by using NCBI BLAST (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) and the Findpatterns program on GCG 10 (Genetics Computer Group, Madison, WI).
SSR locus stability.
Two V. vulnificus strains, v201 from biotype 1 and v214 from biotype 3, were grown and transferred daily for four consecutive days. Two colonies of each of the strains were picked daily from a plate containing an overnight-grown culture, streaked onto a fresh Luria-Bertani plate, and grown at 37°C. Crude and fast DNA extractions from the 62 resulting plates were conducted each day (see below). The relative mutation rate was calculated as the number of mutations per locus per generation (56). The number of generations per colony was determined to be ∼26.6, assuming 108 bacteria per colony (based on an average of counts of viable bacteria from individual colonies).
PCR.
The PCR mixture contained 0.2 mM (each) deoxynucleoside triphosphates, 10 μM (each) forward and reverse primers, 0.5 U Taq polymerase (Super Nova; JMR Holding, Kent, England), 1× PCR buffer (with 1.5 mM MgCl2), and 50 ng of template DNA in a total volume of 25 μl. The reaction was carried out in a PTC-100 thermocycler (MJ Research) as follows: 95°C for 5 min; 5 cycles of 45 s at 95°C, 45 s at the melting temperature (Tm) (see Table SA1 in the supplemental material), and 45 s at 72°C; 20 cycles of 45 s at 95°C, 45 s at the Tm minus 5°C, and 45 s at 72°C; 7 min at 72°C; and cooling to 4°C. PCR amplification products were verified by agarose gel (1.2%) electrophoresis and observed by UV fluorescence.
Detection of specific genes by PCR.
The amplification of vvh loci together with a universal bacterial 16S rRNA subunit was performed in a multiplex PCR assay using a Tm of 57°C. Both sets of primers were previously described for use in amplifying a fragment of 276 bp of the hemolysin/cytolysin gene (58) and 707 bp of the 16S rRNA subunit (49).
DNA sizing.
The sizing of SSR amplification products was performed by 2.5% agarose gel electrophoresis and by capillary electrophoresis on an ABI PRISM 310 automated DNA sequencer using a fluorescence-labeled forward primer. The fluorescent amplification products were diluted from 1:10 to 1:100 in water, according to each amplicon's concentration. One microliter of the diluted product was loaded onto the ABI analyzer together with 10 μl of formamide and 0.6 μl of tetramethylrhodamine (GENESCAN 500; Applied Biosystems Inc.). Results were analyzed with Genescan 3.1 and Genotyper analysis software (PerkinElmer).
Data and statistical analysis. (i) Allele analysis.
A nonparametric analysis of variations in SSR-size alleles was used for SSR loci. An additional allele (a null allele) was counted when there was no amplification product. Each biotype group included a different number of isolates. Therefore, in order to compare allele variations among the different biotypes in a nonbiased manner, the numbers of alleles were normalized. The normalization was done by dividing the observed number of alleles by the number of isolates in a specific biotype group and multiplying the result by the total number of isolates. The statistical analysis of allele variation was performed by using JMP, version 6.0.0 (1989 to 2005; SAS Institute Inc., Cary, NC). The diversity index was calculated as follows: 1 − Σ(Pij)2, where Pij is the frequency of the jth allele at the ith locus. Each of the alleles at a specific locus in all genotypes was scored as present (1) or absent (0) in each isolate.
(ii) Analysis of genetic relationships.
Genetic relationships among strains were inferred from the SSR data by using the Nei coefficient of association and generating the corresponding matrix with SAS 8.02 (1989 to 2005; SAS Institute Inc., Cary, NC). This matrix was used to create dendrograms based on the neighbor-joining method with MEGA 3.1 software (30). Clusters of related bacterial genotypes were inferred using the eBURST V3 algorithm (http://eburst.mlst.net/) (13) on a single data set.
RESULTS AND DISCUSSION
Accurate epidemiological identification in the context of huge microbial diversity requires highly discriminating strain typing technology. The availability of many complete microbial genome sequences allows comparative in silico analyses of strains between and within bacterial species, facilitating the development of DNA-based typing technologies. Among these, SSR- or VNTR-based typing is considered to represent the new generation of strain typing tools. This study presents an SSR-based method for typing and inferring genetic relationships among strains of the bacterium V. vulnificus.
SSR distribution along V. vulnificus genomes.
Genomewide screens of two V. vulnificus strains (YJ016 and CMCP6) revealed tens of thousands of perfect SSR tracts and more than 17,000 mononucleotide repeat (MNR; >5-bp) loci. The SSR scan searched for sequence repeats with a motif larger than 2 bp having a repeat number larger than two (Table 2). These SSR were evenly distributed and highly abundant, showing average frequencies of one SSR tract every 481 and 486 bases in the genomes of the YJ016 and CMCP6 strains, respectively. Similar frequencies of one SSR every 460, 462, 469, and 473 bp were found for E. coli K-12 (6), E. coli O157:H7 (both genomes) (17, 43), and V. cholerae El Tor N16961 (18), respectively. A fine analysis of the latter data showed that there are considerable numbers of longer SSR tracts, with more than 17 bp (58 in strain YJ016 and 54 in strain CMCP6), that may serve as potentially informative markers. In silico comparisons between the two V. vulnificus genomes revealed additional sequence variation within the repeated motifs at some of these sites. Furthermore, the frequency of these long (≥18-bp) SSR in V. vulnificus is up to 8.6 times higher than that in E. coli K-12, 4.5 times higher than that in V. cholerae El Tor N16961, and about 2.7 times higher than that in the two E. coli O157:H7 strains. These findings may suggest a connection between long SSR and pathogenicity (2); however, a deeper analysis including additional species is required. It has been established previously that long SSR, with their elevated mutation rates, can afford the dynamic response needed to withstand changes in the environment or the host organism (2, 36, 39). Interestingly, in V. vulnificus, the majority (53 to 67%) of these SSR have a core motif of 7 bp, compared to 40% in V. cholerae El Tor N16961 and 0% in E. coli K-12 or O157:H7. Approximately 75% of those 7-bp SSR have a common motif, CTAGDNH. Comparison between the two V. vulnificus chromosomes revealed a 6 to 9% higher frequency of SSR tracts in chromosome I, while no significant difference between the two strains was observed. Likewise, results found for V. cholerae show higher frequencies of SSR and MNRs (>8 bp) in chromosome I than in chromosome II (9).
TABLE 2.
Numbers of SSR loci found along the two chromosomes of V. vulnificus (YJ016)
| Chromosome | No. of repeats | No. of SSR core motifs of length (bp)a:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 11 | 14 | ||
| I | 3 | 4,994 | 1,415 | 26 | 1 | 4 | 3 | 2 | 1 | |||
| 4 | 296 | 35 | 0 | 0 | 1 | 4 | 3 | 0 | ||||
| 5 | 11,236 | 13 | 1 | 0 | 0 | 0 | 3 | 2 | 0 | |||
| 6 | 3,153 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | |||
| 7 | 730 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |||
| 8 | 145 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| 9 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | |||
| 10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |||
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | |||
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |||
| 13 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | |||
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |||
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |||
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |||
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |||
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |||
| II | 3 | 3,036 | 713 | 17 | 0 | 0 | 4 | 0 | 0 | 1 | 1 | |
| 4 | 205 | 16 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | ||
| 5 | 6,158 | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | |
| 6 | 1,672 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7 | 355 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 8 | 71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 9 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 33 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |
Numbers in bold correspond to motifs of loci selected for study.
Twelve SSR loci, with core motifs ranging from 6 to 8 nucleotides, were chosen for polymorphism analysis (Table 3). These SSR loci were selected mostly from noncoding regions. All of the tested SSR loci showed primary in silico differences between the two published V. vulnificus genomes (8, 29).
TABLE 3.
Numbers of alleles and diversity values at the 12 SSR loci of 32 clinical and environmental V. vulnificus isolates
| Chromosome | SSR locusa | Core motifc | Position in genome (bp) (gene product) | No. of alleles | Diversity index |
|---|---|---|---|---|---|
| I | VV-0401*b | (TTCTCGC)13 | Noncoding, 397,957 | 11d | 0.81 |
| VV-1044 | (CTCGAAC)12 | Noncoding, 104,6946 | 12 | 0.86 | |
| VV-1151 | (AGAACCT)7 | Noncoding, 1,162,010 | 6 | 0.63 | |
| VV-1368 | (TCATAC)4 | Coding/noncoding, 1,392,009 (DNA binding protein inhibitor) | 2 | 0.17 | |
| VV-2339* | (TAGGTTC)21 | Noncoding, 2,363,752 | 9d | 0.78 | |
| VV-2577* | (CTAGATC)5 | Noncoding, 2,602,460 | 9 | 0.81 | |
| VV-2785* | (GTTTTCTA)13 | Noncoding, 2,839,672 | 9 | 0.75 | |
| VV1-1615 | (TCCTCAA)4 | Noncoding, 1,586,203 | 7 | 0.70 | |
| II | VVA-0305 | (CTAGTAC)4 | Noncoding, 345,619 | 9 | 0.65 |
| VVA-0375* | (TGTTGC)33 | Coding, 441,865 (tetratricopeptide repeat-containing protein) | 16 | 0.90 | |
| VVA-0930* | (TCTAGGG)18 | Noncoding, 1,023,042 | 10 | 0.74 | |
| VVA-1475 | (CTAGGTT)19 | Noncoding, 1,622,343 | 12 | 0.77 |
All information in this table is based on V. vulnificus strain YJ016, except that for locus VV1-1615, which is based on strain CMCP6. *, tested for stability.
This locus was studied previously (44).
Subscript numbers are numbers of repeats of the motifs.
This locus included a null allele (yielding no amplification product) as one of the alleles (see the text).
Polymorphism and size variation at SSR loci.
Multiple alleles were found at each of the 12 SSR loci, revealing a high level of polymorphism among the 32 V. vulnificus isolates. The strain panel included members of the different biotypes from both clinical and environmental sources, 20 of which were isolated in Israel (Table 1). All strains were tested primarily for the presence of vvh gene. Rapid and accurate sizing was achieved using fluorescent capillary electrophoresis. The sizes of the PCR amplification products differed according to the sizes of the repeated motifs, and the number of repeats ranged from 1 to 39 (see Table SA2 in the supplemental material; Fig. 1). Only locus VV-0401 had only one repeat unit in some isolates (as verified by sequencing); the other loci showed a minimum of two alleles in all isolates or yielded no amplification product (a null allele) in some isolates. Each size variant and repeat number was represented by a different allele. SSR loci presented 2 to 16 alleles (Table 3). Most of these loci showed six alleles or more, and only one locus (VV-1368) showed two alleles. Two of the loci gave no amplification product (a null allele) for some of the isolates. Null alleles were taken into account in the analysis of relatedness, thus permitting a more accurate assessment than would be possible by ignoring the presence of null alleles (9, 24, 48, 57). An analysis of the two chromosomes revealed a higher number of alleles per locus in chromosome II than in chromosome I (12 and 8 alleles on average, respectively) (Table 3). These results may imply an overall higher degree of variability in chromosome II than in chromosome I.
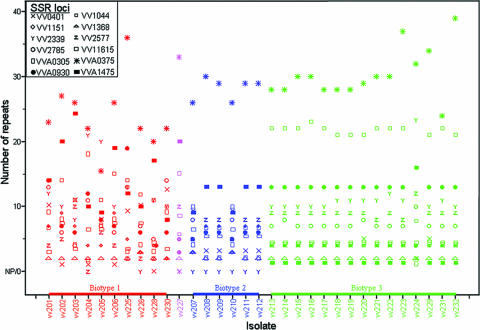
FIG. 1.
Distribution of numbers of repeats at 12 SSR loci as a function of the isolate. NP, no amplification product.
In general, there was a correlation between the number of repeats found in the genome of YJ016 and the number of alleles in the tested panel (R = 0.76; P = 0.0042). Hence, variation in SSR tracts increases with the number of repeats, as found for other bacterial species (e.g., see references 11 and 28). Diversity indices were high as well, ranging from 0.63 to 0.90, with one exception of 0.17 for VV-1368 (Table 3). This low level of diversity at the VV-1368 locus is a result of the presence of one common allele in 29 strains. SSR loci with higher degrees of polymorphism and higher numbers of alleles showed higher corresponding diversity indices (Table 3; Fig. 1). Similarly, Vogler et al. (56) reported a correlation between repeat number and mutation rate in E. coli such that loci with higher numbers of repeats, higher numbers of alleles, and increased diversity exhibit elevated mutation rates.
The relative consistency of SSR markers was verified over 4 days of transfers of colonies of two representative V. vulnificus strains (biotype 1 isolate v201 and biotype 3 isolate v214), each tested for stability at six representatives of the longest SSR loci (Table 3). In all cases, colonies from a specific strain showed amplification of the expected alleles, except at two loci: the VVA-0375 locus, which showed the insertion of one repeat in two colonies, and the VVA-0930 locus, which showed the deletion of one repeat in one colony. The maximal mutation rates for these loci were calculated as 7.6 × 10−4 and 3.8 × 10−4 mutations/locus/generation, respectively. Similar mutation rates were calculated for SSR in B. anthracis (27), E. coli (40, 56), and V. cholerae (9). The rather high maximal mutation rate calculated in the present study is probably an elevated rate estimate as a result of a single mutation event which occurred during the last transfer (40, 56). The slippage of a single repeat within a specific locus in both eukaryotes and prokaryotes has been described previously (e.g., references 19 and 39). Recently, it was concluded that a single-repeat change, either an insertion or a deletion, occurs in approximately 75% of the VNTR mutation events in E. coli (56).
Genetic analyses.
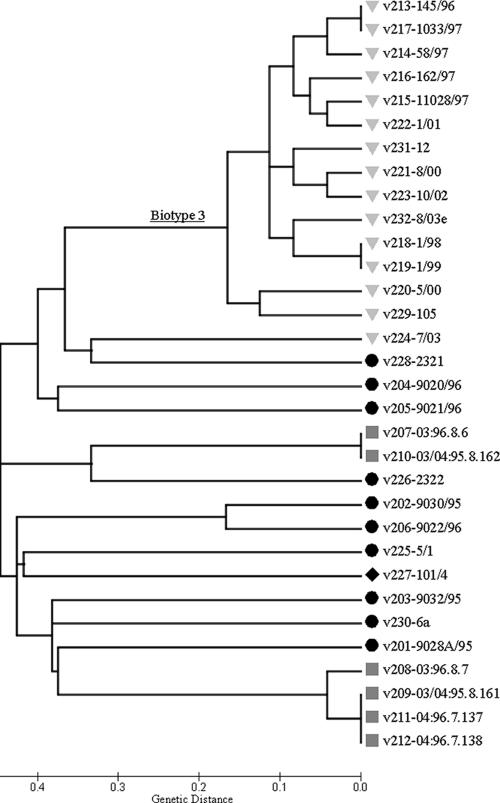
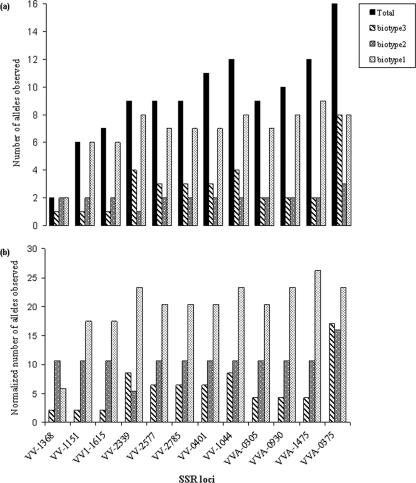
The SSR variation data was used to infer genetic relationships among V. vulnificus isolates. A genetic-distance matrix was generated based on 112 polymorphic points (12 loci times the number of alleles in each locus), followed by cluster analysis using the neighbor-joining method. The resulting dendrogram (Fig. 2) showed that isolates belonging to biotype 3 formed a distinct cluster, with one exception, isolate v224 (see below), whereas isolates of biotypes 1 and 2 showed no significant clustering. We obtained similar results using BURST (13, 50) analysis: isolates of biotype 3 formed a single, clonal complex, supporting previous findings on this group's clonality (4, 5). In addition, no significant separation between clinical and environmental isolates within or among the different biotypes was found; however, there were differences in variation levels, i.e., numbers of alleles, among the three biotypes at most of the SSR loci (Fig. 3). In general, biotype 1 showed the highest level of variation and biotype 3 showed the lowest, as reflected by the numbers of alleles at a specific locus (Fig. 1; see Table SA2 in the supplemental material). These differences in variation levels were even clearer when (to take into consideration the different numbers of isolates of the three biotypes) the numbers of alleles were normalized (Fig. 3). Isolates of the worldwide biotype 1 group showed a normalized average of 20 alleles per locus, and isolates of biotype 2 showed an average of 11 alleles, compared to 6 presented by strains of biotype 3. Similarly, distances between isolates of biotype 3 were rather low (average genetic distance, 0.302 ± 0.197) (Table 4) compared to the high genetic distances (average, 0.820 ± 0.201) found between isolates of biotypes 1 and 2 (Table 4), demonstrating the wide genetic diversity of the latter groups. These results are consistent with previous findings revealing the low level of diversity of biotype 3 compared to biotype 1 isolates (5, 7, 16). Furthermore, distributions of specific alleles across the three biotypes, tested as variations in repeat number, were found to be significantly different at 10 of the tested loci (P, <0.05 for nine loci) (see Table SA3 in the supplemental material). However, in general, no correlations between repeat number and the origin of the isolate (i.e., clinical or environmental) or the biotype group were found, although biotype 3 isolates were more conserved. In recent work, it was suggested that for the heptanucleotide VV-0401 locus, the number of repeats correlates with the isolate source, as lower numbers of repeats were observed mainly in clinical isolates (44). Our results for the VV-0401 locus could not support this assumption; however, a larger number of isolates, equally representing all three groups, should be tested in order to reach a conclusion.
FIG. 2.
Genetic relationships among 32 V. vulnificus isolates based on neighbor-joining cluster analysis of SSR variation. A genetic-distance matrix was generated based on 112 polymorphic points of the 12 SSR loci. Biotype 1, circles; biotype 2, squares; biotype 3, inverted triangles; unidentified biotype, diamond.
FIG. 3.
Alleles observed at 12 polymorphic SSR loci. Shown are the number of alleles (a) and the normalized number of alleles (b) observed at a specific SSR locus in relation to the biotype. The normalization was done by dividing the observed number of alleles by the number of isolates in the specific biotype group and multiplying the result by the total number of isolates.
TABLE 4.
Genetic distances between V. vulnificus strains as calculated based on nonparametric analyses of SSR data
| Biotype(s) | Genetic distancea
|
|||
|---|---|---|---|---|
| Among strains of indicated biotype(s)
|
Between strains of indicated biotype(s) and strain:
|
|||
| Maximum | Avg | v224 (avg) | v228 (avg) | |
| 1 and 2 | 1 | 0.820 ± 0.201 | 0.906 ± 0.077 | 0.885 ± 0.074 |
| 3 | 0.75 | 0.302 ± 0.197 | 0.750 | 0.708 ± 0.043 |
Average values are given with standard deviations.
The variation data for 12 SSR loci enabled the partitioning of the 32 V. vulnificus isolates into 27 different SSR types. We could discriminate all 10 isolates of biotype 1, each of which presented a unique SSR type. Isolate v227, which failed to be classified by biochemical tests, presented a unique SSR type and was associated with the biotype 1 group according to our analysis, consistent with previous multilocus sequence typing (MLST) results (5); however, recent work on eBURST claims that the connection of a single isolate to a clonal complex (similar to that of v227) may sometimes give biased results as an outcome of high recombination rates (54). Here, the six isolates of biotype 2 were divided into three different SSR types, whereas a previous study found only four MLST types among 15 biotype 2 strains (5). Furthermore, the 15 isolates of biotype 3 were divided into 13 SSR types, in contrast to the results of a previous study that used 10 genes to analyze 62 strains, all of which presented the same MLST type (5). Examples of the difference in the discriminatory powers of MLST and SSR analysis were provided by isolates v213, v214, v216, and v228, which were analyzed by both methods and were fully differentiated by the SSR approach. In addition, our SSR analysis allowed the full discrimination of four additional biotype 3 isolates (v214, v215, v216, and v217) that could not be separated using the methods of multilocus enzyme electrophoresis and random amplification of polymorphic DNA (16). These results demonstrate the enhanced discriminatory power of the SSR analysis for closely related strains, found in many bacterial species such as E. coli (O157:H7 and O55:H7) (28), Enterococcus faecalis (52), Leptospira species (45), Bartonella henselae (38), and Burkholderia pseudomallei (42). This high resolving power of the SSR method is further supported by the comparison between VNTR and MLST methods for numerous species such as Neisseria meningitides (46, 61, 62), Staphylococcus aureus (35), Haemophilus influenzae (47), and Mycobacterium tuberculosis (23). Similarly, MNR-MLST of E. coli and V. cholerae could not differentiate among E. coli O157 strains (10) or among pathogenic V. cholerae (O1 and O139 ctxA+) strains (9), in contrast to the SSR (VNTR) analysis (9, 28). However, because of the low mutation rates and high sequence contents of MLST or MNR-MLST, these methods may contribute to the classification of these human pathogens.
In addition, 12 SSR loci of 56 environmental strains were analyzed together with those of our panel of 32 strains, yielding 168 polymorphic points (total number of alleles across the 12 loci). The 56 isolates from five different sampling sites showed 37 different SSR types (level of diversity, 66%), demonstrating again the high resolving power of SSR analysis for V. vulnificus. Furthermore, cluster analysis, presented in Fig. 4, similarly revealed that isolates of biotype 3 formed a distinct node. The only environmental isolate that was clustered in the biotype 3 group was isolate VVyb1-9/04. This isolate was determined biochemically to be a member of biotype 3, while the additional nine environmental isolates tested belonged to biotype 1. Interestingly, all clinical isolates of V. vulnificus collected in Israel in the most recent years have belonged to biotype 3 (according to the Central Laboratories, Ministry of Health, Jerusalem, Israel), although the environmental frequency of biotype 3 seems to be rather low compared to that of biotype 1 (our unpublished data from a preliminary analysis).
FIG. 4.
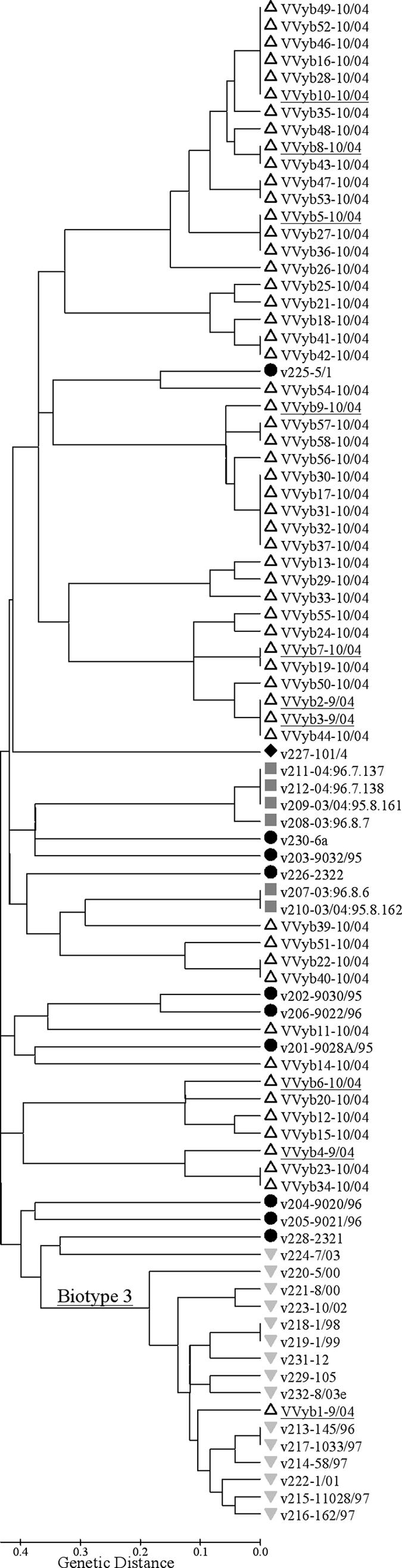
Genetic relationships among 88 V. vulnificus isolates based on neighbor-joining cluster analysis of SSR variation. A set of 56 new environmental isolates were analyzed, together with the panel of 32 strains (Table 1). A genetic-distance matrix was generated based on 168 polymorphic points of 12 SSR loci. Biotype 1, circles; biotype 2, squares; biotype 3, inverted triangles; unidentified biotype, diamond; new environmental isolates, triangles. Underlined isolates: VVyb1, biotype 3; VVyb2 through VVyb10, biotype 1.
An interesting case was presented by isolates v224 and v228, biochemically assigned to biotypes 3 and 1, respectively. Isolate v228 was classified as biotype 3 according to the MLST (5), while no MLST data for v224 were available. Their average genetic distances from other biotype 3 isolates (0.750 and 0.708 ± 0.043) (Table 4) as calculated from the SSR data implied and may suggest a clear separation from the genetically compact biotype 3 cluster. Only 3 (VV-1151, VV-1368, and VV1-1615) of the 12 tested SSR loci presented the same alleles in these isolates as in the other group 3 isolates. Interestingly, these three loci showed the smallest numbers of alleles, with rather low repeat numbers (Table 3), and thus were the most stable SSR of all loci tested (i.e., they had the lowest mutation rates), allowing the assignment of relatively distant genetic relationships. Bisharat et al. (5) suggested that V. vulnificus biotype 3 is a new, highly virulent group, the possible consequence of the hybridization of two populations. Taking together all of the above, we may assume that these specific isolates (v224 and v228) represent an intermediate variant of biotype 3, though more analysis (e.g., MNR-MLST) is required to better support this assumption. If so, the SSR in the present study can be used as a delicate tool for the detailed epidemiological study of a novel pathogen in a spatiotemporal axis.
In conclusion, a high level of genetic diversity is important for bacterial adaptation, especially in pathogenic bacteria. SSR provide a mechanism of site-specific hypermutation along the bacterial chromosome, with the example of contingency loci (2). Here, SSR in V. vulnificus were found to be genetically informative, polymorphic, and relatively stable at the strain level and, therefore, an efficient tool for genetic and epidemiological studies. Despite the clonal nature of the new biotype 3 group, SSR analysis demonstrated high discriminatory power. SSR results enabled the identification of small-scale variations, thus providing insights into the genomic nature of the newly emerged biotype 3 group of V. vulnificus. The fact that all clinical cases of V. vulnificus disease in Israel have been connected to group 3 may indicate that biotype 3 is more virulent than the other biotypes and supports the need for understanding the evolution and adaptation of this emerging group of pathogens.
Supplementary Material
Acknowledgments
We thank Aviv De-Morgan, Technion, Israel, for fruitful discussion and comments, the anonymous reviewer for helpful comments, and the Ministry of Health, Central Laboratories, Jerusalem, Israel, for providing bacterial strains.
This research was supported by the Institute for Future Security Study, Technion; the Grand Water Research Institute, Technion; the Ministry of Health, State of Israel grant number 5492; the Israeli Water Commission; Israel Science Foundation grant number 1005697; and NATO project CBD.MD.SFP 981456.
Footnotes
Published ahead of print on 25 July 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Amaro, C., and E. G. Biosca. 1996. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl. Environ. Microbiol. 62:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayliss, C. D., K. M. Dixon, and E. R. Moxon. 2004. Simple sequence repeats (microsatellites): mutational mechanisms and contributions to bacterial pathogenesis. A meeting review. FEMS Immunol. Med. Microbiol. 40:11-19. [DOI] [PubMed] [Google Scholar]
- 3.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, and J. J. Farmer. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 4.Bisharat, N., C. Amaro, B. Fouz, A. Llorens, and D. I. Cohen. 2007. Serological and molecular characteristics of Vibrio vulnificus biotype 3: evidence for high clonality. Microbiology 153:847-856. [DOI] [PubMed] [Google Scholar]
- 5.Bisharat, N., D. I. Cohen, R. M. Harding, D. Falush, D. W. Crook, T. Peto, and M. C. Maiden. 2005. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 11:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. ColladoVides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Chatzidaki-Livanis, M., M. A. Hubbard, K. Gordon, V. J. Harwood, and A. C. Wright. 2006. Genetic distinctions among clinical and environmental strains of Vibrio vulnificus. Appl. Environ. Microbiol. 72:6136-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. T. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danin-Poleg, Y., L. A. Cohen, H. Gancz, Y. Y. Broza, H. Goldshmidt, E. Malul, L. Valinsky, L. Lerner, M. Broza, and Y. Kashi. 2007. Vibrio cholerae strain typing and phylogeny study based on simple sequence repeats. J. Clin. Microbiol. 45:736-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamant, E., Y. Palti, R. Gur-Arie, H. Cohen, E. M. Hallerman, and Y. Kashi. 2004. Phylogeny and strain typing of Escherichia coli, inferred from variation at mononucleotide repeat loci. Appl. Environ. Microbiol. 70:2464-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farlow, J., D. Postic, K. L. Smith, Z. Jay, G. Baranton, and P. Keim. 2002. Strain typing of Borrelia burgdorferi, Borrelia afzelii, and Borrelia garinii by using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol. 40:4612-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farmer, J. J. 2003. Vibrio, p. 706-716. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, DC.
- 13.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francois, P., A. Huyghe, Y. Charbonnier, M. Bento, S. Herzig, I. Topolski, N. Fleury, D. Lew, P. Vaudaux, S. Harbarth, W. van Leeuwen, A. van Belkum, D. S. Blanc, D. Pittet, and J. Schrenzel. 2005. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gur-Arie, R., C. J. Cohen, Y. Eitan, L. Shelef, E. M. Hallerman, and Y. Kashi. 2000. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res. 10:62-71. [PMC free article] [PubMed] [Google Scholar]
- 16.Gutacker, M., N. Conza, C. Benagli, A. Pedroli, M. V. Bernasconi, L. Permin, R. Aznar, and J. C. Piffaretti. 2003. Population genetics of Vibrio vulnificus: identification of two divisions and a distinct eel-pathogenic clone. Appl. Environ. Microbiol. 69:3203-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 18.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Y. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, S. T., and T. D. Petes. 1992. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2749-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hlady, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 173:1176-1183. [DOI] [PubMed] [Google Scholar]
- 21.Hoi, L., I. Dalsgaard, A. DePaola, R. J. Siebeling, and A. Dalsgaard. 1998. Heterogeneity among isolates of Vibrio vulnificus recovered from eels (Anguilla anguilla) in Denmark. Appl. Environ. Microbiol. 64:4676-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoi, L., J. L. Larsen, I. Dalsgaard, and A. Dalsgaard. 1998. Occurrence of Vibrio vulnificus biotypes in Danish marine environments. Appl. Environ. Microbiol. 64:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwamoto, T., S. Yoshida, K. Suzuki, M. Tomita, R. Fujiyama, N. Tanaka, Y. Kawakami, and M. Ito. 2007. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol. Lett. 270:67-74. [DOI] [PubMed] [Google Scholar]
- 24.Katzir, N., Y. DaninPoleg, G. Tzuri, Z. Karchi, U. Lavi, and P. B. Cregan. 1996. Length polymorphism and homologies of microsatellites in several Cucurbitaceae species. Theor. Appl. Genet. 93:1282-1290. [DOI] [PubMed] [Google Scholar]
- 25.Keim, P., A. M. Klevytska, L. B. Price, J. M. Schupp, G. Zinser, K. L. Smith, M. E. Hugh-Jones, R. Okinaka, K. K. Hill, and P. J. Jackson. 1999. Molecular diversity in Bacillus anthracis. J. Appl. Microbiol. 87:215-217. [DOI] [PubMed] [Google Scholar]
- 26.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keim, P., M. N. Van Ert, T. Pearson, A. J. Vogler, L. Y. Huynh, and D. M. Wagner. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4:205-213. [DOI] [PubMed] [Google Scholar]
- 28.Keys, C., S. Kemper, and P. Keim. 2005. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J. Appl. Microbiol. 98:928-940. [DOI] [PubMed] [Google Scholar]
- 29.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 31.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA-sequence evolution. Mol. Biol. Evol. 4:203-221. [DOI] [PubMed] [Google Scholar]
- 32.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26:2567-2582. [DOI] [PubMed] [Google Scholar]
- 33.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 34.Linkous, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 35.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyszton-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, P., K. Makepeace, S. A. Hill, D. W. Hood, and E. R. Moxon. 2005. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. USA 102:3800-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mead, P. S., L. Slutsker, V. Dietz, L. F. Mccaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monteil, M., B. Durand, R. Bouchouicha, E. Petit, B. Chomel, M. Arvand, H. J. Boulouis, and N. Haddad. 2007. Development of discriminatory multiple-locus variable number tandem repeat analysis for Bartonella henselae. Microbiology 153:1141-1148. [DOI] [PubMed] [Google Scholar]
- 39.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4:24-33. [DOI] [PubMed] [Google Scholar]
- 40.Noller, A. C., M. C. McEllistrem, K. A. Shutt, and L. H. Harrison. 2006. Locus-specific mutational events in a multilocus variable-number tandem repeat analysis of Escherichia coli O157:H7. J. Clin. Microbiol. 44:374-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliver, J. D. 1989. Vibrio vulnificus, p. 569-600. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker Inc., New York, NY.
- 42.Pearson, T., J. M. U'Ren, J. M. Schupp, G. J. Allan, P. G. Foster, M. J. Mayo, D. Gal, J. L. Choy, R. L. Daugherty, S. Kachur, C. L. Friedman, B. Leadem, S. Georgia, H. Hornstra, A. J. Vogler, D. M. Wagner, P. Keim, and B. J. Currie. 2006. VNTR analysis of selected outbreaks of Burkholderia pseudomallei in Australia. Infect. Genet. Evol. 7:416-423. [DOI] [PubMed] [Google Scholar]
- 43.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Limk, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Y. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 44.Rosche, T. M., Y. Yano, and J. D. Oliver. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381-389. [DOI] [PubMed] [Google Scholar]
- 45.Salaun, L., F. Merien, S. Gurianova, G. Baranton, and M. Picardeau. 2006. Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J. Clin. Microbiol. 44:3954-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schouls, L. M., A. van der Ende, M. Damen, and I. van de Pol. 2006. Multiple-locus variable-number tandem repeat analysis of Neisseria meningitidis yields groupings similar to those obtained by multilocus sequence typing. J. Clin. Microbiol. 44:1509-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schouls, L. M., A. van der Ende, I. van de Pol, C. Schot, L. Spanjaard, P. Vauterin, D. Wilderbeek, and S. Witteveen. 2005. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 43:2741-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somer, L., Y. Danin-Poleg, E. Diamant, R. Gur-Arie, Y. Palti, and Y. Kashi. 2005. Amplified intergenic locus polymorphism as a basis for bacterial typing of Listeria spp. and Escherichia coli. Appl. Environ. Microbiol. 71:3144-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somer, L., and Y. Kashi. 2003. A PCR method based on 16S rRNA sequence for simultaneous detection of the genus Listeria and the species Listeria monocytogenes in food products. J. Food Prot. 66:1658-1665. [DOI] [PubMed] [Google Scholar]
- 50.Spratt, B. G., W. P. Hanage, B. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species: the eBURST approach. FEMS Microbiol. Lett. 241:129-134. [DOI] [PubMed] [Google Scholar]
- 51.Tison, D. L., M. Nishibuchi, J. D. Greenwood, and R. J. Seidler. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Titze-de-Almeida, R., R. J. L. Willems, J. Top, I. P. Rodrigues, R. F. Ferreira, H. Boelens, M. C. C. Brandileone, R. C. Zanella, M. S. S. Felipe, and A. van Belkum. 2004. Multilocus variable-number tandem-repeat polymorphism among Brazilian Enterococcus faecalis strains. J. Clin. Microbiol. 42:4879-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Top, J., L. M. Schouls, M. J. M. Bonten, and R. J. L. Willems. 2004. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 42:4503-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner, K. M., W. P. Hanage, C. Fraser, T. R. Connor, and B. G. Spratt. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogler, A. J., C. Keys, Y. Nemoto, R. E. Colman, Z. Jay, and P. Keim. 2006. Effect of repeat copy number on variable-number tandem repeat mutations in Escherichia coli O157:H7. J. Bacteriol. 188:4253-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner, A. P., S. Creel, and S. T. Kalinowski. 2006. Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity 97:336-345. [DOI] [PubMed] [Google Scholar]
- 58.Wang, H. Y., and G. H. Lee. 2003. Rapid identification of Vibrio vulnificus in seawater by real-time quantitative TaqMan PCR. J. Microbiol. 41:320-326. [Google Scholar]
- 59.Wong, H. C., S. Y. Chen, M. Y. Chen, J. D. Oliver, L. I. Hor, and W. C. Tsai. 2004. Pulsed-field gel electrophoresis analysis of Vibrio vulnificus strains isolated from Taiwan and the United States. Appl. Environ. Microbiol. 70:5153-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright, A. C., R. T. Hill, J. A. Johnson, M. C. Roghman, R. R. Colwell, and J. G. Morris. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yazdankhah, S. P., K. Kesanopoulos, G. Tzanakaki, J. Kremastinou, and D. A. Caugant. 2005. Variable-number tandem repeat analysis of meningococcal isolates belonging to the sequence type 162 complex. J. Clin. Microbiol. 43:4865-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yazdankhah, S. P., B. A. Lindstedt, and D. A. Caugant. 2005. Use of variable-number tandem repeats to examine genetic diversity of Neisseria meningitidis. J. Clin. Microbiol. 43:1699-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.