Abstract
Group B streptococci (GBS) comprising three different sets of isolates (31 invasive, 36 noninvasive, and 24 colonizing isolates) were collected in Italy during the years 2002 to 2005. Clonal groups were established by pulsed-field gel electrophoresis (PFGE), and selected isolates were studied by multilocus sequence typing (MLST). GBS isolates were also characterized by classical and molecular techniques for serotyping and protein gene and antibiotic resistance profiling. Some serotypes were significantly associated with a particular isolate population: serotype Ia more frequently corresponded to invasive strains than other strains, serotype V was more frequently encountered among noninvasive strains, and nontypeable strains were more common among isolates from carriers. Four major clonal groups accounted for 52.7% of all isolates: PFGE type 1/clonal complex 1 (CC1) comprised mainly serotype V isolates carrying the alp3 gene, PFGE type 2/CC23 encompassed serotype Ia isolates with the alp1 or alpha gene, PFGE type 3/CC17 comprised serotype III isolates carrying the rib gene, and PFGE type 4/CC19 consisted mainly of serotype II isolates possessing the rib gene. The same serotypes were shared by isolates of different clonal groups, and conversely, isolates belonging to the same clonal groups were found to be of different serotypes, presumably due to capsular switching by the horizontal transfer of capsular genes. Erythromycin resistance (prevalence, 16.5%; 15 resistant isolates of 91) was restricted to strains isolated from patients with noninvasive infections and carriers, while tetracycline resistance was evenly distributed (prevalence, 68.1%; 62 resistant isolates of 91). Most erythromycin-resistant GBS strains were of serotype V, were erm(B) positive, and belonged to the PFGE type 1/CC1 group, suggesting that macrolide resistance may have arisen both by clonal dissemination and by the horizontal transfer of resistance genes.
Streptococcus agalactiae (group B streptococcus [GBS]) is one of the leading causes of neonatal sepsis and meningitis (2, 20, 32). The colonization of the female genital tract with GBS is significantly associated with infections in neonates, and it should be carefully monitored. Moreover, GBS has also been recently recognized as an important pathogen in immunocompromised patients (12, 14, 37). The first-line agent against GBS infection is penicillin, and penicillin resistance among GBS strains has not been reported so far (5). However, for patients allergic to penicillin, macrolides (e.g., erythromycin) and lincosamides (e.g., clindamycin) are the alternative choices for the treatment of GBS infections. In the United States, the frequencies of resistance to erythromycin and clindamycin among GBS isolates have been reported to be approximately 37 and 17%, respectively (16).
Two main mechanisms of erythromycin resistance in GBS isolates have been described previously (17, 25). One mechanism is macrolide-specific efflux encoded by the mef(A)/mef(E) gene (macrolide [M] resistance phenotype). The other mechanism is a ribosomal modification mediated by a methylase encoded by the erm (erythromycin ribosome methylase) genes erm(B) and erm(A) [subclass erm(TR)], which can be either expressed constitutively (constitutive macrolide-lincosamide-streptogramin B [cMLSB] resistance phenotype) or induced by erythromycin (induced MLSB [iMLSB] resistance phenotype) (5).
Capsular serotyping is the classic method for typing GBS in epidemiological studies. Based on their capsular polysaccharides, GBS isolates can be classified into nine different serotypes (Ia, Ib, and II to VIII), with serotypes Ia, II, III, and V being the predominant causes of human GBS diseases (11, 29). In particular, the serotypes most commonly causing neonatal infections are Ia, III, and V (32), with the latter being the main serotype recovered from patients of all age groups, including nonpregnant adults (1, 19).
Some of the best-characterized GBS protein antigens belong to the alpha-like protein (Alp) family, a class of surface proteins characterized by internal long identical tandem repeats. These proteins are named alpha, Alp1, Alp2, Alp3, Alp4, and Rib (28). They are encoded by allelic genes presenting a mosaic structure with conserved 5′ and 3′ ends and a highly variable domain generated by intragenic recombination (24). The possibility that these genes are part of mobile elements cannot be excluded because a surface protein, R28, highly homologous to Alp3 is present in some S. pyogenes strains (28) and a new Alp variant in a clinical S. dysgalactiae subsp. equisimilis isolate has been found previously (9). Although Alp proteins are targets for protective antibodies in a mouse model of infection, the biological role of these proteins is still not fully understood. Recently, it has been demonstrated that alpha protein promotes S. agalactiae invasion of epithelial cells by interaction with host cell glycosaminoglycans (3, 4).
Several molecular typing techniques for defining the clonal relatedness and studying the genetic population structures of GBS strains have been described previously. These include restriction fragment length pattern analysis, pulsed-field gel electrophoresis (PFGE), multilocus enzyme electrophoresis typing, and multilocus sequence typing (MLST) (13, 21, 27, 34). MLST is the most reliable tool for typing strains with unambiguous results, easily allowing the comparison of genetic profiles of different strains recovered from all geographic areas (28, 39). At least two major MLST-defined genetic lineages of GBS isolates of serotype III, sequence types (STs) 17 and 19, have been reported previously (26, 27). Some data support the hypothesis that ST17 complex GBS strains are associated with enhanced invasiveness compared to other serotype III GBS isolates. The finding of more than one genetic lineage among serotype III GBS strains is indicative that the type III capsule of GBS is not associated with pathogenicity, which requires other factors (22).
In this study, we characterized 91 GBS isolates belonging to three different sets, defined by isolation sources, by combining different epidemiological typing systems, such as classical serotyping, molecular serotyping, protein gene profiling, antibiotic resistance gene profiling, PFGE, and MLST.
(This work was presented in part at the VIIth ASM Conference on Streptococcal Genetics, Saint-Malo, France, 18 to 21 June 2006 [abstr. A130].)
MATERIALS AND METHODS
Strains and serotyping.
During the years 2002 to 2005, 91 single-patient GBS isolates were collected from Italian hospitals, the majority as part of an Italian nationwide enhanced surveillance program. Three different sets of strains were considered: 31 invasive isolates, recovered from otherwise sterile body sites or samples, such as blood and cerebrospinal fluid; 36 noninvasive isolates, recovered from wound and abscess specimens, synovial fluid, and samples from patients with respiratory tract and urinary tract infections; and 24 colonizing isolates recovered from vaginal swabs, stools, and urine samples from healthy patients. Twelve subjects (13%) were neonates, all but one presenting with invasive infections. Of the strains from the adult patients, 20, 35, and 24 were invasive isolates, noninvasive isolates, and colonizing isolates, respectively. Clinical samples were plated onto defibrinated sheep blood agar plates, the plates were incubated at 37°C in 5% CO2, and beta-hemolytic colonies were isolated. Bacterial strain identification was confirmed by either the semiautomatic Phoenix system (Becton Dickinson) or the Dryspot streptococcal grouping kit (Oxoid). Serotyping was performed by immunodiffusion with specific rabbit anticapsular antibodies, and nontypeable (NT) GBS isolates were serotyped by molecular methods (30).
Alpha-like protein (Alp) family.
Surface protein markers were inferred by using a multiplex PCR for the direct identification of the Alp protein genes (7). An additional primer (5′ GCTTAAGGATAGCAACGAACCAAAA 3′) to discriminate between the alp2 and alp3 genes was introduced.
Erythromycin, clindamycin, and tetracycline resistance determinants.
Resistance to erythromycin and clindamycin was assessed phenotypically both by the Etest (AB Biodisk, Solna, Sweden) for MIC determination and by the Kirby-Bauer double-disk diffusion method (6) to identify the constitutive, inducible, and M resistance phenotypes (18). The presence of the resistance genes erm(A) [subclass erm(TR)], erm(B), and mef(A) was investigated (8). Tetracycline resistance was determined both phenotypically by using the Etest and genotypically by studying the occurrence of the resistance genes tet(M) and tet(O) as previously described (36).
PFGE analysis.
DNA was extracted as previously described (13) and digested with 40 U of SmaI. PFGE results were interpreted according to the criteria previously reported (35). Isolates with indistinguishable PFGE patterns were assigned to identical PFGE subtypes. Isolates with five or fewer bands of difference were considered to be possibly related and assigned to the same PFGE type but different PFGE subtypes. Isolates with more than five bands of difference were considered to be unrelated and were identified as different PFGE types. Dice's coefficient was used to determine the similarity between each banding pattern, and a dendrogram was constructed by using the unweighted-pair group method with arithmetic averages with a tolerance coefficient of 1.5%. DNA from six strains was not digested by SmaI.
MLST.
Selected isolates representative of the major PFGE clonal clusters were studied by MLST performed as previously described (21). Partial sequences of seven housekeeping genes were obtained, and alleles were classified according to the MLST website criteria (http://pubmlst.org/sagalactiae/). For each isolate, an ST was defined according to the unique allelic profile observed. Isolates were assigned to one of the previously described clonal complexes (CCs) if they shared five or more alleles with the predominant ST in that CC (29).
Statistical analysis.
Categorical data were compared by using the Pearson chi-squared test. The analyses were carried out with the SPSS statistical analysis package (version 14.0 for Windows). P values below 0.05 were considered statistically significant.
RESULTS
Serotyping and surface protein profile.
All 91 GBS isolates were serotyped by conventional phenotypic methods, and in the case of nontypeability, they were subjected to molecular serotyping (30). Nonetheless, 15 strains still yielded the result NT, failing to give any amplification of the cps locus by the molecular method. Overall, the most represented serotypes encountered were types III (21 isolates), Ia (16 isolates), V (14 isolates), and II (13 isolates). Serotypes Ib, IV, and VII were represented by five, three, and four strains, respectively. No strains of serotype VI and VIII were found. Although all serotypes were distributed among the three categories examined, some serotypes were significantly associated with a particular population (Table 1). The incidence of serotype Ia among invasive strains (29%) was higher than that among noninvasive strains (13.9%) or colonizing strains (8.3%; P < 0.05); serotype V was more frequently encountered among noninvasive strains (25%) than among the invasive and colonizing populations (12.9 and 4.2%, respectively; P < 0.05). NT strains were significantly more common among the isolates from carriers (29.1%; P < 0.05), and serotype III was also more common among this group (33.3%) than among the other groups, although not significantly so (P > 0.05).
TABLE 1.
Distribution of the observed serotypes among the studied GBS populationsa
| Serotype | No. of isolates (%) among:
|
||
|---|---|---|---|
| Invasive population | Noninvasive population | Colonizing population | |
| Ia | 9 (29)* | 5 (13.9) | 2 (8.3) |
| Ib | 2 (6.5) | 2 (5.5) | 1 (4.2) |
| II | 6 (19.4) | 4 (11.1) | 3 (12.5) |
| III | 7 (22.6) | 6 (16.7) | 8 (33.3) |
| IV | 1 (3.2) | 1 (2.8) | 1 (4.2) |
| V | 4 (12.9) | 9 (25)* | 1 (4.2) |
| VII | 1 (3.2) | 2 (5.6) | 1 (4.2) |
| NT | 1 (3.2) | 7 (19.4) | 7 (29.1)* |
*, P < 0.05
The presence of a particular surface Alp protein in relation to the serotype was also noted (Table 2). Of the 16 alp1 gene-positive strains isolated, half were of serotype Ia; 12 of 17 GBS alp3 gene-positive strains were of serotype V; 18 of 30 rib gene-positive strains corresponded to serotype III, and 10 corresponded to serotype II.
TABLE 2.
Distribution of the alp genes among the observed GBS serotypes
| Surface protein gene (no. of isolates) | No. of gene-positive isolates (%) of serotype:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VII | NT | |
| alp1 (16) | 8 (50) | 1 (6.2) | 1 (6.2) | 1 (6.2) | 2 (12.5) | 3 (18.9) | ||
| alp2 (7) | 3 (42.8) | 1 (14.3) | 1 (14.3) | 1 (14.3) | 1 (14.3) | |||
| alp3 (17) | 12 (70.6) | 1 (5.9) | 4 (23.5) | |||||
| alpha (21) | 5 (23.8) | 4 (19) | 1 (4.8) | 1 (4.8) | 1 (4.8) | 3 (14.2) | 6 (28.6) | |
| rib (30) | 10 (33.3) | 18 (60.1) | 1 (3.3) | 1 (3.3) | ||||
Conversely, a certain serotype commonly corresponded to a particular Alp protein: serotype Ia possessed the alpha, alp1, and alp2 genes; serotype Ib and IV presented either the alp1 or alpha gene; serotypes II and III carried mainly the rib gene; most serotype V strains (70.6%) possessed the alp3 gene; and serotype VII carried either the alpha or alp3 gene.
PFGE type/ST clonal groups.
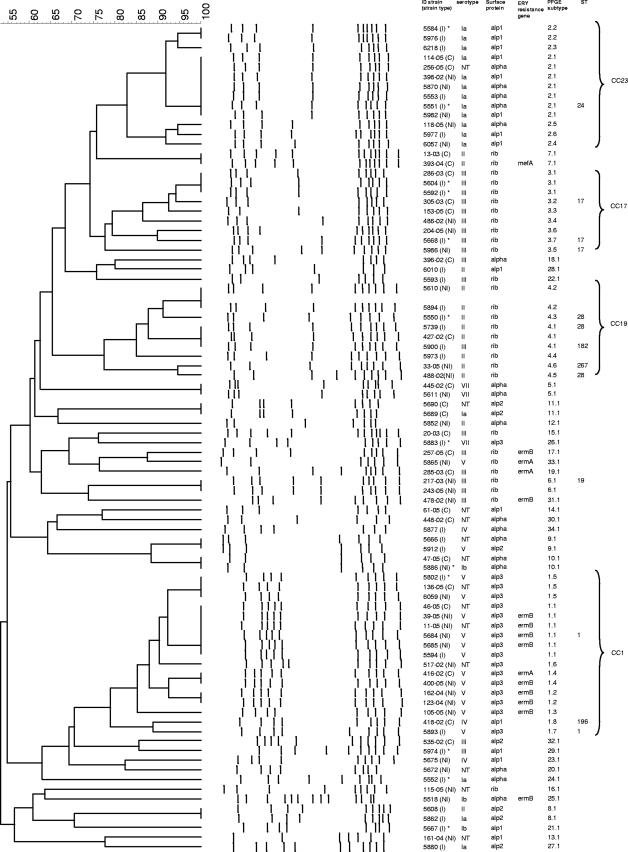
Eighty-five isolates were genotyped by PFGE, and selected isolates representative of the major PFGE clonal clusters were subjected to MLST. A total of 34 different PFGE types were observed, including 11 PFGE types that grouped two or more isolates (Table 3) and 23 unique PFGE types (Fig. 1). One PFGE type consisting of six isolates whose DNA was not digested (PFGE type 35) was also found (data not shown).
TABLE 3.
Characteristics of the multiple-strain clonal clusters of GBS isolates in Italy
| PFGE type (no. of subtypes) | No. of isolates | Strain type(s)a (no. of isolates) | Serotype(s) (no. of isolates) | Surface protein gene(s) (no. of isolates) | ST(s) | Corresponding CC (degree[s] of relatedness)b | Erythromycin resistance phenotype(s)/ genotype(s) (no. of isolates) | Tetracycline resistance genotype(s) (no. of isolates) |
|---|---|---|---|---|---|---|---|---|
| 1 (8) | 17 | I (3), NI (10), C (4) | V (12), NT (4), IV (1) | alp3 (16), alp1 (1) | ST1, ST196 | CC1 (DLV) | cMLSB resistance/erm(B) (8), cMLSB resistance/erm(A) (1) | tet(M) (13), tet(M) tet(O) (4) |
| 2 (6) | 13 | I (6), NI (5), C (2) | Ia (12), NT (1) | alpha (5), alp1 (8) | ST24 | CC23 (DLV) | Sd | tet(M) (12) |
| 3 (7) | 9 | I (3), NI (3), C (3) | III | rib | ST17 | CC17 | S | tet(M) (8) |
| 4 (6) | 9 | I (6), NI (5), C (2) | II (8), III (1) | rib | ST28, ST182, ST267 | CC19 (SLV, DLV) | S | tet(M) (5), tet(M) tet(O) (3) |
| 5 (1) | 2 | NI (1), C (1) | VII | alpha | NDc | ND | S | S |
| 6 (1) | 2 | NI | III | rib | ST19 | CC19 | S | tet(M) (1) |
| 7 (1) | 2 | C | II | rib | ND | ND | M resistance/mef(A) (1) | S |
| 8 (1) | 2 | I | Ia, II | alp2 | ND | ND | S | S |
| 9 (1) | 2 | I | V, NT | alpha, alp2 | ND | ND | S | tet(M) (1) |
| 10 (1) | 2 | NI, C | Ib, NT | alpha | ND | ND | S | tet(M) (2) |
| 11 (1) | 2 | C | Ia, NT | alp2 | ND | ND | S | S |
Three different strain types were represented among the GBS isolates studied: I, invasive strains; NI, noninvasive strains; and C, colonizing strains.
CCs were defined as described in a previous publication (29). The degree of relatedness represents a comparison between different STs exhibited by isolates belonging to the same clonal group and is indicated by classification of the STs as SLVs or DLVs.
ND, not determined.
S, susceptible.
FIG. 1.
Phylogenetic analysis of PFGE profiles obtained for 85 of 91 GBS isolates. DNA from six strains was not digested by the SmaI restriction enzyme. The dendrogram was constructed with PFGE profiles by similarity and clustering analyses using the unweighted-pair group method with arithmetic averages and Dice's coefficient. The levels of genetic similarity in percentages are shown on the left. The strain codes including the strain type (I, invasive; NI, noninvasive; C, colonizing), serotype, surface protein gene, erythromycin (ERY) resistance gene, PFGE subtype, and ST are indicated. The four major CCs are labeled on the right. Marked by an asterisk are isolates from neonates. ID strain, strain identification.
Four PFGE type clonal groups (types 1, 2, 3, and 4) accounted for 48 GBS isolates (52.7%) (Fig. 1 and Table 3). The PFGE type 1 clonal group consisted of eight PFGE subtypes and comprised 17 isolates, of which 12 were of serotype V, 1 was of serotype IV, and 4 were NT. Ten of 17 isolates belonging to this group were from patients with noninvasive infections. All but one isolates harbored the alp3 surface protein gene (Table 3). Two representative isolates of serotype V and the isolate of serotype IV were used for MLST analysis. Serotype V isolates exhibited ST1, and the serotype IV isolate displayed ST196, a double-locus variant (DLV) of ST1 (Fig. 1 and Table 3). Both isolates belonged to CC1 (Table 3). The PFGE type 2 group, with six different PFGE subtypes, was represented by 13 isolates comprising 12 isolates of serotype Ia and 1 NT isolate. Six, five, and two isolates were invasive, noninvasive, and colonizing strains, respectively. Eight isolates carried the alp1 gene, and five isolates carried the alpha gene. These isolates corresponded to the clone ST24, an ST belonging to CC23 (Table 3). The last two major groups, the PFGE type 3 and type 4 groups, comprised nine isolates each. The PFGE type 3 clonal group comprised only serotype III isolates carrying the surface protein gene rib and was subdivided into seven PFGE subtypes sharing ST17 (CC17) (Fig. 1 and Table 3). Three strains were isolated from patients with invasive disease, three were collected from patients with noninvasive disease, and three originated from carriers. The PFGE type 4 clonal group was represented by eight serotype II isolates and one serotype III isolate, with six different PFGE subtypes (Table 3). Five invasive isolates, three noninvasive isolates, and one colonizing isolate were found to belong to this clonal group. All isolates harbored the surface protein gene rib. A total of five representative isolates were studied by MLST (Fig. 1 and Table 3). Three isolates of serotype II displayed ST28, one serotype II isolate exhibited ST267, and the isolate of serotype III presented ST182. ST267 and ST182 were DLVs of ST28. All three different STs corresponded to CC19 (Table 3).
A small clonal group, the PFGE type 6 group, represented by two serotype III strains carrying the surface protein gene rib, was also included in the MLST analysis. A representative isolate of PFGE type 6 exhibited ST19 (Fig. 1 and Table 3).
Among the 11 multiple-strain PFGE groups, 3 contained GBS isolates of different serotypes (Table 3). PFGE type 1 was represented by isolates of serotypes IV and V, PFGE type 4 comprised serotypes II and III, and PFGE type 8 comprised serotypes Ia and II (Table 3). Moreover, five PFGE groups (PFGE types 1, 2, 9, 10, and 11) included both isolates with specific, well-defined serotypes and NT isolates (Table 3).
Among the most prevalent serotypes encountered, serotype III was found to contain the highest degree of genetic variability, with 12 distinct PFGE types among 21 GBS isolates displaying this serotype (Fig. 1).
Macrolide resistance phenotypes and genotypes and tetracycline resistance genes.
Erythromycin resistance was possessed by 16.5% of the strains (15 of 91). Although the macrolide-resistant strains clustered into seven different PFGE groups, 9 of 15 resistant isolates (60%) belonged to the PFGE type 1/CC1 group and were isolated from patients with noninvasive infections (8 isolates) and carriers (1 isolate). The remaining six macrolide-resistant strains were also isolated from patients with noninvasive infections (three isolates; PFGE types 25, 31, and 33) and carriers (three isolates; PFGE types 7, 17, and 19) (Fig. 1 and Table 3). Interestingly, no invasive strain displayed macrolide resistance.
Thirteen of 15 erythromycin-resistant isolates displayed the cMLSB resistance phenotype, while iMLSB and M resistance phenotypes were found in only one isolate each. Eleven GBS strains carried the erm(B) gene, three harbored the erm(A) [subclass erm(TR)] gene, and only one possessed the mef(A) gene (Fig. 1). All erm(B) gene-positive isolates and two erm(A) [subclass erm(TR)] gene-positive strains expressed the cMLSB resistance phenotype. The iMLSB resistance phenotype was found in one isolate containing the erm(A) [subclass erm(TR)] gene. The M resistance phenotype was expressed by the only isolate found to carry the mef(A) gene (Fig. 1 and Table 3).
The pattern of tetracycline resistance was diffuse (frequency, 68.1%; 62 of 91 strains), all tetracycline-resistant isolates harbored the tet(M) gene, and 11 of these isolates also carried the tet(O) gene (data not shown).
DISCUSSION
In the present study, invasive, noninvasive, and colonizing isolates of GBS were serotyped and characterized by their surface-associated proteins and antibiotic resistance genes. The clonal relatedness of isolates was established by using PFGE and MLST. This is the first study dealing with the molecular characterization of GBS isolates in Italy.
GBS strains showed a high degree of genetic diversity, with 34 distinct PFGE types and 1 PFGE profile corresponding to strains with undigested DNA. Nonetheless, 52.7% of the isolates belonged to four major clonal groups, PFGE type 1/CC1, PFGE type 2/CC23, PFGE type 3/CC17, and PFGE type 4/CC19. The major clonal group was PFGE type 1/CC1, represented by 17 isolates and distributed among serotypes V and IV and NT isolates. Representatives of this group displayed ST1, and the only serotype IV isolate exhibited ST196, a DLV of ST1. NT isolates closely related to type V isolates, as determined by PFGE, have been described previously (1), and NT isolates ultimately classified as ST1 in the present study have been previously reported to carry the capsular polysaccharide synthesis-specific gene characteristic of serotype V (35). Indeed, isolates of serotypes IV and V have been previously described as belonging to CC1 (29). The second most prevalent clonal group, PFGE type 2/CC23, was represented by isolates of serotype Ia. These isolates corresponded to ST24, a DLV of ST23, widely described as one of the most important genetic lineages among GBS strains causing human infections and associated mainly with serotype Ia (22, 29). The remaining two major clonal groups were PFGE type 3/CC17 and PFGE type 4/CC19. Isolates belonging to PFGE type 3 were of serotype III and ST17 (CC17). Isolates corresponding to ST17 are considered to be more virulent than other GBS isolates (22, 27, 29). Isolates belonging to PFGE type 4/CC19 were of serotype II (eight strains) and serotype III (one strain). Three different STs, ST28, ST182, and ST267, single-locus variants (SLVs) or DLVs of ST19, were found. ST19 has previously been found mainly among isolates of serotype III but also among isolates of serotype II (29), mostly from colonized infants (27). It has been observed previously that GBS isolates belonging to certain clonal groups, such as ST1, ST17, ST19, and ST23, are the main causes of human infections (29, 35). We also found the corresponding CCs to be the predominant ones recovered from infected or colonized humans in Italy.
The distribution of capsular serotypes has previously been documented to vary geographically (11, 15, 23, 42). Among the 91 GBS strains studied, all capsular serotypes except VI and VIII were found. Although the small number of strains in each population will necessitate future confirmation of the results, a significant association between some serotypes and the source of isolation was noted. Serotype V was more commonly isolated from patients with noninvasive infections, serotype Ia was associated with invasive infections, and NT strains were more frequently encountered among carriers.
The proportion of NT strains encountered in this study was considerable (15 of 91 strains), and these strains comprised mainly carrier isolates. However, by using molecular serotyping methods enabling the specific amplification of only one representative gene for each serotype (33), four NT strains gave an amplicon having reference to serotype V (all belonging to the PFGE type 1/CC1 group), three strains yielded an amplicon indicating serotype Ia, two strains gave products suggestive of serotype Ib, and one strain could be assigned to serotype VII (data not shown).
NT strains presenting the type V capsular polysaccharide synthesis-specific gene, the alp3 gene, and ST1 have been described previously, some as presenting multiple serotypes and insertion elements in the cps locus (35, 36). The possibility that the PFGE type 1/CC1 group in the present study may also include such atypical serotype V strains is intriguing and merits further investigation. However, we believed it was more appropriate to maintain the strains from which a single cps gene was typed in the category of NT strains to distinguish them from those expressing the capsule or possessing a complete cps operon. Serotype III was the predominant type, with 21 GBS isolates. Among the GBS isolates, the same serotypes were shared by isolates of different genetic lineages, and this finding may be ascribed to the phenomenon of horizontal transfer of capsular genes (22, 31). In this investigation, 12 different PFGE types were found for GBS isolates of serotype III. Different genetic lineages of GBS isolates of serotype III have been identified in previous epidemiological studies. Isolates of serotype III have been extensively studied since this serotype has been proven to be associated with infections in infants (19). Similar to serotype III isolates, each of the other serotypes found in this study was distributed among a few divergent groups as defined by PFGE.
Clonal groups shared by different serotypes were also noted. In addition to the five PFGE types shared by isolates with specific serotypes and NT isolates, three PFGE types, 1, 4, and 9, included isolates belonging to different serotypes. In a recent study on the characterization of invasive GBS isolates from Sweden by MLST, the observation of a number of STs corresponding to isolates of different serotypes was explained by the occurrence of a genetic transfer of the capsular genes (29). The capsular switching by the horizontal transfer of capsular operons between GBS isolates of different serotypes is likely driven by the host immune response and supported by the increased fitness acquired by isolates showing specific phenotype-genotype combinations.
It has been hypothesized previously that the diffusion of strains of particular surface protein profiles and serotypes reflects the selection of the best evolutionary lineages by the immune system (28). Our study may confirm this hypothesis: the four major CCs comprised isolates presenting serotype-surface protein gene combinations (type V-alp3, type Ia-alp1 or -alpha, and type II- or III-rib) repeatedly reported worldwide (28, 34). Nonetheless, some less frequent combinations (serotype III-alp1, serotype III-alp2, serotype III-alpha, and serotype Ia- or II-alp2) were observed in this study, suggesting that new successfully selected clones may be emerging.
A total of 15 macrolide-resistant GBS strains (16.5%) were isolated, from both patients with noninvasive infections and carriers. All erythromycin-resistant isolates exhibited one of the resistance genotypes studied, with the erm(B) genotype being the predominant one: it was always associated with the cMLSB resistance phenotype (11 isolates), while isolates carrying the erm(A) [subclass erm(TR)] gene exhibited the cMLSB resistance phenotype (2 isolates) or the iMLSB resistance phenotype (1 isolate).
It could be hypothesized that macrolide resistance has arisen and spread both by clonal dissemination and by the independent acquisition of resistance genes by different genetic lineages. In fact, 9 of 15 erythromycin-resistant isolates were of serotype V and belonged to the PFGE type 1/CC1 clonal group, and all but one carried the erm(B) gene. Interestingly, all nine of these resistant isolates were from patients with noninvasive infections; the serotype V-PFGE type 1/CC1 strains isolated from patients with invasive infections and carriers were sensitive to macrolides.
Serotype V has emerged recently as a cause of human disease with a high propensity to acquire macrolide resistance and spread throughout the population (1, 13). Other studies have also identified serotype V GBS isolates associated with erythromycin resistance (10, 26, 41). Moreover, a study from Korea showed that the most prevalent serotype of the erythromycin-resistant GBS strains examined was type V, with all the isolates harboring the erm(B) gene (40). Conversely, a study aiming at the evaluation of the antimicrobial resistance genes and the distribution of serotypes among human and bovine GBS isolates revealed the clonal spread of serotype V isolates resistant to erythromycin that carried mostly the erm(A) [subclass erm(TR)] gene (11).
The majority of isolates included in this study harbored the tetracycline resistance tet(M) genotype. We observed that all macrolide-resistant GBS isolates of the PFGE type 1 group carrying the erm(B) gene also harbored the tet(M) gene. The contemporary presence of both erm(B) and tet(M) may represent the consequence of a horizontal gene transfer event. Supporting the importance of this mechanism, Seral et al. (38) reported the identification of tet(M) and erm(B) genes on the same conjugative transposon, Tn1545, in S. pneumoniae.
In conclusion, the present study demonstrated that, despite the high degree of genetic diversity observed, more than half of the GBS isolates clustered into four major clonal groups. A strong correlation among clonal types, serotypes, surface-associated proteins, and resistance genes of the GBS isolates could be detected, indicating that, at least among the majority of macrolide-resistant GBS strains, clonal spread has occurred.
Acknowledgments
This work was supported by grant no. 5303 to G.O.
We are indebted to L. De Florio for invaluable assistance in PFGE experiments. We thank the medical staffs of the Italian hospitals who provided strains and descriptions of cases of invasive GBS diseases, particularly G. Amato (Napoli), C. Bonetti (Crema), G. Buonopane, P. Buosi (Cagliari), G. Caffiero (Vercelli), A. Camporese (Pordenone), P. Catalanotti (Napoli), P. Cavalcanti (Cosenza), A. P. Cipolloni (Cesena), A. Colananni (Terni), A. D'Argenio (Avellino), C. Favalli (Roma), P. Gualdi (Rovereto, TN), N. Manca (Brescia), P. Marone (Pavia), L. Migliorini (Siena), L. Pacciani (Roma), A. Palmieri (Sassari), P. Pecile (Firenze), C. Perazzi (Domodossola, VB), M. Pizzolante (Lecce), R. Rescaldani, S. Armitano (Erba, CO), R. Sartori (Trento), M. Sosio (Stradella, PV), C. Sturla (Gallarate, VA), A. Toniolo (Varese), C. Vitovila, and G. Teti (Messina). We are also indebted to the reference coordinators of the National Surveillance Program on Nosocomial Infections, E. A. Debbia, G. C. Schito, B. Posteraro, F. Cavallini, and M. Nicolosi.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Amundson, N. R., A. E. Flores, S. L. Hillier, C. J. Baker, and P. Ferrieri. 2005. DNA macrorestriction analysis of nontypeable group B streptococcal isolates: clonal evolution of nontypeable and type V isolates. J. Clin. Microbiol. 43:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. 2000. Group B streptococcal infections, p. 222-237. In D. Stevens and E. Kaplan (ed.), Streptococcal infections. Oxford University Press, New York, NY.
- 3.Baron, M. J., G. R. Bolduc, M. B. Goldberg, T. C. Auperin, and L. C. Madoff. 2004. Alpha C protein of group B Streptococcus binds host cell surface glycosaminoglycan and enters cells by an actin-dependent mechanism. J. Biol. Chem. 279:24714-24723. [DOI] [PubMed] [Google Scholar]
- 4.Baron, M. J., D. J. Filman, G. A. Prophete, J. M. Hogle, and L. C. Madoff. 2007. Identification of a glycosaminoglycan-binding region of the alpha C protein that mediates entry of group B streptococci into host cells. J. Biol. Chem. 282:10526-10536. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Prevention of perinatal group B streptococcal disease. Morb. Mortal. Wkly. Rep. 51:1-22. [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing: 16th informational supplement. M100-S16, vol. 26 (no. 3). Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Creti, R., F. Fabretti, G. Orefici, and C. von Hunolstein. 2004. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J. Clin. Microbiol. 42:1326-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creti, R., G. Gherardi, M. Imperi, C. von Hunolstein, L. Baldassarri, M. Pataracchia, G. Alfarone, F. Cardona, G. Dicuonzo, and G. Orefici. 2005. Association of group A streptococcal emm types with virulence traits and macrolide-resistance genes is independent of the source of isolation. J. Med. Microbiol. 54:913-917. [DOI] [PubMed] [Google Scholar]
- 9.Creti, R., M. Imperi, L. Baldassarri, M. Pataracchia, G. Alfarone, and G. Orefici. 2007. Lateral transfer of alpha-like protein gene cassettes among streptococci: identification of a new family member in Streptococcus dysgalactiae subsp. equisimilis. Lett. Appl. Microbiol. 44:224-227. [DOI] [PubMed] [Google Scholar]
- 10.Diekema, D. J., J. I. Andrews, H. Huynh, P. R. Rhomberg, S. R. Doktor, J. Beyer, V. D. Shortridge, R. K. Flamm, R. N. Jones, and M. A. Pfaller. 2003. Molecular epidemiology of macrolide resistance in neonatal bloodstream isolates of group B streptococci. J. Clin. Microbiol. 41:2659-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dogan, B., Y. H. Schukken, C. Santisteban, and K. J. Boor. 2005. Distribution of serotypes and antimicrobial resistance genes among Streptococcus agalactiae isolates from bovine and human hosts. J. Clin. Microbiol. 43:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, M. S., and C. J. Baker. 2005. Group B streptococcal infections in elderly adults. Clin. Infect. Dis. 41:839-847. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, J. A., K. D. Farmer, and R. R. Facklam. 1998. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J. Clin. Microbiol. 36:2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 15.Fluegge, K., S. Supper, A. Siedler, and R. Berner. 2005. Serotype distribution of invasive group B streptococcal isolates in infants: results from a nationwide active laboratory surveillance study over 2 years in Germany. Clin. Infect. Dis. 40:760-763. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs, R. S., S. Schrag, and A. Schuchat. 2004. Perinatal infections due to group B streptococci. Obstet. Gynecol. 104:1062-1076. [DOI] [PubMed] [Google Scholar]
- 17.Gigax, S. E., J. A. Schuyler, L. E. Kimmel, J. P. Trama, E. Mordechai, and M. E. Adelson. 2006. Erythromycin and clindamycin resistance in group B streptococcal clinical isolates. Antimicrob. Agents Chemother. 50:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, A. Schuchat, et al. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 20.Heath, P. T., and A. Schuchat. 2007. Perinatal group B streptococcal disease. Best Pract. Res. Clin. Obstet. Gynaecol. 21:411-424. [DOI] [PubMed] [Google Scholar]
- 21.Jones, N., J. F. Bohhsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. M. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, N., K. A. Oliver, J. Barry, R. M. Harding, N. Bisharat, B. G. Spratt, T. Peto, and D. W. Crook for the Oxford Group B Streptococcus Consortium. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B Streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42:915-924. [DOI] [PubMed] [Google Scholar]
- 23.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lachenauer, C. S., R. Creti, J. L. Michel, and L. C. Madoff. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 97:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 26.Lin, F. Y., P. H. Azimi, L. E. Weisman, J. B. Philips III, J. Regan, P. Clark, G. G. Rhoads, J. Clemens, J. Troendle, E. Pratt, R. A. Brenner, and V. Gill. 2000. Antibiotic susceptibility profiles for group B streptococci isolated from neonates, 1995-1998. Clin. Infect. Dis. 31:76-79. [DOI] [PubMed] [Google Scholar]
- 27.Lin, F. Y. C., A. Whiting, E. Adderson, S. Takahashi, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips III, L. E. Weisman, J. Regan, P. Clark, G. G. Rhoads, C. E. Frasch, J. Troendle, P. Moyer, and J. F. Bohnsack. 2006. Phylogenetic lineages of invasive and colonizing strains of serotype III group B streptococci from neonates: a multicenter prospective study. J. Clin. Microbiol. 44:1257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindhal, G., M. Stalhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan, S. L., M. Granlund, M. Sellin, T. Lagergård, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning, S. D., D. W. Lacher, H. D. Davies, B. Foxman, and T. S. Whittam. 2005. DNA polymorphism and molecular subtyping of the capsular gene cluster of group B Streptococcus. J. Clin. Microbiol. 43:6113-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersson, K. 2007. Perinatal infection with group B streptococci. Semin. Fetal Neonatal Med. 12:193-197. [DOI] [PubMed] [Google Scholar]
- 33.Poyart, C., A. Tazi, H. Reglier-Poupet, A. Billoet, N. Tavares, J. Raymond, and P. Trieu-Cuot. 2007. A multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. J. Clin. Microbiol. 45:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramaswamy, S. V., P. Ferrieri, A. E. Flores, and L. C. Paoletti. 2006. Molecular characterization of nontypeable group B streptococcus. J. Clin. Microbiol. 44:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, M. C., Y. Pang, D. E. Riley, S. L. Hillier, R. C. Berger, and J. N. Krieger. 1993. Detection of tetM and tetO tetracycline resistance genes by polymerase chain reaction. Mol. Cell. Probes 7:387-393. [DOI] [PubMed] [Google Scholar]
- 37.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garcia, and R. Gomez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 39.Sun, Y., F. Kong, Z. Zhao, and G. L. Gilbert. 2005. Comparison of a 3-set genotyping system with multilocus sequence typing for Streptococcus agalactiae (group B streptococcus). J. Clin. Microbiol. 43:4704-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uh, Y., I. H. Jang, G. Y. Hwang, M. K. Lee, K. J. Yoon, and H. Y. Kim. 2004. Serotypes and genotypes of erythromycin-resistant group B streptococci in Korea. J. Clin. Microbiol. 42:3306-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Both, U., M. Ruess, U. Mueller, K. Fluegge, A. Sander, and R. Berner. 2003. A serotype V clone is predominant among erythromycin-resistant Streptococcus agalactiae isolates in a southwestern region of Germany. J. Clin. Microbiol. 41:2166-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen, L., Q. Wang, Y. Li, F. Kong, G. L. Gilbert, B. Cao, L. Wang, and L. Feng. 2006. Use of a serotype-specific DNA microarray for identification of group B Streptococcus (Streptococcus agalactiae). J. Clin. Microbiol. 44:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]