Abstract
Two hundred eighty methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates recovered from a tertiary care hospital in Oporto, Portugal, between 2003 and 2005 were studied by a combination of molecular typing techniques in order to investigate the genetic backgrounds associated with the changes in the resistance phenotypes observed since 2001 and compare them to those previously found in the hospital. All MRSA isolates were grouped into resistance profiles for a panel of seven antibiotics and characterized by pulsed-field gel electrophoresis (PFGE) and SCCmec (staphylococcal cassette chromosome mec) typing. Representative isolates of PFGE types were further studied by spa typing and multilocus sequence typing. Our findings clearly document that the increasing isolation of nonmultiresistant MRSA strains was associated with the decline (from 69% in 1996 to 2000 to 12% in 2003 to 2005) and massive replacement of the multiresistant Brazilian clone (ST239-IIIA) by the epidemic EMRSA-15 clone (ST22-IV), in which resistance to antibiotics other than β-lactams is very rare, as the major clone (80% of isolates). The Iberian clone (ST247-IA), a major clone in 1992 to 1993, was represented in the present study by just one isolate. Two other pandemic MRSA clones were detected, as sporadic isolates, for the first time in our hospital: the New York/Japan (ST5-II) and the EMRSA-16 (ST36-II) clones. Furthermore, the pattern of susceptibility of MRSA isolates both to gentamicin and to trimethoprim-sulfamethoxazole was shown to be an excellent phenotypic marker for the discrimination of the EMRSA-15 clone from other nonmultiresistant MRSA clones present in our hospital.
Methicillin-resistant Staphylococcus aureus (MRSA) is currently the most commonly identified antibiotic-resistant nosocomial pathogen worldwide (8, 19, 20, 58). The European Antimicrobial Resistance Surveillance System reported an increase from 16 to 24% between 1999 and 2004 (19) in the overall prevalence of MRSA in all regions of Europe, including countries of low endemicity. According to the most recent report of the European Antimicrobial Resistance Surveillance System, only 7 out of 30 countries still reported a prevalence of MRSA below 3% in 2005, namely, Iceland, Norway, Sweden, and Estonia, with a MRSA prevalence relatively stable over time, and The Netherlands, Denmark, and Finland, with a significant increase since 1999 (20). In summary, the central European countries reported, in 2005, resistance prevalence below 25%, whereas all southern European countries reported higher levels and eight of them reported prevalence rates over 40%, including Portugal with a prevalence of 47% of invasive MRSA isolates (20). However, in two southern European countries, Slovenia and France, the implementation of appropriate long-term infection control measures has resulted in a consistent decrease of the MRSA burden over the past 5 to 6 years, indicating that the control of MRSA in hospitals with high prevalence is not an unachievable goal (20). Data regarding the proportion of MRSA strains among all S. aureus isolates identified at the Laboratory of Microbiology of Hospital Geral de Santo António (HGSA), Oporto, Portugal, since 1992 show that MRSA has been endemic in this hospital with overall MRSA rates near 50% in the years 1992 through 1995. In the following years, the MRSA rates decreased to around 43% in 2004 and then increased significantly to 55% in 2005.
The success of efforts in the control of MRSA transmission within hospitals is largely determined by knowledge of the local epidemiology, in particular the nature and number of clones involved in carriage and infection and the identification of the sources and vehicles of transmission, as well as the identification of the reservoirs of epidemic clones in the population and/or environment (9, 37, 38, 57).
Previous molecular typing of MRSA isolates recovered in HGSA during two periods of surveillance revealed the same scenario documented for several Portuguese hospitals, where the Iberian clone (ST247-IA) was by 1992 and 1993 dominant, represented by 77% of isolates, and drastically declined after the introduction of the Brazilian clone (ST239-IIIA) in 1995. The multidrug-resistant Brazilian clone was then repeatedly identified as the major clone in HGSA over the past several years (1996 to 2000) (7).
The continuous-surveillance study of overall antibiotic resistance in HGSA has shown, at least since 2001, a significant change in the resistance profile of the MRSA isolates recovered. MRSA strains in which resistance to antibiotics, other than β-lactams, was rare emerged more frequently, and at the same time the traditional multiresistant MRSA strains became less frequent. A similar trend of emergence and spread of MRSA isolates with susceptibility to gentamicin (GEN) and other antibiotics has been observed in hospitals in several other European countries, namely, France, Germany, Belgium, and Spain, among others (15, 16, 29, 30, 47, 61, 63).
In this study we characterized a collection of MRSA isolates expressing the most frequent MRSA resistance profiles detected in HGSA during a 3-year period (2003 to 2005) in order to correlate the changes in the resistance profiles with the clonal nature of MRSA in our institution and compare them with those previously detected in 1992 to 1993 and 1996 to 2000 (7). A combination of molecular typing methods, including pulsed-field gel electrophoresis (PFGE), spa typing, multilocus sequence typing (MLST), and SCCmec (staphylococcal cassette chromosome mec) typing, was used to detect shifts in the prevalence of major clones, the emergence of new epidemic clones, and the possible spread of previously detected minor/sporadic clones.
MATERIALS AND METHODS
HGSA and wards/units included in the study.
HGSA, Oporto, Portugal, is a 700-bed tertiary care teaching hospital with 26 wards; three intensive care units; and 34 outpatient service units, including one specific diabetic foot unit and one surgery unit. The numbers of patients hospitalized in 2003, 2004, and 2005 were 21,620, 22,208, and 24,282, respectively. In a retrospective analysis of data regarding the HGSA wards with the highest number of MRSA isolates, it was observed that in the vascular surgery and endocrinology wards there was an important rise of almost two-and-a-half- and fivefold, respectively, in the number of MRSA strains isolated from 2001 to 2002. These wards are the major destination of diabetic foot outpatients when admitted to HGSA as inpatients and were selected, together with the diabetic foot outpatient unit (DFOU) of HGSA, for a preintervention study to better understand the epidemiology of MRSA strains circulating in the hospital and their possible connections to the community served by HGSA. In the vascular surgery ward 25 beds and 45 healthcare workers (HCWs) were included, and in the endocrinology ward those figures were 17 and 38, respectively. The DFOU of HGSA is a non-scheduled-appointment outpatient unit with a weekly average attendance of 120 diabetic patients from northern Portugal. From January 2003 through December 2005 averages of 2,026 initial and 13,568 follow-up visits per year were recorded.
Clinical isolates.
A total of 280 MRSA isolates representative of nasal colonization samples (n = 92) and infection samples (n = 188) were included in the present work; a single MRSA isolate per patient was studied. Colonization screenings were performed during 1 week three times a year (February, June, and November) in the three units included in this study. All admitted patients and outpatients attending the DFOU during the week of the survey were screened, and HCWs were also asked to participate. Also included in the colonization collection were 10 strains isolated from patients and HCWs from a high-risk-patient ward, the neonatal intensive care unit. One nasal swab (Culturette EZ; Becton Dickinson) was taken from both anterior nares and tested for the presence of S. aureus in the laboratory within 2 hours. Nasal swabs were inoculated directly onto 5% sheep blood agar (bioMérieux, France) and mannitol salt agar plates (Biogerm, Portugal) and incubated for 24 to 48 h at 35°C; colonies suspected to be S. aureus were tested with the Slidex StaphPlus agglutination test (bioMérieux, France). Identification and susceptibility testing of all S. aureus isolates were done with Vitek 2 system cards (bioMérieux, France).
MRSA isolates associated with infection were recovered from several clinical sources, including pus (n = 159), blood (n = 14), tissue (n = 4), bronchial secretions (n = 3), catheter (n = 2), bone (n = 1), urine (n = 1), and other diverse clinical sources (n = 4).
The 280 MRSA isolates were recovered between 2003 and 2005 (85, 78, and 117 isolates in 2003, 2004, and 2005, respectively) from colonization or infection samples of 255 patients (single MRSA isolates) and from colonization samples of 25 HCWs. The majority of isolates, 56% (146 out of 280), were recovered in the vascular surgery ward, followed by DFOU with 30% (85 out of 280) and the endocrinology ward with 14% (39 out of 280).
Antimicrobial susceptibility testing.
The identification and antimicrobial susceptibility testing of MRSA isolates were performed with the Vitek 2 system (cards GP and AST-P536, respectively) following the manufacturer's recommendations (bioMérieux, France). Inducible clindamycin (CLI) resistance was detected using the disk approximation test according to CLSI guidelines, and the phenotypes of resistance to macrolides, lincosamides, and type B streptogramins (MLSB) were further assigned as follows: constitutive (cMLSB), erythromycin (ERY) and CLI resistant; inducible (iMLSB), ERY resistant with CLI resistance inducible by ERY (flattening of the CLI zone adjacent to the ERY disk); or M phenotype, ERY resistant and CLI susceptible (no flattening of the CLI zone) (12). MRSA isolates were further grouped into resistance profiles (RPs) according to the pattern of susceptibility to a panel of seven antibiotics: GEN, ERY, CLI, tobramycin (TOB), tetracycline (TET), rifampin (RIF), and trimethoprim-sulfamethoxazole (SXT).
PFGE.
PFGE was performed as described by Chung et al. (10) for all 280 isolates. The patterns obtained were interpreted by visual inspection according to the criteria of McDougal et al. (31) and further deposited in a database and analyzed with the Bionumerics software, version 4.6 (Applied-Maths, Kortrijk, Belgium). PFGE patterns were identified using a dendrogram generated by the unweighted-pair group method with arithmetic mean based on Dice coefficients, where optimization and band position tolerance were set at 0.5 and 1.3%, respectively. A similarity coefficient of 80% was selected to define the patterns (31).
SCCmec typing and ccrAB typing.
SCCmec typing was performed by a multiplex PCR strategy, previously described by Oliveira and de Lencastre (41), in which a specific amplification pattern for each structural type of SCCmec is generated. SCCmec type assignments were confirmed for a subset of 99 representative strains by ccrAB typing as previously described by Okuma et al. (40). Isolates classified as nontypeable or negative for ccrAB by PCR were selected for ccrB sequencing, according to the work of Oliveira et al. (42).
spa typing and MLST.
spa typing and MLST were performed as previously described (14, 17, 27, 54). Sequences of both strands were determined at Macrogen (Seoul, South Korea), and sequence analysis was carried out using DNAStar software, version 5.07 (Lasergene, Madison, WI). Ridom StaphType software, version 1.4 (Ridom GmbH, Würzburg, Germany), was also used for spa type analyses. The new spa type assignments were obtained through the Ridom SpaServer (http://spa.ridom.de/index.shtml). MLST sequence types were assigned through the MLST database (http://www.mlst.net).
Statistical analysis.
Statistical analysis was performed with SPSS software, version 15.0. The proportions were compared using the chi-squared test or Fisher's exact test, as appropriate. A P value of <0.05 was defined as statistically significant; all statistical tests were two-tailed.
RESULTS
Phenotypic and genotypic characterization of MRSA isolates.
Of the 280 MRSA isolates studied, 10% were resistant to GEN, 82.5% to ERY, 82.5% to CLI, 16.4% to TOB, 14.3% to TET, 14.6% to RIF, and 12.8% to SXT. Among the 231 isolates resistant to CLI, resistance was constitutive (cMLSB) in 63.2% (n = 146) of isolates and inducible (iMLSB) in 36.8% (n = 85); no isolate showed the M phenotype.
The MRSA isolates were grouped into 12 RPs with the majority belonging to RP-I (susceptibility to all antibiotics) and RP-II (resistance to ERY and CLI) with 16% (46 out of 280) and 64% (179 out of 280) of isolates, respectively (Table 1). The remaining MRSA isolates were included in RP-III (resistance to RIF), RP-IV (resistance to SXT), RP-V (resistance to ERY, CLI, and TET), RP-VI (resistance to ERY, CLI, and RIF), RP-VII (resistance to ERY, CLI, and TOB), RP-VIII (resistance to ERY, CLI, TOB, and RIF), RP-IX (resistance to ERY, CLI, TET, RIF, and SXT), RP-X (resistance to all but GEN), RP-XI (resistance to all but SXT), and RP-XII (resistance to all antibiotics) (Table 1).
TABLE 1.
Resistance profiles of the 280 MRSA strains studied and associated PFGE types
| RP | Susceptibility toa:
|
Total no. of isolates | PFGE type(s) (no. of isolates) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GEN | ERY | CLI | TOB | TET | RIF | SXT | |||
| I | S | S | S | S | S | S | S | 46 | J (40); M (5); E (1) |
| II | S | R | R | S | S | S | S | 179 | J (178); M (1) |
| III | S | S | S | S | S | R | S | 1 | J (1) |
| IV | S | S | S | S | S | S | R | 2 | D (2) |
| V | S | R | R | S | R | S | S | 4 | J (4) |
| VI | S | R | R | S | S | R | S | 1 | J (1) |
| VII | S | R | R | R | S | S | S | 8 | M (2); C (6) |
| VIII | S | R | R | R | S | R | S | 3 | M (2); J (1) |
| IX | S | R | R | S | R | R | R | 1 | B (1) |
| X | S | R | R | R | R | R | R | 7 | B (7) |
| XI | R | R | R | R | R | R | S | 2 | A (1); F (1) |
| XII | R | R | R | R | R | R | R | 26 | B (25); G (1) |
MIC breakpoints, expressed in μg/ml, used for the definition of categories of susceptible (S), intermediate (I), and resistant (R), were those recommended by CLSI: GEN, ≤4, 8, and ≥16; ERY, ≤0.5, 1 to 4, and ≥8; CLI, ≤0.5, 1 to 2, and ≥4; TOB, ≤4, 8, and ≥16; TET, ≤4, 8, and ≥16; RIF, ≤1, 2, and ≥4; SXT, ≤2/38, — (intermediate breakpoint not applicable to SXT per CLSI), and ≥4/76, respectively.
The 280 MRSA isolates were classified into nine PFGE types: J (80.3%; n = 225), B (11.8%; n = 33), M (3.5%; n = 10), C (2.1%; n = 6), D (0.7%; n = 2), and A, E, F, and G, with one isolate each (Table 2). The major type could be further divided into 15 subtypes with the majority of the isolates (79%, n = 178) belonging to a single subtype (J189). PFGE pattern B could be also divided into 15 subtypes with 36% of isolates (12 out 33) belonging to a single subtype (B234). Type M was divided into three subtypes.
TABLE 2.
Summary of the molecular typing data for 280 MRSA strains, January 2003 to December 2005, HGSA
| PFGE type; no. (%) of strains | SCCmec type; no. of strains | Ridom spa typingb
|
MLSTb
|
||
|---|---|---|---|---|---|
| spa type | Sequence of repeats | Allelic profile | STc | ||
| J; 225 (80.3) | IV; 224 | t032 | 26-23-23-13-23-31-29-17-31-29-17-25-17-25-16-28 | 7-6-1-5-8-8-6 | 22 |
| t747 | 26-23-23-31-29-17-31-29-17-25-17-25-16-28 | ||||
| t849 | 26-23-23-29-17-25-17-25-16-28 | ||||
| t022 | 26-23-13-23-31-29-17-31-29-17-25-17-25-16-28 | ||||
| IV vara; 1 | t032 | 26-23-23-13-23-31-29-17-31-29-17-25-17-25-16-28 | |||
| B; 33 (11.8) | IIIA; 33 | t037 | 15-12-16-02-25-17-24 | 2-3-1-1-4-4-3 | 239 |
| M; 10 (3.5) | I; 4 | t002 | 26-23-17-34-17-20-17-12-17-16 | 1-4-1-4-12-1-10 | 5 |
| II; 1 | t045 | 26-17-20-17-12-17-16 | |||
| IV; 2 | |||||
| IVA; 3 | t002 | 26-23-17-34-17-20-17-12-17-16 | 1-43-1-4-12-1-10 | 146 | |
| t509 | 26-23-17-20-17-12-17-16 | ||||
| C; 6 (2.1) | II; 6 | t018 | 15-12-16-02-16-02-25-17-24-24-24 | 2-2-2-2-3-3-2 | 36 |
| D; 2 (0.7) | IV; 2 | t186 | 07-12-21-17-13-13-34-34-33-34 | 22-1-14-23-12-4-31 | 88 |
| A; 1 (0.4) | IA; 1 | t051 | 11-19-21-12-21-17-34-24-34-22-25 | 3-3-1-12-4-4-16 | 247 |
| E; 1 (0.4) | IVA; 1 | t645 | 14-44-13-12-17-23-18-17 | 6-5-6-2-7-14-5 | 121 |
| F; 1 (0.4) | NTd; 1 | t860 | 08-16-02-25-17 | 2-3-1-1-4-4-3 | 239 |
| G; 1 (0.4) | IIIA; 1 | t021 | 15-12-16-02-16-02-25-17-24 | 2-3-1-1-4-4-3 | 239 |
IV var, dcs negative, ccrAB type 2.
spa typing and MLST were performed on representative strains of all PFGE types and subtypes.
Clones correspond to sequence types as follows: EMRSA-15 clone, ST22-IV; Brazilian clone, ST239-IIIA; New York/Japan clone, ST5-II; pediatric clone, ST5-IV; EMRSA-16 clone, ST36-II; Iberian clone, ST247-IA.
NT, nontypeable.
No correlation could be established between single resistance profiles and PFGE types, since the same resistance profile could be found among isolates presenting distinct PFGE types (Table 1). Nevertheless, MRSA isolates belonging to the major PFGE type J demonstrated a uniform susceptibility to GEN and SXT and were all susceptible to TOB, RIF, and TET, except for one, three, and four isolates, respectively; the majority of the isolates (82%) were resistant to ERY and CLI (57.6% with the constitutive phenotype and 42.4% with the inducible phenotype) (Table 1). In contrast, 100% of MRSA isolates associated with PFGE type B were resistant to ERY, CLI, TET, RIF, and SXT, and all but one were resistant to TOB; 87.9% of MRSA isolates resistant to CLI showed the constitutive phenotype (cMLSB). A heterogeneous resistance profile for GEN (76%; 25 out of 33) was observed (Table 1).
For the seven minor PFGE types found, which represented 8% (22 out of 280) of isolates, a greater heterogeneity in resistance profiles was observed. Nevertheless, two general groups could be distinguished: one with MRSA isolates resistant to β-lactams only or to β-lactams plus one to three antibiotics (PFGE types C, D, E, and M) and a second group with MRSA isolates resistant to all or nearly all seven antibiotics (PFGE types A, F, and G) (Table 1).
Furthermore, no correlation could be established between single PFGE types and the phenotypes of macrolide resistance, though 91.8% (n = 78) of MRSA isolates with the inducible phenotype were characterized by PFGE type J, 4.7% (n = 4) by PFGE type B, and 3.5% (n = 3) by PFGE type M.
spa typing, MLST, and SCCmec typing.
spa typing and MLST were performed on representative strains from each group as defined by PFGE (Table 2). The predominant PFGE type J was characterized by four different spa types (t022, t032, t747, and t849), sequence type 22 (ST22), and SCCmec type IV (clone ST22-IV, EMRSA-15), except for one strain classified as a SCCmec type IV variant. PFGE type B isolates were characterized by spa type t037, ST239, and SCCmec type IIIA (clone ST239-IIIA, Brazilian). Two isolates, with closely related spa types (types t860 and t021) but unique patterns (types F and G) were also characterized by ST239.
All the 10 PFGE type M isolates belong to clonal complex 5 (CC5) (ST5 or its single-locus variant ST146) and have closely related spa types (t002, t045, and t509). However, four different SCCmec types were detected (I, II, IV, and IVA), so that these strains were assigned to different clonal types: the New York/Japan clone (ST5-II), the pediatric clone (ST5-IV), and clone ST146-IVA. ST146-IVA may have evolved from the pediatric clone, resulting from a single point mutation (aroE allele 4 → 43), plus the integration of pUB110 in the SCCmec type IV cassette. This clone has been already described in association with the hospital environment in Spain (47).
Molecular typing identified two other previously described epidemic MRSA clones: EMRSA-16 (ST36-II) and the Iberian/Archaic clone (ST247-I), which accounted for 2.1% (6 out of 280) and 0.4% (1 out of 280) of the isolates, respectively. Two other clones, ST88-IV and ST121-IVA, were also found in two strains and one strain, respectively.
SCCmec typing results of the 280 MRSA isolates are summarized in Table 2. All isolates except one were assigned to a SCCmec type based on the SCCmec multiplex strategy (41). The single isolate nontypeable by the SCCmec multiplex strategy did not present a typical pattern; despite the fact that some amplifications were specific for SCCmec III, when ccrAB typing was performed, only the control primers presented amplification products (40). ccrAB typing was performed on 99 representative isolates and confirmed the SCCmec type assignment for all strains except two, previously assigned to SCCmec type II by the multiplex strategy. The two strains nontypeable by ccrAB typing were further evaluated by ccrB typing according to the work of Oliveira et al. (42). One strain could not be characterized either by ccrAB or by ccrB typing, and further studies are needed in order to characterize the SCCmec type present; ccrB typing results confirmed the SCCmec multiplex result for the other strain.
DISCUSSION
Epidemiological studies using different molecular typing techniques have demonstrated that most hospital-acquired MRSA infections are due to a relatively small number of epidemic MRSA clones spread worldwide, namely, the Iberian (ST247-IA), the Brazilian/Hungarian (ST239-III), the Berlin (ST45-IV), the New York/Japan (ST5-II), the pediatric (ST5-IV), the EMRSA-15 (ST22-IV), and the EMRSA-16 (ST36-II) clones (18, 43, 44).
Previous characterization of MRSA isolates recovered in our hospital from two periods of surveillance allowed us to detect important shifts in the clonal nature of endemic MRSA in our hospital since 1992 (7). The most relevant change was the replacement of the Iberian clone (ST247-IA) by the Brazilian clone (ST239-IIIA) in the mid-1990s. Besides these two multiresistant MRSA clones, which were also the most frequent in several other Portuguese hospitals during the last decade, minor or sporadic clones also with epidemic potential were identified at HGSA, such as the pediatric clone (ST5-IV) (22, 51).
Several molecular techniques are available for MRSA typing, allowing the discrimination of epidemiologically related or clonal isolates from unrelated isolates. However, criteria for the application of each technique will be different according to the purpose of the study, dealing for example either with local outbreak investigations or with large-scale surveillance studies (55, 60). Although PFGE is a powerful technique for evaluating the relatedness of isolates for epidemiological purposes, it might not be the best option for evolutionary studies. MLST has been proven to be a typing tool suitable for long-term and global epidemiology, allowing related MRSA strains recovered over extended periods of time and different countries to be readily identified (18). In our study the application of a combination of methods such as PFGE, spa typing, MLST, and SCCmec typing allowed us to better identify the clonal nature of MRSA strains circulating in HGSA over 3 years (2003 to 2005) and compare them with those observed in 1992 to 1993 and 1996 to 2000.
Changes in the genetic background of MRSA isolates from HGSA over time.
Data regarding the MRSA prevalence among all S. aureus isolates identified at the Laboratory of Microbiology, HGSA, since 1992 show that MRSA has been endemic in this hospital with an overall MRSA rate around 45% (minimum, 36%; maximum, 55%) for the last 14 years, without significant changes over time. Nevertheless, the present study has revealed major shifts in the clonal nature of these isolates, namely, the massive replacement of the Brazilian clone (ST239-IIIA, PFGE type B) by the EMRSA-15 (ST22-IV, PFGE type J) clone as the major clone (80% of all the 280 MRSA isolates studied) in HGSA. As a matter of fact, the Brazilian clone, which in 1996 to 2000 was the major clone (69%) at HGSA, in 2003 to 2005 was represented by only 12% of the MRSA strains isolated. The EMRSA-15 clone was found to be widely disseminated, with no statistically significant difference, in the three wards and the outpatient unit evaluated (P = 0.61). Furthermore, no difference was observed concerning the type of clinical sample (infection versus colonization; P = 0.73) or whether MRSA was recovered from patients or HCWs (P = 0.60). However, we found an interesting and statistically significant association (P = 0.005) of this clone with patients who usually attend the DFOU versus those patients who do not attend the DFOU. Our findings support the data recently published by Alcoceba et al. reporting the spread of the EMRSA-15 clone more frequently among nonhospitalized patients (emergency room and outpatient departments) than among hospitalized patients from the three main public Majorcan hospitals (6). Nevertheless and despite the suggestion by Alcoceba et al. about the movement of EMRSA-15 occurring mainly from the community to the hospital setting, the MRSA isolates recovered from the outpatients attending the DFOU were classified as hospital acquired since all of the patients had had previous visits to the DFOU, an average of four in the previous 12 months, and 70% had been previously admitted as inpatients to HGSA. In contrast to the observations by Alcoceba et al., the proportion of the EMRSA-15 isolates among blood samples was not statistically different from other samples (P = 0.49), though 71.4% of MRSA isolates from blood samples belonged to the EMRSA-15 clone; nevertheless, further studies with a large number of blood samples would be necessary to clarify this issue.
EMRSA-15 was first reported in 1991 in southeast England and the Midlands, rapidly spread across a large number of hospitals in the United Kingdom, and became one of the two most dominant epidemic clones, accounting for 70% of MRSA strains isolated in England and Scotland (25, 36, 48). The EMRSA-15 clone is one of the pandemic MRSA clones (18) and has been identified in several countries such as The Netherlands (18, 62), Denmark (21), Germany (63), Ireland and Sweden (18), the Czech Republic (32, 33), Australia and New Zealand (23, 46), Kuwait (59), and Spain (6, 35, 47).
Another relevant finding of the present study concerns the Iberian clone (ST247, SCCmec type IA, PFGE type A), which almost disappeared from HGSA, being represented by just one isolate. It is important to note that the Iberian clone was in 1992 to 1993 the major clone (77%) in HGSA, as well as in several other Portuguese hospitals. With the emergence of the Brazilian clone around 1996, the representation of the Iberian clone drastically declined to 17% in our hospital during 1996 to 2000. Our results shows that the pediatric clone (ST5-IV, PFGE type M), which was one of the minor clones (1.7% of isolates) detected at HGSA in 1992 to 1993, was unable to spread over time, remaining in 2003 to 2005 at a stable low level (less than 2% of isolates).
Two other important pandemic MRSA clones were detected for the first time in our hospital in 2005, the New York/Japan (ST5-II, PFGE type M) clone and the EMRSA-16 (ST36-II, PFGE type C) clone, represented by one and six isolates, respectively. The New York/Japan clone was found to be a predominant clone in the United States (31, 49, 50), Canada (56), Japan (3), and Korea (26) and was also detected in Finland, Ireland, and the United Kingdom (18). To our knowledge, this is the first report of the New York/Japan clone in a Portuguese hospital. An interesting observation was the heterogeneity of SCCmec types associated with CC5 (ST5 and ST146). Notwithstanding the fact that SCCmec types II to IV have been already reported associated with ST5 (2), it is worthwhile to note that PFGE profile type M was first described in HGSA in 2003 associated with ST146, a single-locus variant of ST5, and SCCmec type IVA and that at the same time, a group of methicillin-susceptible S. aureus strains, characterized by the same PFGE profile and spa type but by ST5, were also isolated in the same wards (data not shown). Since different SCCmec types have been found associated with CC5 strains (ST5 and ST146), we may be in the presence of multiple acquisitions of the SCCmec (types I, II, IV, and IVA) by a single background (CC5). The EMRSA-16 clone, initially associated with an outbreak in 1992 in Northamptonshire, United Kingdom, is nowadays together with EMRSA-15 one of the major clones in the United Kingdom, where the two are responsible for more than 95% of MRSA bacteremia (13, 25, 36). This clone was also found in Mexico (4), Canada (56), and several European countries such as Greece (1); Sweden (53); Finland (52); Denmark, Switzerland, and Belgium (39); and Spain (35, 47).
Recent studies from two hospitals in Tenerife, Spain, reported the abrupt replacement, after 2000, of the Iberian clone (ST247-IA) by EMRSA-16 (ST36-II) as the major clone (35, 47). The presence of the EMRSA-15 clone was also reported, although it represented only 2 to 3% of MRSA isolates recovered from these hospitals. A different picture was observed in our hospital, where EMRSA-15 was shown to have a clear advantage over EMRSA-16, which emerged later as a minor clone (5% of isolates). However, future follow-up studies of the clonal nature of MRSA in our hospital will allow us to trace the spread of EMRSA-16, which seems to have the same epidemicity as the EMRSA-15 clone as shown in hospitals in the United Kingdom and other European countries.
Clone ST88-IV, which has been already reported in Asia (26) and associated with community-associated MRSA strains in Switzerland (24), was also identified in our study. Although only two strains were found, both were from the same ward, one isolated from a newborn (blood) and the other from an HCW (nasal swab). Although the two strains were not isolated at the same time, this may demonstrate a possible route for colonization/infection in the hospital due to the HCWs, since no other ST88-IV strains were found during the study period. Similar observations were already made in previous studies (11, 34).
Clone ST121 has been most often associated with methicillin-susceptible S. aureus from carriage, as previously described (5, 28), and according to the data available at the MLST database (http://www.mlst.net). However, in our study we identified a single ST121 MRSA strain. An ST121 MRSA strain, isolated from sputum, was also identified in a Chinese hospital in 2006 (http://www.mlst.net), and another ST121-IV MRSA strain, associated with nasal carriage, was detected in 2005 in the United States (45). The emergence in our hospital of ST121 MRSA strains may be a result of a local and recent SCCmec acquisition by a susceptible S. aureus background.
Relationship between the narrowing of resistance phenotypes and the spread of new epidemic clones.
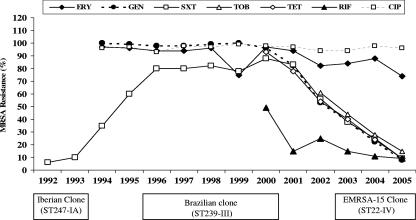
In contrast with MRSA strains isolated in HGSA during the last decade (1990 to 2000), which were typically multiresistant, a significant change was observed, at least since 2001, with the increasing isolation of MRSA strains susceptible to multiple antibiotics such as GEN, SXT, TOB, TET, and RIF. The proportion of all MRSA strains isolated in HGSA that were resistant to these antibiotics drastically decreased from 2001 to 2005, as shown in Fig. 1. Our data revealed an excellent correlation between the susceptibility both to GEN and to SXT and the EMRSA-15 clone (ST22-IV), as 100% of MRSA strains of this clone were susceptible to those antibiotics. Globally, 86% (242 out of 280) of all MRSA isolates were susceptible to GEN and SXT, with 93% (225 out of 242) of these belonging to PFGE type J; the remaining 7% were distributed among three minor clones (PFGE types C, M, and E). Furthermore, the great majority of EMRSA-15 isolates were shown to be susceptible to other antibiotics, namely, TOB (224 out of 225 isolates), RIF (222 out of 225), and TET (221 out of 225). The data presented here document that the observed changes in the resistance phenotypes of MRSA isolates recovered in HGSA since 2001 have been directly associated with the epidemic spread of the EMRSA-15 clone within the institution.
FIG. 1.
Resistance phenotypes of all MRSA isolates recovered from HGSA during the last 14 years and major MRSA clones found among the collections studied.
Our findings confirmed the susceptibility patterns observed by others for EMRSA-15, which is commonly susceptible to GEN, TET, and RIF, among other antibiotics, but with the great majority of isolates resistant to CIP, ERY, and CLI (6, 63). The most frequent MLSB phenotype detected was the constitutive (cMLSB) phenotype expressed by almost 60% of the EMRSA-15 isolates resistant to CLI, in contrast with data recently published by Alcoceba et al. (iMLSB phenotype in 67% of isolates) (6). Resistance to SXT was confirmed to be an important phenotypic marker for the identification of the Brazilian clone (ST239, SCCmec type IIIA, PFGE type B) among the multiresistant MRSA isolates, since 92% (33 out of 36) of the MRSA isolates found to be resistant to SXT presented PFGE type B. Therefore, the decrease in the number of MRSA isolates resistant to SXT observed at HGSA, at least since 2001, is related to the replacement of the Brazilian clone by EMRSA-15, which is susceptible to SXT, as the major clone in HGSA.
In this study, we characterized, by a combination of molecular typing techniques, a collection of MRSA isolates to investigate the genetic backgrounds associated with the changes in the resistance phenotypes of MRSA isolates from a Portuguese hospital during a 3-year period. Our results clearly document that the decrease of resistant traits of MRSA strains isolated in HGSA is associated with the decline and replacement of the multiresistant Brazilian clone (ST239-III) as the major MRSA clone until 2000 and the astonishing spread of the epidemic EMRSA-15 (ST22-IV) clone, at least since 2003, in which resistance to antibiotics other than β-lactams is very rare. Furthermore, the pattern of susceptibility of MRSA isolates to both GEN and SXT was shown to be an excellent phenotypic marker for the identification and discrimination of the EMRSA-15 clone from other nonmultiresistant MRSA clones present in our hospital. We believe that EMRSA-15 emerged in our institution around 2001, attesting to the significant changes in the resistance phenotypes of MRSA isolates observed, though a single-locus variant of ST22, the allelic profile ST79, was previously identified in 2000. The finding that the overwhelming majority (80%) of MRSA isolates recovered from infection and colonization samples, from patients and HCWs, belonged to the EMRSA-15 clone, mainly to only one PFGE subtype that was found among 79% of the EMRSA-15 isolates, emphasizes its epidemic nature and remarkable success in spreading within an institution. These results enhanced our understanding of the epidemiology of MRSA in the selected wards in terms of reservoirs and routes of transmission, providing valuable information for the development of intervention strategies to control MRSA infections. An additional finding of this study of interest and concern was the high prevalence of the EMRSA-15 clone (77%) among the MRSA isolates recovered from the nasal samples of outpatients attending the DFOU, who may represent an important reservoir and vector for MRSA dissemination within the hospital, when admitted as inpatients and most likely within the community.
Acknowledgments
We thank all HCWs from the wards and units included in this study, namely, the vascular surgery ward, the endocrinology ward, the DFOU, and the neonatal intensive care unit, for their collaboration in the sample collection procedure; Alice Moreira and Ernestina Aires from the Infection Control Team for their helpful assistance in the nasal screenings of patients and HCWs; technicians from the microbiology laboratory for the microbiological procedures on clinical samples; Paulo Pinto for helping with the collection of the epidemiological data from nasal screenings; and Lucinda Vilar for her helpful assistance in the collection and registration of all epidemiological data.
This work made use of the MLST website (http://www.mlst.net), which is hosted at Imperial College London.
Partial support for this study was provided by project POCTI/BIA-MIC/58416/2004 from Fundação para a Ciência e Tecnologia, Lisbon, Portugal, and by project 55068 from Fundação Calouste Gulbenkian, Lisbon, Portugal, both awarded to H. de Lencastre. N.A.F. and D.C.O. were supported by grants SFRH/BD/19208/2004 and SFRH/BPD/9374/2002, respectively, from Fundação para a Ciência e Tecnologia, Lisbon, Portugal.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Aires de Sousa, M., C. Bartzavali, I. Spiliopoulou, I. S. Sanches, M. I. Crisostomo, and H. de Lencastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., and H. de Lencastre. 2003. Evolution of sporadic isolates of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and their similarities to isolates of community-acquired MRSA. J. Clin. Microbiol. 41:3806-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., H. de Lencastre, I. Santos Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6:253-258. [DOI] [PubMed] [Google Scholar]
- 4.Aires de Sousa, M., M. Miragaia, I. S. Sanches, S. Avila, I. Adamson, S. T. Casagrande, M. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aires-de-Sousa, M., T. Conceicao, and H. de Lencastre. 2006. Unusually high prevalence of nosocomial Panton-Valentine leukocidin-positive Staphylococcus aureus isolates in Cape Verde Islands. J. Clin. Microbiol. 44:3790-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcoceba, E., A. Mena, M. Cruz Perez, E. Ruiz de Gopegui, E. Padilla, J. Gil, A. Ramirez, C. Gallegos, A. Serra, J. L. Perez, and A. Oliver. 2007. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Majorcan hospitals: high prevalence of the epidemic clone EMRSA-15. Clin. Microbiol. Infect. 13:599-605. [DOI] [PubMed] [Google Scholar]
- 7.Amorim, M. L., M. Aires de Sousa, I. S. Sanches, R. Sa-Leao, J. M. Cabeda, J. M. Amorim, and H. de Lencastre. 2002. Clonal and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus (MRSA) from a Portuguese hospital over time. Microb. Drug Resist. 8:301-309. [DOI] [PubMed] [Google Scholar]
- 8.Anonymous. 2004. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32:470-485. [DOI] [PubMed] [Google Scholar]
- 9.Boyce, J. M. 1992. Methicillin-resistant Staphylococcus aureus in hospitals and long-term care facilities: microbiology, epidemiology, and preventive measures. Infect. Control Hosp. Epidemiol. 13:725-737. [DOI] [PubMed] [Google Scholar]
- 10.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 11.Cimiotti, J. P., F. Wu, P. Della-Latta, M. Nesin, and E. Larson. 2004. Emergence of resistant staphylococci on the hands of new graduate nurses. Infect. Control Hosp. Epidemiol. 25:431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing, 17th informational supplement. M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29:87-106. [DOI] [PubMed] [Google Scholar]
- 14.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denis, O., A. Deplano, R. De Ryck, C. Nonhoff, and M. J. Struelens. 2003. Emergence and spread of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus in Belgian hospitals. Microb. Drug Resist. 9:61-71. [DOI] [PubMed] [Google Scholar]
- 16.Donnio, P. Y., L. Preney, A. L. Gautier-Lerestif, J. L. Avril, and N. Lafforgue. 2004. Changes in staphylococcal cassette chromosome type and antibiotic resistance profile in methicillin-resistant Staphylococcus aureus isolates from a French hospital over an 11 year period. J. Antimicrob. Chemother. 53:808-813. [DOI] [PubMed] [Google Scholar]
- 17.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.European Antimicrobial Resistance Surveillance System. 2005. EARSS annual report 2004. http://www.rivm.nl/earss/.
- 20.European Antimicrobial Resistance Surveillance System. 2006. EARSS annual report 2005. http://www.rivm.nl/earss/.
- 21.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 43:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes, A. R., I. S. Sanches, M. Aires de Sousa, E. Castaneda, and H. de Lencastre. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Colombian hospitals: dominance of a single unique multidrug-resistant clone. Microb. Drug Resist. 7:23-32. [DOI] [PubMed] [Google Scholar]
- 23.Gosbell, I. B., T. Barbagiannakos, S. A. Neville, J. L. Mercer, A. M. Vickery, F. G. O'Brien, G. W. Coombs, M. J. Malkowski, and J. C. Pearson. 2006. Non-multiresistant methicillin-resistant Staphylococcus aureus bacteraemia in Sydney, Australia: emergence of EMRSA-15, Oceania, Queensland and Western Australian MRSA strains. Pathology 38:239-244. [DOI] [PubMed] [Google Scholar]
- 24.Harbarth, S., P. Francois, J. Shrenzel, C. Fankhauser-Rodriguez, S. Hugonnet, T. Koessler, A. Huyghe, and D. Pittet. 2005. Community-associated methicillin-resistant Staphylococcus aureus, Switzerland. Emerg. Infect. Dis. 11:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. Wale, M. Warner, D. M. Livermore, and B. D. Cookson. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48:143-144. [DOI] [PubMed] [Google Scholar]
- 26.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Layer, F., B. Ghebremedhin, W. Konig, and B. Konig. 2006. Heterogeneity of methicillin-susceptible Staphylococcus aureus strains at a German university hospital implicates the circulating-strain pool as a potential source of emerging methicillin-resistant S. aureus clones. J. Clin. Microbiol. 44:2179-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lelievre, H., G. Lina, M. E. Jones, C. Olive, F. Forey, M. Roussel-Delvallez, M. H. Nicolas-Chanoine, C. M. Bebear, V. Jarlier, A. Andremont, F. Vandenesch, and J. Etienne. 1999. Emergence and spread in French hospitals of methicillin-resistant Staphylococcus aureus with increasing susceptibility to gentamicin and other antibiotics. J. Clin. Microbiol. 37:3452-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaitre, N., W. Sougakoff, A. Masmoudi, M. H. Fievet, R. Bismuth, and V. Jarlier. 1998. Characterization of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus involved in nosocomial spread. J. Clin. Microbiol. 36:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melter, O., M. Aires de Sousa, K. Laskafeldova, P. Urbaskova, M. Wunschova, and H. de Lencastre. 2004. Delineation of the endemic and sporadic methicillin-resistant Staphylococcus aureus clones in a Czech hospital. Microb. Drug Resist. 10:218-223. [DOI] [PubMed] [Google Scholar]
- 33.Melter, O., P. Urbaskova, V. Jakubu, B. Mackova, and H. Zemlickova. 2006. Emergence of EMRSA-15 clone in hospitals throughout the Czech Republic. Euro Surveill. 11:E060803.6. [DOI] [PubMed] [Google Scholar]
- 34.Milisavljevic, V., F. Wu, J. Cimmotti, J. Haas, P. Della-Latta, E. Larson, and L. Saiman. 2005. Genetic relatedness of Staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Am. J. Infect. Control 33:341-347. [DOI] [PubMed] [Google Scholar]
- 35.Montesinos, I., T. Delgado, D. Riverol, E. Salido, M. A. Miguel, A. Jimenez, and A. Sierra. 2006. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus associated with the emergence of EMRSA-16 at a university hospital. J. Hosp. Infect. 64:257-263. [DOI] [PubMed] [Google Scholar]
- 36.Moore, P. C., and J. A. Lindsay. 2002. Molecular characterisation of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 51:516-521. [DOI] [PubMed] [Google Scholar]
- 37.Mulligan, M. E., and R. D. Arbeit. 1991. Epidemiologic and clinical utility of typing systems for differentiating among strains of methicillin-resistant Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 12:20-28. [DOI] [PubMed] [Google Scholar]
- 38.Mulligan, M. E., K. A. Murray-Leisure, B. S. Ribner, H. C. Standiford, J. F. John, J. A. Korvick, C. A. Kauffman, and V. L. Yu. 1993. Methicillin-resistant Staphylococcus aureus: a consensus review of the microbiology, pathogenesis, and epidemiology with implications for prevention and management. Am. J. Med. 94:313-328. [DOI] [PubMed] [Google Scholar]
- 39.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliveira, D. C., C. Milheirico, S. Vinga, and H. de Lencastre. 2006. Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J. Antimicrob. Chemother. 58:23-30. [DOI] [PubMed] [Google Scholar]
- 43.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 44.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 45.Pan, E. S., B. A. Diep, E. D. Charlebois, C. Auerswald, H. A. Carleton, G. F. Sensabaugh, and F. Perdreau-Remington. 2005. Population dynamics of nasal strains of methicillin-resistant Staphylococcus aureus and their relation to community-associated disease activity. J. Infect. Dis. 192:811-818. [DOI] [PubMed] [Google Scholar]
- 46.Pearman, J. W., G. W. Coombs, W. B. Grubb, and F. O'Brien. 2001. A British epidemic strain of methicillin-resistant Staphylococcus aureus (UK EMRSA-15) in Western Australia. Med. J. Aust. 174:662. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Roth, E., F. Lorenzo-Diaz, N. Batista, A. Moreno, and S. Mendez-Alvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 42:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson, J. F., and S. Reith. 1993. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 25:45-52. [DOI] [PubMed] [Google Scholar]
- 49.Roberts, R. B., M. Chung, H. de Lencastre, J. Hargrave, A. Tomasz, D. P. Nicolau, J. F. John, Jr., and O. Korzeniowski. 2000. Distribution of methicillin-resistant Staphylococcus aureus clones among health care facilities in Connecticut, New Jersey, and Pennsylvania. Microb. Drug Resist. 6:245-251. [DOI] [PubMed] [Google Scholar]
- 50.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 51.Sa-Leao, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salmenlinna, S., O. Lyytikainen, and J. Vuopio-Varkila. 2002. Community-acquired methicillin-resistant Staphylococcus aureus, Finland. Emerg. Infect. Dis. 8:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seeberg, S., L. Larsson, C. Welinder-Olsson, T. Sandberg, E. Skyman, B. Bresky, A. Lindqvist, and M. van Raalte. 2002. How an outbreak of MRSA in Gothenburg was eliminated: by strict hygienic routines and massive control-culture program. Lakartidningen 99:3198-3204. (In Swedish.) [PubMed] [Google Scholar]
- 54.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shopsin, B., and B. N. Kreiswirth. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 7:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simor, A. E., M. Ofner-Agostini, E. Bryce, A. McGeer, S. Paton, and M. R. Mulvey. 2002. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: results of 5 years of national surveillance, 1995-1999. J. Infect. Dis. 186:652-660. [DOI] [PubMed] [Google Scholar]
- 57.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 58.Tiemersma, E. W., S. L. Bronzwaer, O. Lyytikainen, J. E. Degener, P. Schrijnemakers, N. Bruinsma, J. Monen, W. Witte, and H. Grundman. 2004. Methicillin-resistant Staphylococcus aureus in Europe, 1999-2002. Emerg. Infect. Dis. 10:1627-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Udo, E. E., N. Al-Sweih, and B. Noronha. 2006. Characterisation of non-multiresistant methicillin-resistant Staphylococcus aureus (including EMRSA-15) in Kuwait hospitals. Clin. Microbiol. Infect. 12:262-269. [DOI] [PubMed] [Google Scholar]
- 60.van Belkum, A. 2000. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains: state of affairs and tomorrow's possibilities. Microb. Drug Resist. 6:173-188. [DOI] [PubMed] [Google Scholar]
- 61.Vindel, A., P. Trincado, E. Gomez, R. Cabrera, T. Boquete, C. Sola, S. Valdezate, and J. A. Saez-Nieto. 2006. Prevalence and evolution of methicillin-resistant Staphylococcus aureus in Spanish hospitals between 1996 and 2002. J. Clin. Microbiol. 44:266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wannet, W. J., M. E. Heck, G. N. Pluister, E. Spalburg, M. G. van Santen, X. W. Huijsdans, E. Tiemersma, and A. J. de Neeling. 2004. Panton-Valentine leukocidin positive MRSA in 2003: the Dutch situation. Euro Surveill. 9:28-29. [PubMed] [Google Scholar]
- 63.Witte, W., C. Braulke, C. Cuny, D. Heuck, and M. Kresken. 2001. Changing pattern of antibiotic resistance in methicillin-resistant Staphylococcus aureus from German hospitals. Infect. Control Hosp. Epidemiol. 22:683-686. [DOI] [PubMed] [Google Scholar]