Abstract
In the present study, we have investigated processing and maturation of the envelope glycoprotein (GP) of Ebola virus. When GP expressed from vaccinia virus vectors was analyzed by pulse–chase experiments, the mature form and two different precursors were identified. First, the endoplasmic reticulum form preGPer, full-length GP with oligomannosidic N-glycans, was detected. preGPer (110 kDa) was replaced by the Golgi-specific form preGP (160 kDa), full-length GP containing mature carbohydrates. preGP was finally converted by proteolysis into mature GP1,2, which consisted of two disulfide-linked cleavage products, the amino-terminal 140-kDa fragment GP1, and the carboxyl-terminal 26-kDa fragment GP2. GP1,2 was also identified in Ebola virions. Studies employing site-directed mutagenesis revealed that GP was cleaved at a multibasic amino acid motif located at positions 497 to 501 of the ORF. Cleavage was blocked by a peptidyl chloromethylketone containing such a motif. GP is cleaved by the proprotein convertase furin. This was indicated by the observation that cleavage did not occur when GP was expressed in furin-defective LoVo cells but that it was restored in these cells by vector-expressed furin. The Reston subtype, which differs from all other Ebola viruses by its low human pathogenicity, has a reduced cleavability due to a mutation at the cleavage site. As a result of these observations, it should now be considered that proteolytic processing of GP may be an important determinant for the pathogenicity of Ebola virus.
Keywords: proteolytic processing
Ebola and Marburg viruses, the two species within the family Filoviridae (1), are among the most pathogenic agents causing fulminant hemorrhagic fever in humans and nonhuman primates. Yet, we are only beginning to understand the interactions of these viruses with their hosts, and our knowledge on genetics, pathogenicity, and natural history is still limited. Although outbreaks have so far always been self-limiting, our ignorance concerning the natural reservoir and the lack of immunoprophylactic and chemotherapeutic measures makes these infections a matter of high concern in biomedical science. The chronology of human epidemics and epizootics in nonhuman primates proves that filoviruses are prototypes of emerging/re-emerging pathogens. The recent emergence of a new Ebola subtype in Cote d’Ivoire (2) and the re-emergence of Ebola subtype Zaire in former Zaire (3) and Gabon (4, 5) once again showed that these viruses have altered from exotic agents to pathogens of serious public health concerns.
The genomes of filoviruses display linear arranged genes on a single negative-stranded RNA molecule that encode the seven structural proteins in the order nucleoprotein, virion structural protein (VP) 35, VP40, glycoprotein (GP), VP30, VP24, and RNA-dependent RNA polymerase (L) (6–8). In general, filoviral genes are transcribed into monocistronic subgenomic RNA species (mRNA) (6, 9). In contrast to all other filoviral genes, including the GP gene of Marburg virus (10), the organization and transcription of the fourth gene (GP) of EBOV is unusual and involves transcriptional editing, which is needed to express GP. A nonstructural small glycoprotein (sGP) is synthesized from the unedited GP mRNA, which is extensively secreted from infected cells (11, 12).
EBOV GP is a type I transmembrane glycoprotein 676 amino acids in length. The middle region of GP is variable, extremely hydrophilic, and carries the bulk of the glycosylation sites for N- and O-glycans that account for approximately one-third of the molecular weight (11, 13). Experimental data on GP function do not exist. However, the fact that GP is the only surface protein of virions suggests a function in receptor binding and fusion with cellular membranes.
The fusogenic properties of many viral glycoproteins require posttranslational proteolytic processing (14). Most of these glycoproteins are activated by subtilisin-like eukaryotic endoproteases (proprotein convertases) at multibasic cleavage sites (for review, see ref. 15). This type of processing has never been demonstrated with filovirus glycoproteins, and their functions were thought to be associated with noncleaved GP. We show here, however, that maturation of EBOV GP involves posttranslational cleavage of a precursor at the C-terminal end of the sequence R-T-R-R501 into the disulfide-linked fragments GP1 and GP2. The proprotein convertase furin has been identified as a cleavage enzyme. Lower susceptibility to furin cleavage has been observed with the less pathogenic subtype Reston, suggesting that cleavability of GP may be a determinant of pathogenicity.‡
MATERIALS AND METHODS
Viruses and Cell Cultures.
Strain Eckron of the Zaire subtype (provided by Institute Voor Tropische Geneeskunde, Antwerp, Belgium) and strain Pennsylvania of the Reston subtype of EBOV (provided by A. Sanchez, CDC, Atlanta, GA) were propagated on Vero-E6 cells (ATCC CRL 1586) and purified from tissue culture medium as described previously (16). The recombinant vaccinia viruses vTF7-3, expressing T7 polymerase (provided by B. Moss. National Institutes of Health, Bethesda, MD), VVHfur, expressing human furin (provided by G. Thomas, Oregon Health Sciences University, Portland, OR), and vSCGP8, expressing EBOV GP (11), were propagated in Vero-E6 and/or CV-1 cells. CV-1, HeLa, RK13, and Vero-E6 cells were maintained in Dulbecco’s medium (GIBCO) containing 10% fetal calf serum (GIBCO). LoVo cells (human colon adenocarcinoma cells, ATCC CCL 229) were maintained in F-12 HAM medium (Sigma) containing 10% fetal calf serum.
Metabolic Labeling.
Vero-E6 cells (75-cm2 flasks) were inoculated with EBOV strain Eckron at a multiplicity of infection (moi) of 1 plaque-forming unit (pfu)/cell for 1 h. Cells were washed 3–4 days postinfection with Dulbecco’s medium, starved in methionine-cysteine-free or glucose-free medium for 1 h, and labeled with 100 μCi/ml [35S]Promix (Cys-Met) (Amersham) or 500 μCi/ml [3H]glucosamine (Amersham), respectively. Virus was harvested 12 h postlabeling and partially purified from culture medium by centrifugation through a 20% sucrose cushion (120 min at 100,000 × g). Pelleted virus was resuspended in PBS and lysed with 2% SDS, 1% Nonidet P-40, and 0.4% deoxycholate.
Pulse–Chase Experiments.
HeLa or RK13 cells (3 × 106) were infected with either vSCGP8 or vTF7-3 (transient expression) at an moi of 10 pfu/cell. For transient expression, the inoculum was replaced after 1 h by transfection medium containing recombinant plasmid DNA [9 μg; 15 μl of lipofectin (BRL)]. Cells were washed 6 h postinfection, starved in methionine-cysteine-free medium for 1 h, and labeled with 100 μCi/ml [35S]Promix (Cys-Met) (Amersham). After a 20-min pulse, the inoculum was replaced with medium for chase. Labeled cells were lysed immediately in lysis buffer (1% Nonidet P-40/0.4% sodium deoxycholate/0.5% BSA/5 mM EDTA/100 mM NaCl/20 mM Tris⋅HCl, pH 7.6/25 mM iodoacetamide/1 mM phenylmethylsulfonyl fluoride) at 4°C. Immunoprecipitation was performed using goat anti-EBOV Igs.
Generation of EBOV Reston GP.
A cDNA fragment containing the edited GP-ORF of the Pennsylvania strain of subtype Reston was generated from GP-specific mRNA by reverse transcriptase-PCR using strain Reston-derived oligonucleotides from a sequence determined previously (EVU23152, GenBank). The cDNA product was cloned into pGEM 3Zf(+) and is referred to as pGEM-RP8. The sequence was determined and submitted to GenBank (AF034645).
Oligonucleotide-Directed Mutagenesis.
PCR mutagenesis using the Quick-change kit (Stratagene) according to the instructions of the manufacturer was performed to generate the Zaire (Z) and Reston (R) GP cleavage site mutants (pGEM-Z/F1, pGEM-Z/F2, pGEM-R/K, pGEM-R/R) and the Zaire GP tail elongation mutant (pGEM-GPLT). Clones with mutated sequences were verified by sequence determination.
Endoproteolytic Cleavage Inhibition Assay.
For cleavage inhibition studies, cells infected with vSCGP8 were incubated during starvation, pulse, and chase periods either without or with the decanoylated peptidyl chloromethylketone decRVKR-cmk at concentrations of 25 or 80 μM.
Glycosidase Treatment.
Analyses were performed on recombinant GP expressed from vTF7-3-infected and plasmid (pGEM-GP8 or pGEM-GPLT)-transfected HeLa cells (1 × 106). Immunoprecipitated proteins were treated under reducing and denaturing conditions according to the instructions of the supplier (BioLabs, Germany).
RESULTS
EBOV GP Undergoes Posttranslational Proteolytic Cleavage into Two Disulfide-Linked Fragments.
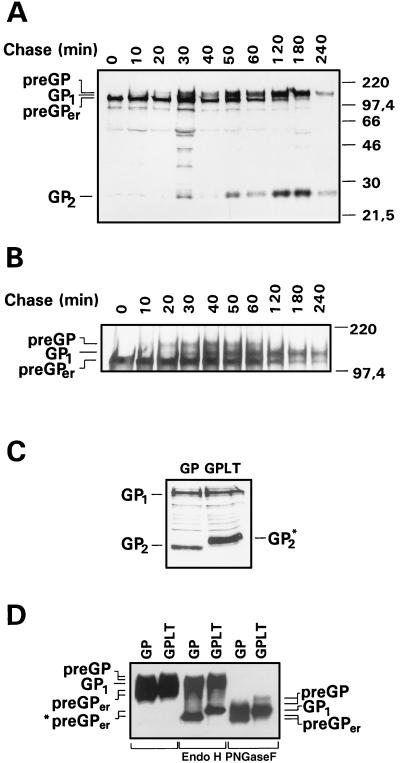
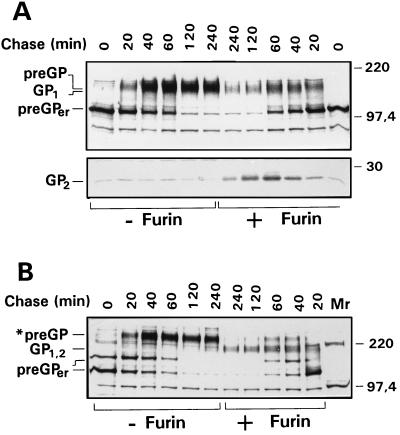
EBOV GP is a complex membrane glycoprotein. To better understand the sequence of maturation steps, pulse–chase labeling experiments were performed on vector-expressed GP. Cells infected with the recombinant vaccinia virus vSCGP8 were pulse-labeled for 20 min with [35S]methionine/cysteine and chased for different time intervals prior to lysis; proteins were then immunoprecipitated using anti-EBOV Igs. Three high molecular weight forms of GP could be discriminated (Fig. 1A). Immediately after pulse labeling, GP was detected in a 110-kDa form that had previously been identified as the precursor present in the endoplasmic reticulum due to its endo H sensitivity (11) and has therefore now been designated preGPer. From chase periods of about 20 min onward, there was a gradual decrease of preGPer and a concomitant increase of a 160-kDa form that, as shown below, contains complex N-glycans and represents therefore a precursor form present in the Golgi apparatus (preGP). Within the next 20 to 30 min, preGP completely disappeared, giving rise to a 140-kDa form designated GP1. Concomitant with GP1, a 26-kDa glycoprotein (GP2) was detected after chase periods of 20–30 min, migrating slightly faster than an unidentified protein that was seen throughout the pulse–chase experiment. GP1 and GP2 increased with the same kinetics up to 4 h postlabeling. These observations are compatible with the concept that preGPer is converted into preGP, which is subsequently cleaved into GP1 and GP2.
Figure 1.
Synthesis, processing, and transport of EBOV GP. Pulse–chase analyses of EBOV GP were done in RK13 cells infected with vSCGP8 (A) or in HeLa cells infected with vTF7-3 and transfected with pGEM-GPLT (B). At 6 h postinfection, cells were pulse-labeled for 20 min (lane 0) and chased for the indicated time intervals. Lysed cells were immunoprecipitated with goat anti-EBOV Igs. Precipitated proteins were separated on 15% (A, C) or 8% (B) polyacrylamide gels, and proteins were visualized by fluorography. (D) Endoglycosidase digestion of the tail elongation mutant GPLT expressed in HeLa cells. Cells were infected with vTF7-3 and subsequently transfected with pGEM-mGP8 expressing wild-type GP (GP) or pGEM-GPLT expressing GPLT. At 18 h posttransfection, cells were lysed and subjected to endoglycosidase treatments. Digested samples were separated by SDS/PAGE and blotted onto a poly(vinylidene difluoride) membrane. Detection of GP-specific proteins was performed with goat anti-EBOV Igs. The positions of the precursors (preGPer and preGP) and the proteolytically cleaved subunit GP1 before and after endoglycosidase treatment are indicated.
The data shown in Fig. 1A demonstrate that the difference in the electrophoretic mobilities of preGP and GP1 is small. To improve the resolution between both proteins, we have constructed a tail elongation mutant designated GPLT, which contained 36 additional amino acids at the carboxyl-terminal end. PAGE analysis now allowed a clear discrimination of preGP and GP1, and the pulse-chase experiment shown in Fig. 1B confirmed the kinetics of GP processing, with preGPer appearing first, followed by preGP, and finally by GP1 and GP2. GP2 of the tail elongation mutant showed an increase in size when compared with wild-type GP2 and the unidentified protein mentioned above, again indicating that the latter one is different from GP2 (Fig. 1C). Additional information on the precursor-product relationships was obtained when wild-type and tail elongation mutant GPLT were compared after glycosidase treatment (Fig. 1D). As expected, preGPer and preGP showed differential molecular weights with mutant and wild type, whereas GP1 did not. preGPer was sensitive to endo H as well as to PNGase F, indicating that it contains oligomannosidic N-glycans. preGP and GP1 were only sensitive to PNGase F, indicating maturation of the N-glycans to the complex form. The observation that GP1 had a higher molecular weight after PNGase F treatment than preGPer is consistent with the presence of O-glycans on GP1. The data support the concept that preGPer is GP present in the endoplasmic reticulum, whereas preGP and GP1 are Golgi and trans-Golgi forms, respectively.
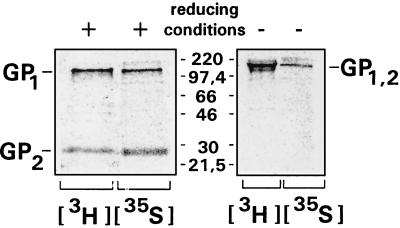
The data described so far indicate that proteolytic processing is the last step in GP maturation. It was therefore of interest to see if cleaved GP can also be identified in virus particles. Vero-E6 cells were infected with EBOV and metabolically labeled 4 days postinfection. Proteins from purified virions were immunoprecipitated and subsequently analyzed on 15% polyacrylamide gels under reducing and under nonreducing conditions (Fig. 2). After treatment with β-mercaptoethanol, the 140- and 26-kDa bands of GP1 and GP2, respectively, were detected. Labeling with [3H]glucosamine and immunoprecipitation with a GP-specific antiserum proved that GP2 is distinct from VP24, which has a similar molecular weight. In the absence of β-mercaptoethanol, a band designated GP1,2 was detected, indicating that GP1 and GP2 form a disulfide-linked complex under nonreducing conditions. Taken together, these data indicate that GP undergoes proteolytic processing after vector expression as well as after EBOV infection and that GP is present in virions in the cleaved form.
Figure 2.
Virion GP of EBOV consists of two cleavage subunits. EBOV GP metabolically labeled with [3H]glucosamine or with [35S]methionine-cysteine was immunoprecipitated from purified virions grown in Vero E6 cells and analyzed by 15% SDS/PAGE under reducing or nonreducing conditions. The positions of the mature GP (GP1,2), and the two cleavage subunits GP1 and GP2, are indicated.
EBOV GP Is Cleaved by Furin.
EBOV GP contains the sequence R-R-T-R-R501, which is a potential proprotein convertase recognition motif. To test whether this motif is indeed used as a cleavage site, the sequence was modified by site-specific mutagenesis. In mutant Z/F1, arginine at position 501 was substituted by lysine (R-R-T-R-R → R-R-T-R-K), and in mutant Z/F2 the arginine residues at positions 500 and 501 were changed to asparagine and methionine, respectively (R-R-T-R-R → R-R-T-N-M) (Fig. 3). Both mutants were evaluated for proteolytic processing by transient expression in VTF7-3-infected HeLa cells. Unlike wild-type GP, which was processed into subunits GP1 and GP2, both mutants expressed only the uncleaved preGP. This result indicates that GP is cleaved at the carboxyl-terminal side of arginine 501 and that the cleavage site has the classical consensus sequence R-X-K/R-R recognized by ubiquitous proprotein convertases, such as furin. The observation that mutant Z/F1 displaying the consensus sequence R-X-X-R500 is not cleaved may be explained by the dislocation of the motif within the GP sequence or by the fact that this motif is less frequently used as a cleavage site (17). In addition, the results obtained with these mutants indicate that another multibasic sequence RKIR302 is not used as a cleavage site of the EBOV GP.
Figure 3.
Intracellular processing of the cleavage site mutants Z/F1 and Z/F2. HeLa cells were infected with vTF7-3 and transfected with the plasmids pGEM-mGP8 (wild-type GP, WT), pGEM-Z/F1 (mutant Z/F1), and pGEM-Z/F2 (mutant Z/F2). At 6 h postinfection, cells were pulse-labeled for 20 min and chased for 240 min. Immunoprecipitated proteins were separated under reducing conditions on 8% (Upper gel) or 15% (Lower gel) polyacrylamide gels. The positions of the noncleaved precursor (preGP) of both mutants and the cleavage subunits GP1 and GP2 are indicated. The sequences at the cleavage sites are shown at Top.
Peptidyl chloromethylketones of the general structure decR-X-K/R-R-cmk have been shown to act as potent inhibitors of furin by covalently blocking the substrate binding site of the enzyme (15). It was therefore of interest to find out if such inhibitors were able to prevent cleavage of preGP. Experiments were performed on RK13 cells infected with vSCGP8 (Fig. 4). Six-hour postinfection cells were treated with 25 or 80 μM of decRVKR-cmk, pulse-labeled, and incubated for a 4-h chase. Proteins were immunoprecipitated from cell lysates using anti EBOV Ig, treated with 2% SDS with or without β-mercaptoethanol, and subsequently analyzed on SDS/PAGE. The inhibitor completely blocked cleavage at both concentrations as indicated by the persistence of preGP and the absence of GP1 and GP2 (Fig. 4). An interesting observation was made when uncleaved GP that had accumulated after expression in the presence of decRVKR-cmk was compared with cleaved GP by electrophoresis under nonreducing conditions. Whereas GP1,2 showed the expected molecular mass of about 160 kDa, uncleaved GP had a significantly reduced electrophoretic mobility with an apparent molecular mass well above 220 kDa (*preGP). preGPer was partly also present in a form with reduced electrophoretic mobility (*preGPer). The most likely explanation for these differences in migration are variations in protein folding. It therefore seems that the conformation of uncleaved GP differs from that of cleaved GP.
Figure 4.
Effect of decanoylated R-V-K-R chloromethylketone on cleavage of EBOV GP. RK13 cells were infected with vSCGP8. At 6 h postinfection, cells were pulse-labeled for 20 min and chased for various times in the absence (0 μM) or presence (25 μM, 80 μM) of inhibitor. Immunoprecipitated samples were separated by PAGE under reducing conditions on 15% gels (Top) and 8% gels (Middle) and under nonreducing conditions on 8% gels (Bottom). The positions of the precursors (preGPer and preGP) as well as the cleavage subunits GP1 and GP2 are indicated. Under nonreducing conditions, high Mr forms of both precursors are seen (*preGP and *preGPer).
To further support the concept that GP is cleaved by furin, we have expressed GP in LoVo cells, which lack the active form of this enzyme (18). Unlike HeLa (Figs. 1B and 3), RK13 (Figs. 1A and 4), and Vero cells (Fig. 2), LoVo cells were unable to cleave GP (Fig. 5A, lanes 1–6). To definitely prove that GP is cleaved by furin, LoVo cells were co-infected with recombinant vaccinia viruses expressing GP (vSCGP8) and human furin (VVhfur). As also shown in Fig. 5A (lanes 7–12), GP was now cleaved in LoVo cells. This experiment demonstrates clearly that GP is cleaved by furin but not by other proprotein convertases expressed in LoVo cells, such as PACE4 or PC7. Thus, it seems that furin plays a prime role in GP cleavage. When analyzed under nonreducing conditions, the *preGP and *preGPer forms of uncleaved GP were also detected in LoVo cells (Fig. 5B), again supporting the concept that uncleaved and cleaved GP differ in conformation.
Figure 5.
Synthesis and processing of EBOV GP in LoVo cells with or without coexpression of furin. LoVo cells were either infected with vSCGP8 alone (moi of 10 pfu/cell) or coinfected with vSCGP8 and VVHfur (each with a moi of 10 pfu/cell). Infected cells were pulse-labeled for 20 min at 6 h postinfection and chased for different time intervals as indicated. Immunoprecipitated samples were separated on 8% (Upper) and 15% (Lower) polyacrylamide gels under reducing (A) and an 8% polyacrylamide gel under nonreducing (B) conditions. The positions of the precursors (preGPer and preGP) as well as the cleavage subunits GP1 and GP2 are indicated. Under nonreducing conditions, the uncleaved precursor showed an Mr higher than 220 kDa (*preGP).
Subtype Reston GP Shows Reduced Cleavability.
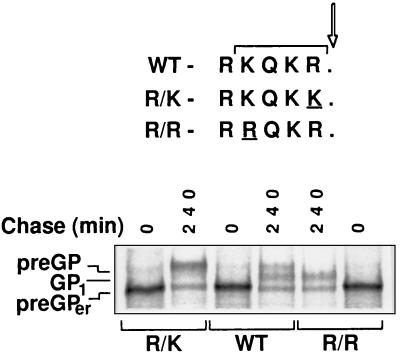
Subtype Reston differs from subtype Zaire by an altered cleavage site displaying the sequence R-K-Q-K-R (EVU23152, EV23416, EV23417; GenBank), which is not a classical furin recognition motif. To analyze the effect of this sequence variation on processing, the GP gene of the Pennsylvania strain of EBOV Reston was cloned, modified by site-directed mutagenesis at the cleavage site, and expressed using the vaccinia virus T7 system. As shown in Fig. 6, both precursors as well as the cleaved form could be identified with Reston GP (lanes 3 and 4). However, comparison with mutant R/R that has a classical furin cleavage site similar to Zaire GP (lanes 5 and 6) indicated that cleavage kinetics were significantly decreased with Reston GP. The results obtained with mutant R/K demonstrated that cleavage was even further reduced when R at position −1 was replaced by K (lanes 1 and 2). Thus, we could show that proteolytic processing of EBOV GP can be modulated by altering the cleavage site and that Reston GP has a lower cleavability then Zaire GP.
Figure 6.
Processing of wild-type and cleavage site mutants of EBOV subtype Reston. HeLa cells were infected with vTF7-3 and transfected with the plasmids pGEM-PR8 (wild-type Reston GP, WT), pGEM-R/K (mutant R/K), and pGEM-R/R (mutant R/R). At 6 h postinfection, cells were pulse-labeled for 20 min and chased for 240 min. Immunoprecipitated proteins were separated under reducing conditions on an 8% polyacrylamide gel. The positions of the noncleaved precursors (preGPer and preGP) and the cleavage subunit GP1 are indicated. The sequences at the cleavage sites are shown at Top.
DISCUSSION
Maturation of the EBOV GP involves a complex sequence of co- and posttranslational processing events. In the present study, mature GP and two different precursor forms have been identified. The first precursor form is detected after pulse labeling as a 110-kDa glycoprotein that is converted by endo H treatment into a 75-kDa polypeptide (11). The data indicate that this precursor is full-length GP containing oligomannosidic N-glycans. It represents precursor GP present in the endoplasmic reticulum and has therefore been designated preGPer. The second precursor, designated preGP, is first observed after a 10–20-min chase period as an endo H-resistant glycoprotein with a molecular mass of about 160 kDa. This form represents full-length GP containing mature carbohydrates and is present in the Golgi apparatus. Mature GP is GP1,2, which consists of two disulfide-linked cleavage products, the amino-terminal 140-kDa fragment GP1 and the carboxyl-terminal 26-kDa fragment GP2 (Fig. 7). GP1,2 is the virion form of GP, and it is also present in cells, presumably in the trans Golgi network and on the cell surface. These observations demonstrate that the filovirus glycoprotein is modified by extensive N-glycosylation and O-glycosylation when transported through the exocytotic pathway. We show here that the glycoprotein of a filovirus undergoes posttranslational proteolytic cleavage by a proprotein convertase.
Figure 7.
Scheme of EBOV GP, cleavage sites of different subtypes, and a model for the structure of the mature GP monomer. Signal peptide sequence, the carboxyl-terminal transmembrane domain, and the putative fusion domain are indicated by gray boxes. The altered amino acid in the cleavage site of EBOV subtype Reston is marked by a box. Asterisk, potential N-glycosylation site; C, cysteine residue; GP1, larger cleavage product of GP; GP2, smaller cleavage product of GP; S-S, disulfide bridge.
Although it has been known for some time from sequence analyses that EBOV GP contains a potential recognition site for proprotein convertases (7, 19), it has not been observed until now that cleavage does indeed occur. The failure to detect cleavage seems to be largely due to the fact that preGP, GP1,2, and GP1 have very similar migration rates on polyacrylamide gels. Furthermore, GP2 tends to escape detection because it contains relatively little carbohydrate (Fig. 7) and co-migrates with VP24, another structural protein of EBOV. The use of the tail elongation mutant proved therefore to be very important in the present study because it allowed a clear discrimination between the precursor and the large cleavage fragment. It should be mentioned that Marburg virus GP has also a multibasic cleavage site at position 432 to 435, and preliminary data show that it is cleaved, too (unpublished data).
EBOV GP is cleaved by furin. This is indicated by the observation that cleavage did not occur when GP was expressed in the furin-defective LoVo cell line but that it was restored in these cells by vector-expressed furin. The finding that cleavage was inhibited by a sequence-specific peptidyl chloromethylketone or by mutation of the cleavage site supports this concept. Furin belongs to the proprotein convertases, a family of subtilisin-like eukaryotic endoproteases that includes also PC1/PC3, PC2, PC4, PACE4, PC5/PC6, and LPC/PC7 (20). These enzymes are differentially expressed in cells and tissues, and they display similar but not identical specificity for basic motifs, such as R-X-K/R-R, at the cleavage site of their substrates. Furin seems to be expressed in most cells. It is a processing enzyme of the constitutive secretory pathway, as seems to be the case with PACE4, PC5/PC6, and LPC/PC7. The expression of PC1/PC3 and PC2 is restricted to the regulated secretory pathway of neuroendocrine cells. Furin is localized predominantly in the trans Golgi network (21, 22), but it is also secreted from cells in a truncated form (23, 24). Proprotein convertases activate numerous cellular proteins (25) and surface proteins of enveloped viruses. Furin seems to be the key enzyme in virus activation (26), but PC5/PC6 (27) and LPC/PC7 (28) are also involved. Thus, LPC/PC7 may be responsible for cleavage of the HIV glycoprotein in the furin-deficient LoVo cells. The observation that EBOV GP is not cleaved in these cells is interesting in this context. It is also noteworthy that furin, although ubiquitous, is particularly rich in hepatocytes and endothelial cells, which are both prime targets of Ebola virus (29). These observations stress the importance of furin as a processing enzyme of GP, but it remains to be seen in future studies if other proprotein convertases can substitute as cleaving enzymes.
Processing by protein convertases is an important control mechanism for the biological activity of viral surface proteins (26, 30). Cleavage occurs often next to a protein domain involved in fusion, and it has long been known that in these cases proteolytic cleavage is necessary for fusion activity. Proteolytic cleavage is the first step in the activation of these fusion proteins and is followed by a conformational change resulting in the exposure of the fusion domain (31–33). The conformational change may be triggered by low pH in endosomes, as is the case with influenza virus (34), or by the interaction with a secondary receptor protein at the cell surface, as is the case with HIV (35). We have so far not been able to demonstrate that cleavage of GP has an effect on fusion activity or on infectivity of EBOV. However, it is interesting to see that GP2 contains a sequence of 16 uncharged and hydrophobic amino acids at a short distance (22 amino acids) from the cleavage site, which bears some structural similarity to the fusion peptides of retroviruses and has therefore been thought to play a role in EBOV entry (19). Furthermore, it seems from the present study that cleaved and uncleaved GP differ in folding, as indicated by their differential electrophoretic mobilities under nonreducing conditions. These observations are compatible with the view that proteolytic cleavage is a priming mechanism that renders GP susceptible for a conformational change required for fusion.
Finally, it has to be pointed out that proteolytic activation of viral glycoproteins is an important determinant for pathogenicity. Cleavage by furin and other ubiquitous proprotein convertases has been shown to be responsible for systemic infection caused by highly pathogenic strains of avian influenza and Newcastle disease virus (36). It is therefore tempting to speculate that cleavage by furin is also an important factor for the pantropism of EBOV and its rapid dissemination through the organism. Furthermore, variations at the cleavage site of GP may account for differences in the pathogenicity of EBOV. Our data show that the pathogenic strains Zaire, Sudan, and Ivory Coast, which have the canonical furin motif R-X-K/R-R at the cleavage site, are highly susceptible to cleavage, whereas the Reston strain, which seems to be apathogenic for humans and only moderately pathogenic for at least some monkey species (37), has reduced cleavability because of the suboptimal cleavage site sequence K-Q-K-R. That highly pathogenic variants may suddenly emerge from Reston-like strains by mutations restricted to the cleavage site is an intriguing hypothesis. On the other hand, it may be possible to obtain EBOV mutants with even lower cleavability than the Reston strain, and such viruses may have a potential as life vaccines. Because furin cleavage can be inhibited not only by peptidyl chloromethylketones as described here but also by less toxic components (38), inhibition of proteolytic cleavage may be a novel concept for treatment of EBOV infections.
Acknowledgments
We thank Carina Eckel for excellent technical assistance, Aleksandr Chepurnov (Russian National Research Center of Virology and Biotechnology, Koltsovo, Russia) for providing the anti-Ebola (Zaire) virus Igs, and Wolfgang Garten (Institut für Virologie, Philipps-Universität Marburg, Germany) for critical review of the manuscript and helpful discussions. The work was supported by grants of the Deutsche Forschungsgemeinschaft (SFB 286, SFB 535, Fe 286/4-1), BMBF (01KV9503), the Hoechst AG, and the Kempkes-Stiftung (21/95).
ABBREVIATIONS
- EBOV
Ebola virus
- GP
glycoprotein
- moi
multiplicity of infection
- pfu
plaque-forming unit
- VP
virion structural protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF034645).
This work was presented in part at the Tenth International Conference on Negative Strand Viruses, Dublin, Ireland, September 21–26, 1997.
References
- 1.Murphy, F. A., Fauquet, C. M., Bishop, D. H. L., Ghabrial, S. A., Jarvis, A. W., Martelli, G. P., Mayo, M. A. & Summers, E. D. (1995) Arch. Virol. 1, Suppl., 265–292.
- 2.LeGuenno B, Formentry P, Wyers M, Gounon P, Walker F, Boesch C. Lancet. 1995;345:1271–1274. doi: 10.1016/s0140-6736(95)90925-7. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Wkly Epidemiol Rec. 1995;70:241–242. [Google Scholar]
- 4.Georges-Courbot M C, Sanchez A, Lu C Y, Baize S, Leroy E, Lansout-Soukate J, Tevi-Benissan C, Georges A J, Trappier S G, Zaki S R, Swanepoel R, Leman P A, Rollin P E, Peters C J, Nichol S T, Ksiazek T G. Emerg Infect Dis. 1997;3:59–62. doi: 10.3201/eid0301.970107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volchkov V E, Volchkova V, Eckel C, Klenk H-D, Bouloy M, LeGuenno B, Feldmann H. Virology. 1997;23:139–144. doi: 10.1006/viro.1997.8529. [DOI] [PubMed] [Google Scholar]
- 6.Feldmann H, Mühlberger E, Randolf A, Will C, Kiley M P, Sanchez A, Klenk H-D. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 7.Volchkov V E, Blinov V M, Netesov S V. FEBS Lett. 1992;305:181–184. doi: 10.1016/0014-5793(92)80662-z. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez A, Kiley M P. Virology. 1987;157:414–420. doi: 10.1016/0042-6822(87)90283-2. [DOI] [PubMed] [Google Scholar]
- 10.Will C, Mühlberger E, Linder D, Slenczka W, Klenk H-D, Feldmann H. J Virol. 1993;67:1203–1210. doi: 10.1128/jvi.67.3.1203-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H-D. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez A, Trappier S G, Mahy B W, Peters C J, Nichol S T. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldmann H, Nichol S T, Klenk H-D, Peters C J, Sanchez A. Virology. 1994;199:469–473. doi: 10.1006/viro.1994.1147. [DOI] [PubMed] [Google Scholar]
- 14.Choppin P W, Scheid A. Rev Infect Dis. 1980;2:40–61. doi: 10.1093/clinids/2.1.40. [DOI] [PubMed] [Google Scholar]
- 15.Garten W, Hallenberger S, Ortmann D, Schäfer W, Vey M, Angliker H, Shaw E, Klenk H-D. Biochimie (Paris) 1994;76:217–225. doi: 10.1016/0300-9084(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 16.Feldmann H, Will C, Schikore M, Slenczka W, Klenk H-D. Virology. 1991;182:353–356. doi: 10.1016/0042-6822(91)90680-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vey M, Orlich M, Adler S, Klenk H-D, Rott R, Garten W. Virology. 1992;188:408–413. doi: 10.1016/0042-6822(92)90775-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi S, Nakagawa T, Kasai K, Banno T, Duguay S J, Van de Ven W J M, Murakami K, Nakayama K. J Biol Chem. 1995;27:26565–26569. doi: 10.1074/jbc.270.44.26565. [DOI] [PubMed] [Google Scholar]
- 19.Gallaher W R. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 20.Seidah N G, Hamelin J, Mamarbachi M, Dong W, Tadro H, Mbikay M, Chretien M, Day R. Proc Natl Acad Sci USA. 1996;93:3388–3393. doi: 10.1073/pnas.93.8.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molloy S S, Thomas L, van Slyke J K, Stenberg P E, Thomas G. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schäfer W, Stroh A, Berghöfer S, Seiler J, Vey M, Kruse M L, Kern H F, Klenk H-D, Garten W. EMBO J. 1995;14:2424–2435. doi: 10.1002/j.1460-2075.1995.tb07240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vey M, Schäfer W, Reis B, Ohuchi R, Britt W, Garten W, Klenk H-D, Radsak K. Virology. 1995;206:746–749. doi: 10.1016/s0042-6822(95)80002-6. [DOI] [PubMed] [Google Scholar]
- 24.Wise R J, Barr P J, Wong P A, Kiefer M, Brake A J, Kaufman R J. Proc Natl Acad Sci USA. 1990;87:9378–9382. doi: 10.1073/pnas.87.23.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barr P J. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- 26.Klenk H-D, Garten W. In: Cellular Receptors for Animal Viruses. Wimmer E, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 241–280. [Google Scholar]
- 27.Horimoto T, Nakayama K, Smeekens S P, Kawaoka Y. J Virol. 1994;68:6074–6078. doi: 10.1128/jvi.68.9.6074-6078.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallenberger S, Moulard M, Sordel M, Klenk H-D, Garten W. J Virol. 1997;71:1036–1045. doi: 10.1128/jvi.71.2.1036-1045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geisbert T W, Jahrling P B, Hanes M A, Zack P M. J Comp Pathol. 1992;106:137–152. doi: 10.1016/0021-9975(92)90043-t. [DOI] [PubMed] [Google Scholar]
- 30.Klenk H-D, Garten W. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 31.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 32.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 33.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 34.Skehel J J, Bayley P M, Brown E B, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 36.Klenk H-D, Rott R. Adv Virus Res. 1988;34:247–281. doi: 10.1016/S0065-3527(08)60520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher-Hoch S P, Brammer T L, Trappier S G, Hutwagner L C, Farrar B B, Ruo S L, Brown B G, Hermann L M, Perez-Oronoz G I, Goldsmith C S, Hanes M A, McCormick J B. J Infect Dis. 1992;166:753–763. doi: 10.1093/infdis/166.4.753. [DOI] [PubMed] [Google Scholar]
- 38.Anderson E D, Thomas L, Hayflick J S, Thomas G. J Biol Chem. 1993;268:24887–24891. [PubMed] [Google Scholar]