Abstract
Cryptosporidium hominis and Cryptosporidium parvum are associated with massive disease outbreaks worldwide. Because these two species have different transmission cycles, identification of these parasites to the species level in clinical samples may provide laboratory data of crucial importance in epidemiologic investigations. To date, the most reliable way to differentiate C. hominis and C. parvum is based on DNA sequencing analysis of PCR amplicons. Although this approach is very effective for differentiation of Cryptosporidium species, it is labor-intensive and time-consuming compared with methods that do not require DNA sequencing analysis as an additional step and that have been successfully used for specific identification of a number of pathogens. In this study, we describe a novel Luminex-based assay that can differentiate C. hominis from C. parvum in a rapid and cost-effective manner. The assay was validated by testing a total of 143 DNA samples extracted from clinical specimens, environmental samples, or samples artificially spiked with Cryptosporidium oocysts. As few as 10 oocysts per 300 μl of stools could be detected with this assay. The assay format includes species-specific probes linked to carboxylated Luminex microspheres that hybridize to a Cryptosporidium microsatellite-2 region (ML-2) where C. hominis and C. parvum differ by one nucleotide substitution. The assay proved to be 100% specific when samples that had been characterized by direct fluorescent antibody test (DFA) and DNA sequencing analysis were tested. In addition, the assay was more sensitive than DFA and provided species identification, which is an advantage for epidemiologic studies.
The apicomplexan genus Cryptosporidium comprises 15 valid species that cause intestinal disease in humans and animals (14-16, 32). Among these species, Cryptosporidium hominis and Cryptosporidium parvum are associated with massive diarrhea outbreaks worldwide, generally caused by exposure to drinking or recreational water or direct contact with infected persons through the oral-fecal route (14, 17). The largest cryptosporidiosis outbreak ever reported occurred in 1993 in Milwaukee, Wisconsin, affected 400,000 individuals, and created major economic disruption (24). To date, all outbreaks in the United States for which identification of Cryptosporidium to the species level was possible were associated with infection by C. hominis or C. parvum, previously known as C. parvum genotype 1 or C. parvum genotype 2, respectively (28). These two species have distinct epidemiologic cycles, with C. hominis infecting mainly humans (30). C. parvum, the most prevalent zoonotic species of the genus Cryptosporidium, infects a large number of animal species, as well as humans (9, 18, 27, 29). Therefore, diagnostic differentiation of these two species is very helpful in epidemiologic investigations by providing laboratory data that helps to identify the source of infection and link cases of infections to outbreaks. Morphologically, C. hominis and C. parvum are identical. Currently, no antigen detection or serologic assay that allows differentiation of these two species exists, but DNA-based approaches can provide this level of identification. Although a number of PCR protocols with or without DNA sequencing for differentiation of C. hominis and C. parvum have been reported (5, 21, 23, 26, 33), PCR followed by DNA sequencing analysis is a robust method for identification of Cryptosporidium species and also genotypes, depending on the genes analyzed, as reported in a large number of studies (2, 4, 9, 15-19, 27, 28, 31). However, sequencing is currently costly, labor-intensive, and time-consuming and requires an experienced staff to operate the DNA sequencer and process the data, which makes it less adequate for a rapid diagnostic response.
In this study, we describe a novel, more rapid technology that allows detection and identification of C. hominis and C. parvum in clinical samples. The assay is based on amplification of the microsatellite locus, microsatellite-2 region (ML-2), with biotinylated primers followed by hybridization of the amplicons with probes that differentiate between a single nucleotide substitution in the C. hominis and C. parvum sequences. The two capture probes are covalently bound to spectrally distinct populations of fluorescent microspheres that are reacted with streptavidin-phycoerythrin and analyzed in a unique, compact flow cytometer, the Luminex 100.
The assay was validated with a total of 143 DNA extracts, and the entire procedure can be performed within 5 h of the time a specimen is received by the laboratory. This assay is significantly less expensive than PCR amplification followed by DNA sequencing analysis and proved to be 100% specific and more sensitive than the direct fluorescent antibody test (DFA), which cannot differentiate C. parvum from C. hominis. With this technique we were able to detect two cases of mixed infections by C. hominis and C. parvum, which were confirmed with an alternate molecular method and would be of critical importance to an outbreak investigation.
MATERIALS AND METHODS
Samples.
A total of 143 DNA extracts from human stools or zoonotic or environmental samples were used to develop and optimize the Luminex assay. A total of 72 specimens were from individuals infected with C. hominis or C. parvum associated with outbreaks or sporadic cases of infections. In these samples, the DFA test was used as the gold standard for the determination of the presence of Cryptosporidium sp. A total of 30 samples were from studies that identified Cryptosporidium spp. in animal stools, flies, and mussels (18-20). Twenty of 72 human stools and all 30 animal samples were confirmed to be positive for either C. parvum or C. hominis by PCR using primers for Cryptosporidium oocyst wall protein (COWP) and 18S rRNA genes and ML-1 locus followed by DNA sequencing analysis of the amplicons (3, 22, 33, 34). Seventeen negative controls were also used; DNA extracted from stool samples was confirmed positive by PCR for Cyclospora cayetanensis (n = 4), Enterocytozoon bieneusi (n = 4), Encephalitozoon intestinalis (n = 2), Toxoplasma gondii (n = 2), Giardia intestinalis (n = 1), Entamoeba histolytica (n = 2), Entamoeba dispar (n = 1), and Encephalitozoon cuniculi (n = 1). To ascertain the lowest concentration of C. parvum oocysts that could be detected by this technique, we artificially created 24 samples by spiking human stools with C. parvum oocysts. The concentration of oocysts was determined visually by using a hemacytometer in a preparation purified from cow stools experimentally infected with C. parvum. Initially, three 1.7-ml tubes were filled with 300 μl of the same stool preparation obtained from one individual that was free of Cryptosporidium sp. (determined by DFA examination). Each tube was spiked with 1 × 107 of C. parvum oocysts, and each preparation was 10-fold serially diluted in tubes that contained 300 μl of the same stool preparation used initially. Three duplicate samples of seven serial dilutions that contained from 1 to 107 oocysts were prepared. One additional 1/10 dilution from the tubes containing one oocyst was done to create a set of samples to represent the endpoint. DNA was extracted from each dilution point as described below, amplified with ML-2 primers, and hybridized with DNA probes bound to microspheres. Doing the same with C. hominis oocysts was not possible because large concentrations cannot be produced efficiently in vitro or in vivo in zoonotic hosts.
DFA procedure.
The DFA test was performed using the MERIFLUOR Cryptosporidium/Giardia kit (Meridian Bioscience, Inc., Cincinnati, OH), following the manufacturer's instructions. Briefly, stool specimens were processed by ethyl acetate sedimentation. Four replicate direct wet smears were prepared from each of the specimens, air dried, and then processed separately by the fluorescent antibody test of the MERIFLUOR test kit (Meridian Diagnostic, Cincinnati, OH). Slides were examined for Cryptosporidium oocysts using fluorescence microscopy.
DNA extraction.
Total genomic DNA was extracted from 300 to 500 μl of spiked and clinical samples using a modified version of the FastDNA method (MP Biomedicals, Solon, OH) as previously described (8). Samples were disrupted in an FP120 cell disruptor (MP Biomedicals) at a speed of 5.5 for 10 seconds. Potential inhibitors were removed by further purification with the QIAquick PCR purification kit (QIAGEN Inc., Valencia, CA) per the manufacturer's instructions. Purified DNA was stored at 4°C until used in PCRs.
PCR amplification.
PCRs were performed using the primer pair M15/M16 (CAATGTAAGTTTACTTATGATTAT and CGACTATAAAGATGAGAGAAG, respectively) that amplify a specific fragment from the ML-2 locus, a microsatellite region of the Cryptosporidium genome (4). For Luminex detection, the reverse primer M16 was synthesized with biotin at the 5′ extremity to allow detection of hybridized amplicons with streptavidin-phycoerythrin. All PCRs were performed using AmpliTaq Gold PCR Master Mix (Applied Biosystems [ABI], Foster City, CA) and 0.3 μM of each primer in a final 50-μl reaction volume. Cycling parameters were 95°C for 5 min to activate the AmpliTaq Gold, followed by 35 cycles of 95°C for 15 s, 50°C for 15 s, and 72°C for 1 min, with a final extension at 72°C for 7 min. Amplified products were analyzed by electrophoresis on 2% SeaKem GTG agarose (catalog no. 50074; FMC Bioproducts, Rockland, ME), stained with ethidium bromide, and visualized on an UV transilluminator.
Amplification of fragments from 18S rRNA, COWP, and ML-1 was performed as described elsewhere (9, 17, 19), using primers CPBDIAGF/CPBDIAGR (22), CRY15/CRY9 (34), and GAGF/GAGR (4), respectively.
DNA sequencing reactions.
DNA sequencing reactions were performed by cycle sequencing with BigDye v.3.1 chemistry (ABI). Sequencing data were obtained using the ABI Prism 3100 sequence analyzer equipped with data collection software v. 2.0 and DNA Sequence Analysis Software v. 5.1 sequences were assembled, edited, and aligned in DNASTAR SeqMan (DNASTAR, Inc., Madison, WI), as well as in the GeneStudio suite (GeneStudio, Inc., Suwanee, GA).
Probe design.
The ML-2 sequences of C. hominis and C. parvum deposited in GenBank under accession numbers AY342297 and AY342296, respectively, were used to design the specific hybridization probes. Sequence AY342297 was for C. hominis type ML2-179 and sequence AY342296 for C. parvum ML2-176. These sequences were downloaded and aligned in the Wisconsin GCG package (Accelrys Software, San Diego, CA). Preselection of the length, specificity, and sequence of the probes was facilitated by application of the BLAST program (National Center for Biotechnology Information, Bethesda, MD; www.ncbi.nlm.nih.gov) to test 22- to 17-oligomers designed with the single mismatched base located in the middle of the probe sequences. The optimal sequence for the C. hominis-specific probe was TTA ATA AGA GTT TTA ACA, and the C. parvum probe was TTA ATA AGA ATT TTA ACA (the single mismatched base is shown in bold italic type), positions 107 to 125 and 105 to 123 of sequences AY342297 and AY342296, respectively. The selected sequences were amino modified at the 5′ end and attached to a 12-carbon linker, previously determined to be functionally optimal for microsphere-based hybridization assays (6). Each probe was covalently linked to a different Luminex microsphere and tested for specificity with a 5′ biotinylated, complementary oligonucleotide in a hybridization assay, as described below, before final conditions for the assay were optimized. Signals were generated only when biotinylated sequences bound to the complementary probe on the respective microsphere population.
Probe coupling.
The capture probes were covalently coupled to carboxylated microspheres (Luminex Corp., Austin, TX) using a carbodiimide coupling procedure. For individual sets of microspheres and capture probes, 5 × 106 microspheres were sonicated, vortexed, and pelleted in 1.5-ml microtubes (catalog no. 1415-2500; USA Scientific, Ocala, FL). The microspheres were then resuspended in 50 μl of 0.1 M MES (2-N-morpholino-ethanesulfonic acid) (Sigma, St Louis, MO), pH 4.5, and mixed by vortexing with 2 nM (2 μl of 1 mM) of amino-modified oligonucleotide suspended in 0.1 M MES. To the microsphere-capture probe mixture, 10 μl of 30 mg/ml of N-(3-dimethylaminodipropyl)-N′-ethylcarbodiimide (EDC) (Pierce, Rockford IL) prepared fresh was added and vortexed immediately. After incubation for 30 min with mixing, a fresh 10-μl aliquot of EDC was added, and the incubation was repeated. Coupled microspheres were washed with 0.1% sodium dodecyl sulfate, washed with 0.02% Tween 20, and resuspended in 500 μl TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) (10, 12, 35). Microsphere stocks were counted on a Beckman Coulter Z2 (Hialeah, FL) to determine the concentration of microspheres and stored in the dark at 4°C in TE buffer.
Hybridization assay procedure.
The hybridization assay was based on the binding of the complementary biotinylated PCR product to the capture probes. The assay was performed in a 96-well (conical well) plate (catalog no. 6509; Costar, Corning, NY). The total reaction volume was 50 μl, which included 33 μl of microsphere mixture and 17 μl of the amplified product or TE buffer (blank). To prepare the microsphere mixture, a calculated volume of each microsphere set was added to 1.5× TMAC buffer (1.5× TMAC buffer is 4.5 M tetramethylammonium chloride, 75 mM Tris-HCl, pH 8.0, 6 mM EDTA, and 0.15% Sarkosyl) to achieve a concentration of 1,500 to 2,500 microspheres per set in 33 μl. PCR product was added, the titer plate was sealed, and the amplified DNA was denatured at 95°C for 5 min followed by incubation for 50 min at 42°C in a thermocycler (PTC 200; MJ Research, Bio-Rad, Hercules, CA). Post-PCR purification was not required prior to hybridization. After incubation, the plate was centrifuged at 3,580 × g for 5 min, 25 μl of supernatant was carefully removed, and 75 μl of a 1:40 dilution of streptavidin-R-phycoerythrin (SA-PE) (Molecular Probes, Eugene, OR) was added. For comparison, we also performed this step without centrifugation and removal of 25 μl of the supernatant. At this point, mixing by hand pipetting was required. The plate was incubated for 15 min at 42°C and read on a heated plate at the same temperature on the Luminex 100 platform. This platform analyzes polystyrene microspheres of 5.6 μm that are internally dyed with two distinct fluorochromes mixed in different ratios to generate microsphere populations with specific spectral addresses. These microspheres are classified by two lasers, a red diode laser to detect the internal dyes of the microspheres and a 532-nm laser that excites the preferred reporter molecule, R-phycoerythrin (PE), to a high intensity emission at 578 nm. For data acquisition, BioPlex Manager Software v. 3.0 or 4.0 (Bio-Rad, Hercules, CA) or MiraiBio CT software (Alameda, CA) was used. Each sample was run in duplicate or triplicate with four blanks per plate. The median fluorescence intensity (MFI) of the SA-PE conjugate bound to 100 of each microsphere population was reported and considered statistically significant.
Calculations of results.
For the purpose of assigning the presence or absence of the Cryptosporidium species in each sample, the ratios of the signal/blank MFIs for the two microsphere populations were calculated and designated as the signal ratio (SR). To calculate the SR for any sample, the highest signal/blank ratio for the two probes was divided by the lowest. Based on experimental results, an SR of ≥1.5 indicated that the sample was positive for the probe with the higher signal/blank MFI. An SR value of <1.5 indicated a Cryptosporidium-negative or mixed (C. hominis plus C. parvum) sample but did not discriminate between the two possibilities. The negative samples had the lowest MFIs, similar to the blanks, while the mixed samples had high MFIs for both species, suggesting positive signal for both species. The validity of the SR in making the species-specific call was cross validated by the available sequencing data or PCRs using primers for COWP, 18S rRNA, or ML1 sequences followed by sequence analysis.
RESULTS
Optimization conditions for the hybridization assay.
Individual samples positive for either Cryptosporidium species were used for optimization experiments. Samples were PCR products amplified from stool specimens that had been tested by DFA and evaluated by an alternative PCR method using primers for other genes (e.g., COWP and 18S rRNA), followed by DNA sequencing analysis. Equal sample volumes were used for both species, and each optimization experiment was performed at least twice with duplicate data points.
Hybridization temperature.
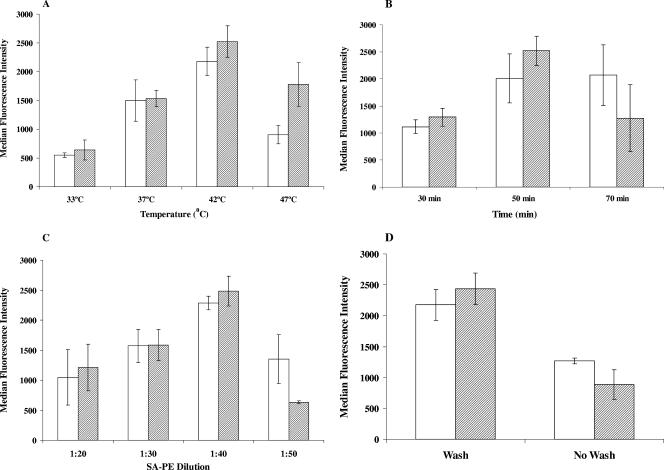
The hybridization assay was performed at four different hybridization temperatures: 33°C, 37°C, 42°C, and 47°C. Hybridization time was 50 min. SA-PE was diluted 1:40 in 1× TMAC buffer, and the washing of the plate was performed as described in Materials and Methods. A hybridization temperature of 42°C was found to be optimal for the assay, generating the highest MFIs for both C. hominis and C. parvum samples (Fig. 1A).
FIG. 1.
Effects of hybridization temperature (A), time (B), streptavidin-R-phycoerythrin (SA-PE) concentrations (C), and wash conditions (D) on hybridization of C. hominis (hatched bars) and C. parvum (white bars) capture probes to ML-2 amplicons. The bars represent means and standard deviation values of the median fluorescence intensity (MFI) from two and more experiments, each with duplicate data points. Equal volumes of PCR products were used to determine the optimal conditions for hybridization. Panels A, B, C, and D demonstrate that maximal MFI values were obtained with a hybridization temperature of 42°C, hybridization time of 50 min, SA-PE at a dilution of 1:40, and when samples were washed, respectively.
Hybridization time.
Hybridization times of 30, 50, and 70 min were tested with a hybridization temperature of 42°C and SA-PE dilution of 1:40. The optimal time of hybridization was observed to be 50 min (Fig. 1B).
Concentration of the reporter fluorophore.
Four dilutions of SA-PE (1:20, 1:30, 1:40, and 1:50 in 1× TMAC buffer) were examined. Figure 1C shows that SA-PE at a dilution of 1:40 produced maximum signals with minimum background. Reduced signals were observed at both higher and lower dilutions of SA-PE.
Wash conditions.
Samples that were treated by hybridization followed by centrifugation and removal of the 25-μl supernatant resulted in higher MFI values than samples that were not centrifuged (Fig. 1D). The hybridization time and temperatures were 50 min and 42°C, respectively, and the SA-PE dilution was 1:40.
Lowest concentration of C. parvum detected in spiked stool samples.
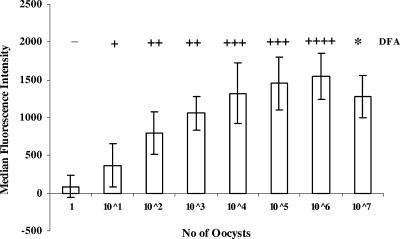
C. parvum ML-2 amplicons from stool specimens spiked with known concentrations of oocysts were analyzed (Fig. 2). The assay could successfully detect the equivalent of 10 oocysts. The ML-2 assay correlated directly with DFA, except that DFA was not performed at the highest concentration tested. For 107 oocysts, the ML-2 assay demonstrated the “hook effect,” as described by others (10). Essentially, the upper detection limit of the assay is reached, as the capture probes become oversaturated and further binding is sterically inhibited.
FIG. 2.
Detection limit for the Luminex ML-2 assay compared to the direct fluorescent antibody (DFA) test. DNA was extracted from uninfected stool samples spiked with different concentrations of C. parvum oocysts, ranging from 1 × 107 to 1 × 100, and PCR amplified with ML-2 PCR. Amplicons were hybridized to the C. hominis or C. parvum probe on different sets of fluorescent microspheres, reacted with SA-PE, and analyzed on a Luminex 100. The graph summarizes data from five separate experiments, each with triplicate data points. The DFA results are shown above the bars as follows: −, no oocysts detected by DFA; +, oocytes detected (the number of plus signs corresponds to the number of oocysts detected); *, not done.
Specificity and sensitivity of the Luminex ML-2 assay for clinical and environmental samples.
Table 1 includes the Luminex ML-2 assay results for a set of human stools and zoonotic samples. These samples were confirmed to contain C. hominis or C. parvum by DNA sequence analysis of amplicons subsequent to PCR amplification with primers for the COWP gene, 18S rRNA, or ML-1 coding regions as described above. The data shown in Table 1 are not comparative, since not all samples were tested with all markers, with the exception of the samples identified as C. parvum by the Luminex ML-2 assay, which were all tested by COWP PCR followed by DNA sequencing analysis. These data demonstrated that the assay was 100% specific at the species level.
TABLE 1.
Comparison of the Luminex ML-2 assay with PCR followed by DNA sequencing analysis of fragments amplified from COWP gene, ML-1, and 18S rRNA coding regions for identification of C. hominis and C. parvuma
| Species identified | No. of samples sequenced | No. of samples tested by DNA sequencing analysis with:
|
No. of samples tested by the Luminex ML-2 hybridization assay | ||
|---|---|---|---|---|---|
| COWP DNA | ML-1 DNA | 18S rRNA | |||
| C. hominis | 25 | 10 | 15 | 6 | 25 |
| C. parvum | 26 | 26 | 16 | 7 | 26 |
Samples displayed in this table were from studies done previously at CDC. Samples tested included human stools and a variety of samples of animal origin. Thirty-five human stools and 16 samples of animal origin were used. Six samples were confirmed to be C. hominis by using ML-1 and 18S rRNA loci. Sixteen samples were confirmed to be C. parvum by using COWP and ML-1 DNA. Of these 16 samples, 7 were also confirmed to be C. parvum by using 18S rRNA. DNA extracted from stool samples was obtained from individuals with intestinal symptoms and with positive results by DFA. The data shown are not comparative, since not all samples were tested with all markers, with the exception of the samples identified as C. parvum by the Luminex ML-2 assay, which were all tested by COWP PCR followed by DNA sequencing analysis.
The Luminex assay was subsequently used to detect the presence of C. hominis and C. parvum in stool samples that were associated with sporadic cases of infection or outbreak transmission (Table 2). All samples in this group were evaluated by DFA for Cryptosporidium and Giardia. In some cases, an alternative PCR followed by DNA sequencing analysis was used to confirm results obtained by the Luminex ML-2 assay. Samples that were negative by DFA but positive by the Luminex ML-2 assay (n = 5) were confirmed to be positive by PCR using primers for other loci, such as COWP or 18S rRNA. The Luminex ML-2 assay identified two samples that contained C. hominis and C. parvum. These samples were confirmed to be mixed by sequencing analysis of cloned COWP N-terminal amplicons.
TABLE 2.
Validation of the Luminex ML-2 assay with 42 clinical stool samples from sporadic cases of infections and outbreaksa
| Total no. of specimens tested | Total no. of specimens positive by DFA | No. of samples with the following result by the Luminex ML-2 assay:
|
Total no. of samples positive by the ML-2 Luminex assay | Total no. of discrepant results | |
|---|---|---|---|---|---|
| C. parvum | C. hominis | ||||
| 42 | 27 | 11b | 23b | 32 | 5 |
In this evaluation, DFA, a technique that cannot discriminate between Cryptosporidium species, was used as the gold standard. Two samples contained both C. parvum and C. hominis, and five samples negative by DFA were positive for C. parvum (n = 2) or C. hominis (n = 3). The five DFA-negative/Luminex ML-2 assay-positive samples were confirmed positive for either C. hominis or C. parvum by other molecular methods, including PCR-based methods and DNA sequencing analysis of COWP amplicons.
Two samples were positive for C. hominis and C. parvum.
Samples negative for Cryptosporidium (n = 27), whether positive or negative for other intestinal parasites (n = 17), did not produce signals above background.
DISCUSSION
Fast and accurate diagnostic response with early confirmatory identification of C. parvum and C. hominis is important for implementation of control measures and support of epidemiologic investigations. While a simple inexpensive morphology-based identification can be used to detect Cryptosporidium species in stool samples, only molecular approaches guarantee identification to the species level. This is important because C. hominis and C. parvum have distinct epidemiologies, and identification to the species level is important for determination of the source of the infection, and the assay described in this study will be specially useful in these investigations (28, 30).
To date, the major drawback to identification of C. hominis and C. parvum has been the requirement for PCR and DNA sequencing analysis of amplicons (1-5, 31). This approach is expensive and time-consuming and available only in select laboratories. Direct identification of C. hominis and C. parvum without the use of DNA sequencing analysis has been reported in some studies, but the methods were not thoroughly validated as diagnostic tests (23, 26, 33). In addition, the differential identification of C. parvum and C. hominis using such tools was not very robust compared with methods using PCR with DNA sequencing analysis. Nevertheless, such an approach would be of great value, especially during outbreak investigations. Based on this assumption, we were prompted to develop a technique that would be as robust as DNA sequencing analysis. The Luminex platform proved to be reliable and robust for this purpose and allowed rapid identification of C. hominis and C. parvum without cross-reaction with DNA from a number of distinct intestinal parasites. For this study, we chose the M15/M16 primers for amplification of the Cryptosporidium microsatellite locus ML-2 region because previous studies showed the usefulness of these primers for identification of C. hominis and C. parvum (1, 4). In addition, no ML-2 sequences for Cryptosporidium species other than C. hominis and C. parvum have been deposited thus far in GenBank, and these PCR primers were very sensitive in our hands compared with others described previously for detection of Cryptosporidium sp. (data not shown).
By allying the ML-2 locus amplification with hybridization-based Luminex differentiation, we were able to produce a sensitive assay that can distinguish C. hominis from C. parvum without the need for DNA sequencing analysis within approximately 6 h from receipt of the sample. The assay was validated using 143 samples, including 50 that were associated with cases of cryptosporidiosis that occurred in the United States or abroad. The assay described here is not only faster but is less expensive than sequencing of PCR amplicons. The total cost of one test using this approach is less than $0.16 (11, 35) compared with the traditional automated dye terminator DNA sequencing method, which is not less than $4.00 per reaction. These estimations do not include costs associated with lab personnel and specific equipment. Few DNA-based Luminex applications are available for detection and identification of parasites. A recent study demonstrated the usefulness of this approach for the simultaneous, semiquantitative identification of the four species of Plasmodium associated with the transmission of human malaria (25).
Most of the organism-specific assays are specific and sensitive only for the one particular organism that is suspected to be the infectious agent; confirmation requires subsequent experimentation and/or genotypic analysis. The Luminex system has the capacity to multiplex a number of targets at the same time and can identify multiple organisms or different genotypes of one particular organism in the same reaction well utilizing very little volume. Several DNA assays developed on the Luminex platform over the years have been used for identification and genotyping of infectious agents, such as Escherichia coli and Mycobacterium, Trichosporon, Salmonella, Listeria, and Candida spp. (6, 7, 10, 13). Most of these molecular methods are based on direct or competitive DNA hybridization for identification of PCR products.
For the present study, direct DNA hybridization was used. Design of short complementary probes with a single base mismatch placed at the center of the sequence was crucial for species identification. The extensive repetitive base sequences of the ML-2 microsatellites, combined with numerous insertions and deletions that varied the genotypes significantly, limited probe selection to a more stable region at the 5′ ends where a single base difference was found to be conserved between the species. By varying the length of the probes and the exact location of the base mismatch near the center, we were able to design probes with high specificity. Just as critically, TMAC buffer facilitated the use of more-stringent reaction conditions than those predicted by the melting temperatures of the probes and allowed the experimental discrimination between the one base difference in the PCR products. Tetramethylammonium chloride binds to the A+T-rich regions of the genome and significantly reduces the difference in the melting temperature of the A-T and G-C pairs. Probes with different melting temperatures thus melt irrespective of G+C content and more respective of probe length, therefore permitting the use of relatively short probes to discriminate minor differences in sequence (36).
Other single-nucleotide polymorphism-specific methods, such as allele-specific primer extension, single base chain extension, and oligonucleotide ligation, which involve sequence-specific enzymatic reactions, are also applicable for genotyping microorganisms (35). These methods require expensive DNA polymerases, ligases, and labeled dideoxynucleoside triphosphates and in some cases post-PCR purification of the amplified product using enzymes that significantly increase the cost and time for completion of the assay.
In summary, the sensitivity of the Luminex biplex assay was greater than that of DFA, a method routinely used for identification of Cryptosporidium and Giardia species in clinical laboratories. All five samples that were DFA negative and Luminex ML-2 positive were confirmed to contain C. hominis or C. parvum by other standard molecular methods used to identify Cryptosporidium sp., including DNA sequencing analysis of amplified fragments from genes, such as COWP. The detection limit of the assay was as low as 10 oocysts per 300 μl of stools spiked with C. parvum oocysts. The DNA samples used to validate the assay were previously identified by DNA sequencing analysis, and comparative results showed that the Luminex ML 2 assay was 100% specific for this set of samples. Interestingly, the Luminex ML-2 assay also proved to be an excellent tool for identification of samples from individuals infected with both C. hominis and C. parvum. As the Luminex platform is becoming more common in U.S. public health and clinical laboratory settings, this rapid, inexpensive, and relatively simple method may prove to be a very useful diagnostic tool for rapid identification of C. hominis and C. parvum. In the near future, we plan to expand this assay to detect a variety of intestinal pathogens in a multiplex fashion.
Acknowledgments
This study received funds from the National Food Safety Initiative. The work of Kakali Bandyopadhyay, Iaci Moura, and Maria Cristina Casaqui Carollo at the Atlanta VA Medical Center was supported in part by the Atlanta Research and Education Foundation.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The use of trade names is for identification only and does not imply endorsement by the Public Health Service or by the U.S. Department of Health and Human Services.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Alves, M., A. M. Ribeiro, C. Neto, E. Ferreira, M. J. Benoliel, F. Antunes, and O. Matos. 2006. Distribution of Cryptosporidium species and subtypes in water samples in Portugal: a preliminary study. J. Eukaryot. Microbiol. 53(Suppl. 1):S24-S25. [DOI] [PubMed] [Google Scholar]
- 2.Bornay-Llinares, F. J., A. J. da Silva, I. N. Moura, P. Myjak, H. Pietkiewicz, W. Kruminis-Lozowska, T. K. Graczyk, and N. J. Pieniazek. 1999. Identification of Cryptosporidium felis in a cow by morphologic and molecular methods. Appl. Environ. Microbiol. 65:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caccio, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120:237-244. [DOI] [PubMed] [Google Scholar]
- 4.Caccio, S., F. Spano, and E. Pozio. 2001. Large sequence variation at two microsatellite loci among zoonotic (genotype C) isolates of Cryptosporidium parvum. Int. J. Parasitol. 31:1082-1086. [DOI] [PubMed] [Google Scholar]
- 5.Chalmers, R. M., C. Ferguson, S. Caccio, R. B. Gasser, Y. G. Abs El-Osta, L. Heijnen, L. Xiao, K. Elwin, S. Hadfield, M. Sinclair, and M. Stevens. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35:397-410. [DOI] [PubMed] [Google Scholar]
- 6.Cowan, L. S., L. Diem, M. C. Brake, and J. T. Crawford. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 42:474-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, S., T. M. Brown, K. L. Kellar, B. P. Holloway, and C. J. Morrison. 2006. DNA probes for the rapid identification of medically important Candida species using a multianalyte profiling system. FEMS Immunol. Med. Microbiol. 46:244-250. [DOI] [PubMed] [Google Scholar]
- 8.da Silva, A. J., F. J. Bornay-Llinares, I. N. Moura, S. B. Slemenda, J. L. Tuttle, and N. J. Pieniazek. 1999. Fast and reliable extraction of protozoan parasite DNA from fecal specimens. Mol. Diagn. 4:57-64. [DOI] [PubMed] [Google Scholar]
- 9.da Silva, A. J., S. Caccio, C. Williams, K. Y. Won, E. K. Nace, C. Whittier, N. J. Pieniazek, and M. L. Eberhard. 2003. Molecular and morphologic characterization of a Cryptosporidium genotype identified in lemurs. Vet. Parasitol. 111:297-307. [DOI] [PubMed] [Google Scholar]
- 10.Diaz, M. R., and J. W. Fell. 2004. High-throughput detection of pathogenic yeasts of the genus Trichosporon. J. Clin. Microbiol. 42:3696-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar, S. A. 2006. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 363:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, S. A., and J. W. Jacobson. 2000. Application of the Luminex LabMAP in rapid screening for mutations in the cystic fibrosis transmembrane conductance regulator gene: a pilot study. Clin. Chem. 46:1498-1500. [PubMed] [Google Scholar]
- 13.Dunbar, S. A., C. A. Vander Zee, K. G. Oliver, K. L. Karem, and J. W. Jacobson. 2003. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J. Microbiol. Methods 53:245-252. [DOI] [PubMed] [Google Scholar]
- 14.Fayer, R. 2004. Cryptosporidium: a water-borne zoonotic parasite. Vet. Parasitol. 126:37-56. [DOI] [PubMed] [Google Scholar]
- 15.Fayer, R., M. Santin, and L. Xiao. 2005. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). J. Parasitol. 91:624-629. [DOI] [PubMed] [Google Scholar]
- 16.Fayer, R., J. M. Trout, L. Xiao, U. M. Morgan, A. A. Lai, and J. P. Dubey. 2001. Cryptosporidium canis n. sp. from domestic dogs. J. Parasitol. 87:1415-1422. [DOI] [PubMed] [Google Scholar]
- 17.Goncalves, E. M., A. J. da Silva, M. B. Eduardo, I. H. Uemura, I. N. Moura, V. L. Castilho, and C. E. Corbett. 2006. Multilocus genotyping of Cryptosporidium hominis associated with diarrhea outbreak in a day care unit in Sao Paulo. Clinics 61:119-126. [DOI] [PubMed] [Google Scholar]
- 18.Graczyk, T. K., A. J. Da Silva, M. R. Cranfield, J. B. Nizeyi, G. R. Kalema, and N. J. Pieniazek. 2001. Cryptosporidium parvum genotype 2 infections in free-ranging mountain gorillas (Gorilla gorilla beringei) of the Bwindi Impenetrable National Park, Uganda. Parasitol. Res. 87:368-370. [DOI] [PubMed] [Google Scholar]
- 19.Graczyk, T. K., B. H. Grimes, R. Knight, A. J. Da Silva, N. J. Pieniazek, and D. A. Veal. 2003. Detection of Cryptosporidium parvum and Giardia lamblia carried by synanthropic flies by combined fluorescent in situ hybridization and a monoclonal antibody. Am. J. Trop. Med. Hyg. 68:228-232. [PubMed] [Google Scholar]
- 20.Graczyk, T. K., D. J. Marcogliese, Y. de Lafontaine, A. J. Da Silva, B. Mhangami-Ruwende, and N. J. Pieniazek. 2001. Cryptosporidium parvum oocysts in zebra mussels (Dreissena polymorpha): evidence from the St. Lawrence River. Parasitol. Res. 87:231-234. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, J., and L. Xiao. 2003. An evaluation of molecular diagnostic tools for the detection and differentiation of human-pathogenic Cryptosporidium spp. Eukaryot. Microbiol. 50(Suppl.):542-547. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limor, J. R., A. A. Lal, and L. Xiao. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacKenzie, W. R., W. L. Schell, K. A. Blair, D. G. Addiss, D. E. Peterson, N. J. Hoxie, J. J. Kazmierczak, and J. P. Davis. 1995. Massive outbreak of waterborne Cryptosporidium infection in Milwaukee, Wisconsin: recurrence of illness and risk of secondary transmission. Clin. Infect. Dis. 21:57-62. [DOI] [PubMed] [Google Scholar]
- 25.McNamara, D. T., L. J. Kasehagen, B. T. Grimberg, J. Cole-Tobian, W. E. Collins, and P. A. Zimmerman. 2006. Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am. J. Trop. Med. Hyg. 74:413-421. [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 27.Morgan, U. M., L. Xiao, P. Monis, I. Sulaiman, I. Pavlasek, B. Blagburn, M. Olson, S. J. Upton, N. V. Khramtsov, A. Lal, A. Elliot, and R. C. Thompson. 2000. Molecular and phylogenetic analysis of Cryptosporidium muris from various hosts. Parasitology 120:457-464. [DOI] [PubMed] [Google Scholar]
- 28.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 29.Navarro-i-Martinez, L., F. J. Bornay-Llinares, C. Rueda, C. del Aguila, A. J. da Silva, A. Oleaga, V. Ramajo, S. Fenoy, and N. J. Pieniazek. 2003. Molecular characterization of Cryptosporidium sp. from animals in Spain. J. Eukaryot. Microbiol. 50(Suppl.):553-554. [DOI] [PubMed] [Google Scholar]
- 30.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. MacKenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. da Silva, I. N. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan, U. M., P. Monis, H. L. Enemark, I. Sulaiman, B. Samarasinghe, C. Read, R. Buddle, I. Robertson, L. Zhou, R. C. Thompson, and L. Xiao. 2004. Cryptosporidium suis n. sp. (Apicomplexa: Cryptosporidiidae) in pigs (Sus scrofa). J. Parasitol. 90:769-773. [DOI] [PubMed] [Google Scholar]
- 33.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209-217. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, J. D., D. Briley, Q. Nguyen, K. Long, M. A. Iannone, M. S. Li, F. Ye, A. Afshari, E. Lai, M. Wagner, J. Chen, and M. P. Weiner. 2001. Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. BioTechniques 30:661-666, 668-669. [DOI] [PubMed] [Google Scholar]
- 36.Wood, W. I., J. Gitschier, L. A. Lasky, and R. M. Lawn. 1985. Base composition-independent hybridization in tetramethylammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc. Natl. Acad. Sci. USA 82:1585-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]