Abstract
Antigen sequence typing (ST) of FetA is part of the molecular typing scheme of Neisseria meningitidis. Among invasive meningococcal isolates from 2,201 patients in Germany, we identified 11 strains lacking the fetA gene because of deletions mediated by repeat arrays flanking the gene, i.e., Correia elements, repeat sequence 13 (RS13), and duplicated RS3. Geographic mapping and multilocus ST of invasive isolates revealed that fetA deletion was a sporadic event without genetic fixation. Among 821 carrier strains, 12 strains lacked fetA, suggesting that fetA is maintained during asymptomatic carriage. Interestingly, most of these isolates belonged to the multilocus ST-35 clonal complex (cc). ST-35 cc strains and the recently published ST-192 strains from Burkina Faso may benefit from loss of fetA, but their infrequent occurrence among invasive isolates currently does not affect fetA antigen ST.
Antigen sequence typing (ST) is now widely used for highly discriminatory and precise typing of Neisseria meningitidis (8). Finetyping data are used to monitor epidemiological changes over time and space and to unravel the plethora of circulating antigen types for the sake of subcapsular vaccine development. We recently demonstrated the benefits of including FetA antigen typing in the typing regime at the German National Reference Laboratory for Meningococci (5). With regard to the investigation of infections in 1,616 patients, the addition of FetA antigen typing increased the number of finetypes 2.3-fold compared to the use of serogrouping in conjunction with PorA typing but without FetA antigen typing.
The ferric enterochelin receptor FetA (formerly FrpB) is an immunodominant protein present in outer membrane vesicle vaccines (14, 17). During the preparation of the manuscript, Marsh et al. electronically published ahead of print the identification of three fetA-negative isolates among 768 invasive isolates collected in the United States between 1990 and 2006 (10). FetA deletion was mediated by duplicated repeat sequence 3 (dRS3) repetitive elements. Deletion via flanking repeats is reminiscent of what has been observed in strains lacking the porin gene porA (18). FetA deletion was also a trait of ST-192 strains isolated from cases and healthy carriers in Burkina Faso (7).
In this paper, we report the frequency of fetA deletion in Germany, provide evidence by geographic mapping and multilocus ST (MLST) (9) that fetA deletion is a sporadic event, and report a detailed genetic analysis of deletion events.
MATERIALS AND METHODS
Meningococcal strains.
Strains were collected at the German National Reference Laboratory for Meningococci as part of the nationwide laboratory surveillance of meningococcal disease. The majority of strains have been described recently (5). For genetic studies, we include four fetA-negative strains recently identified among noninvasive isolates, which are sent to the reference laboratory on an unsystematic basis or as part of a carriage study (11). Furthermore, the recently published Bavarian carrier strain collection was screened for fetA deletions (4). Finally, a representative ST-192 strain was analyzed for the genetic mechanism of its fetA deletion, described recently (7). This strain was isolated in Burkina Faso among a series of clonally related capsule null locus strains.
FetA antigen ST and MLST.
FetA typing and MLST were performed as described previously (9, 16). Allele and ST assignments were achieved by comparison with the databases at www.neisseria.org. The fetA gene was routinely amplified using the primers S13-2 (5′-CAT ACC CAA ATC ACC ACT CG-3′) and S15 (5′-TTG CAG CGC GTC RTA CAG GCG-3′). In cases of negative results, the primers S1 (5′-CGG CGC AAG CGT ATT CGG-3′) and S8 (5′-CGC GCC CAA TTC GTA ACC GTG-3′) were tested. If no amplification of fetA was achieved, the presence of fetA was determined by Southern blot hybridization using a fetA probe generated from a fetA-positive strain (MC58) by PCR with the primers S1 and S8. The carrier isolates from the Bavarian carriage study were analyzed by dot blot hybridization according to recently published protocols (3), using the S1/S8 probe. Negative results were confirmed by S1/S8 PCR. In cases of fetA-negative strains, the remnants of the fetA chromosomal locus were investigated either by two PCR assays employing flanking primers (CM21 [5′-AAC GCA TCG AAA TCC ACA GC-3′] and CM24 [5′-CTT TGA GGT TGG CGG TAT CG-3′] or CM28 [5′-ATG CCC GCA ATC TCA AAT CC-3′] and CM26 [5′-GGT CGG ACA AAC CGG AAC G-3′], respectively) and subsequent sequencing of the PCR products or by cloning of the locus. Briefly, chromosomal DNA of fetA-negative meningococcal strains was restricted with MluI, which cuts in the genes adjacent to fetA, and analyzed by Southern blot hybridization using probes flanking fetA (i.e., PCR product CM21/CM22 [5′-GGT CGT CCG CCG TAA ACT C-3′] and PCR product CM23 [5′-TTT GAC TGC TTT AGC CGT ATG-3′]/CM24). MluI fragments hybridizing with both probes were cloned into the EcoRI site of pBluescript SK (Stratagene, The Netherlands) and subsequently sequenced by primer walking.
Geographic maps.
Latitude and longitude coordinates (map date, WSG 84) of zip codes of the year 2003 were obtained from GfK Macon (Waghäusel, Germany). The Regiograph 8 (GfK Macon) and Fireworks MX Macromedia (Adobe Systems Inc., San Jose, CA) programs were used to generate and edit maps.
Nucleotide sequence accession numbers.
The sequences of the intergenic regions between NMB1987 and NMB1989 or NMB1990, respectively, identified in the fetA-negative strains have been deposited in the EMBL nucleotide sequence database under accession numbers AM748025 to AM748034 and AM748728 to AM748731.
RESULTS AND DISCUSSION
We report the frequency, geographic distribution, and lineages of fetA-negative isolates in Germany. Furthermore, we provide a detailed dissection of the genetic mechanisms giving rise to loss of the gene.
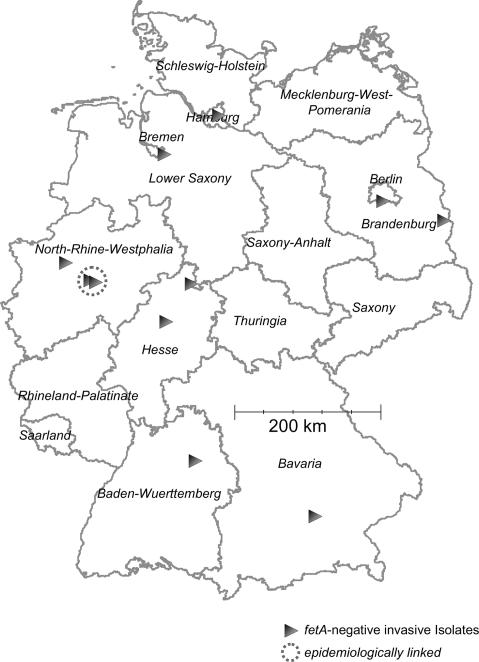
Among strains from 2,201 patients suffering from invasive meningococcal disease in Germany between 2001 and 2007, we identified 11 fetA-negative strains (0.5%). The strains were almost randomly distributed throughout Germany (Fig. 1). This finding suggests sporadic emergence of invasive fetA-negative strains. All invasive strains belonged to different STs, with the exception of two isolates from an outbreak of ST-11 disease among schoolchildren and two ST-35 cases which were not epidemiologically linked (Table 1). The genetic diversity, together with the geographic distribution, indicates that, in Germany, there are no pathogenic genetic lineages regularly lacking fetA. The above-mentioned outbreak due to ST-11 strains comprised fetA-negative and fetA-positive strains, which otherwise proved to be identical, as confirmed by MLST, fumC single-nucleotide polymorphism typing, and IS1301 restriction fragment length polymorphism typing (6). Interestingly, van der Ende et al. reported an outbreak of seven cases due to ST-11 strains, of which five were caused by strains with a stable point mutation in the porA gene resulting in the loss of expression of PorA (19). The authors hypothesized that the strains were transmitted between cases and were unlikely to have emerged during infection.
FIG. 1.
Geographic distribution of cases of invasive meningococcal disease in Germany caused by strains lacking the fetA gene. Time period: December 2001 through February 2007.
TABLE 1.
Characteristics of Neisseria meningitidis strains lacking the fetA genea
| Strain | Epidemiology of isolate | Serogroup | Serogenogroup | Allele of PorA variable regionb:
|
ST | Clonal complex | |
|---|---|---|---|---|---|---|---|
| 1 | 2 | ||||||
| DE8926 | Invasive | C | C | 5 | 2 | 8 | 8 |
| DE9260 | Invasive | C | C | 5-1 | 10-8 | 11 | 11 |
| DE9301 | Invasive | C | C | 5-1 | 10-8 | 11 | 11 |
| DE9489 | Invasive | B | B | 22-1 | 14 | 35 | 35 |
| DE9516 | Invasive | B | B | 22 | 14 | 162 | 162 |
| DE9598 | Invasive | B | B | 18-1 | 3 | 4615 | |
| DE9734 | Invasive | B | B | 22-1 | 14 | 35 | 35 |
| DE9848 | Invasive | B | B | 19 | 15 | 44 | 41/44 |
| DE9872 | Invasive | B | B | 7-2 | 13-2 | 180 | 41/44 |
| DE10260 | Invasive | B | B | 7 | 16 | 32 | 32 |
| DE10383 | Invasive | B | B | 17 | 9 | 5802 | |
| DE9313 | Carriage | Y | Y | 5-8 | 10-4 | 785 | 92 |
| DE9991 | Carriage | Acapsular | X | 7-1 | 1 | 5799 | 22 |
| DE10920 | Carriage | * | * | 22 | 9 | 5901 | 35 |
| γ30 | Carriage | Acapsular | Y | 5-2 | 10-1 | 23 | 23 |
| 10115V3B | Carriage | Acapsular | cnl | 18-11 | 42 | 192 | |
| α43 | Carriage | Acapsular | C | 22-1 | 14 | 806 | 35 |
| α53 | Carriage | Acapsular | C | 12-1 | 35 | 35 | |
| α58 | Carriage | B | B | 17-1 | 23 | 1132 | 35 |
| α193 | Carriage | Acapsular | B | 18-1 | 3 | 799 | |
| α323 | Carriage | Acapsular | B | 7-2 | 16 | 801 | 32 |
| α458 | Carriage | Acapsular | cnl | 18 | 25-19 | 845 | |
| α496 | Carriage | Acapsular | cnl | 7 | 30-1 | 53 | 53 |
| α501 | Carriage | Acapsular | C | 22-1 | 14 | 810 | 35 |
| α604 | Carriage | * | * | 22-1 | 14 | 160 | 35 |
| α718 | Carriage | B | B | 7-2 | 16 | 32 | 32 |
| α725 | Carriage | Acapsular | B | 7-1 | 1 | 44 | 41/44 |
| α788 | Carriage | * | * | 22-1 | 14 | 35 | 35 |
Strains include 11 invasive isolates (prefix “DE,” National Reference Laboratory for Meningococci) and 17 carriage isolates (prefix “α,” Bavarian carriage study [4]; prefix “γ,” Sangerhausen carriage study [11]; strain 10115V3B, Burkina Faso [7]). cnl, capsule null locus (3); *, not serogroup and serogenogroup B, C, W-135, and Y, as determined by slide agglutination and specific PCR, respectively.
Allele assignment was achieved by comparison with the PorA database at www.neisseria.org.
We determined precisely the mechanisms of fetA loss by DNA sequence analysis not only in the 11 invasive strains from the national laboratory surveillance and 12 carrier strains from the Bavarian carrier strain collection but also in five additional fetA-negative isolates from other sources. The results are depicted in Fig. 2. Marsh et al. elucidated the responsibility of dRS3 repeats for fetA loss in the three strains analyzed (10). We now demonstrate a variety of mechanisms of intragenomic recombination or rearrangement leading to loss of fetA and, in some instances, also of the downstream gene fetB. This includes (i) RS13-mediated recombination, in one case in conjunction with the uptake of the insertion sequence IS1655; (ii) recombination mediated by Correia elements; and (iii) recombination via dRS3 repeats.
FIG. 2.
Genetic analysis of strains lacking the fetA gene with emphasis on repeat elements. Strains Z2491 and MC58 were included as reference strains (13, 15). Strains with prefix “NM” were deduced from a recent publication describing three fetA-negative strains from the United States (10). Strains with the prefix “DE” are from the National Reference Center for Meningococci and mostly represent invasive isolates. Strains with the prefix “α” were isolated in the course of the Bavarian carriage study (4). Strain γ30 was isolated during a swabbing campaign whose results were published recently (11). Strain 10115V3B is an ST-192 capsule null locus isolate from Burkina Faso (7).
Strain 10115V3B was a representative of 17 fetA-negative ST-192 strains isolated in Burkina Faso between 2003 and 2004 (7). The intergenic region was not sequenced in all strains; however, PCR analysis revealed its identical length, suggesting a single mechanism of fetA deletion in all ST-192 isolates. According to our knowledge, the ST-192 clone is the first clone to be uniformly associated with a fetA deletion.
FetA is a variable gene whose antigenic variability might be at least partially driven by flanking noncoding repeat elements, as postulated recently by Bentley et al. (1). These repeat arrays, however, apparently also support gene loss. FetA, formerly FrpB, is an iron-repressed TonB-dependent receptor which binds ferric enterobactin. In gonococci, FetA expression can be down-regulated by phase variation (2). Inspection of the corresponding homopolymeric tracts within the meningococcal fetA promoter sequence of strain MC58 (15) suggests that phase variation is possible in this species also (data not shown). Mutation of FetA is nonlethal, iron uptake is not affected, and serum sensitivity is only mildly enhanced (14). FetA is expressed during invasive infection, but a clear role in the bloodstream needs to be defined. The fact that fetA-negative strains emerge now and then and cause invasive disease is noteworthy with regard to the use of outer membrane vesicle vaccines harboring FetA. It will be interesting to see whether the use of such vaccines in New Zealand (12) will enhance the frequency of such strains.
We next focused on the proportion of fetA deletion in carriage isolates. We have recently demonstrated the high rate of capsule synthesis gene-silencing events in carried meningococci, which, however, are not fixed in the population (20). We have now analyzed the same strain collection for the frequency of fetA deletion by using dot blot hybridization and, in the case of negative results, by PCR. A total of 821 strains were included, and 12 independent strains were fetA negative, in comparison to 11 independent strains of 2,201 for the invasive disease cases. Herein, we defined dependence of strains as the spatiotemporal linkage of identical STs. In a chi-square test, the difference between the collections with regard to the number of fetA-negative strains was significant (P = 0.01). However, one has to consider that the Bavarian carrier strain collection was established in one federal state of Germany in winter 1999/2000, thus, before the collection of the invasive isolates. It was noteworthy that 6 of 12 strains belonged to the ST-35 complex. Thus, the ST-35 complex might tolerate fetA deletions similarly to the ST-192 complex (7). This finding is supported by the fact that Marsh et al. also found ST-35 in 2 of 3 strains with fetA deletion (10). Nevertheless, in the Bavarian carrier strain collection, only 6 of 45 ST-35 clonal complex strains were fetA negative.
It cannot be excluded for certain that fetA deletions might have occurred during laboratory passage in some cases. We estimate a number of four passages on artificial media prior to typing and storage. However, the observation of two epidemiologically linked fetA-negative strains within the ST-11 outbreak described above, as well as of the uniformly fetA-negative genotype of ST-192 strains from Burkina Faso (7), supports the view of a within-host evolution of fetA deletion variants.
In conclusion, fetA typing is not hampered by rare and sporadic gene deletion events. Deletion is supported by flanking, noncoding repeat arrays. The reasons for selection against loss of fetA are unclear. Due to the geographic and temporal independence of the strain collections, our study does not have the power to clearly demonstrate that the carrier state is more permissive for fetA deletion than invasive disease, but we could demonstrate a trend for more fetA deletions in carriage than in invasive disease. This phenomenon could be explained either by the importance of the ferric enterochelin receptor for systemic spread during infection or by the differences of clonal lineages and their biology which have recently been epidemiologically demonstrated (21).
Acknowledgments
The National Reference Center for Meningococci is supported by the German Ministry of Health via the Robert Koch-Institute. This study was part of the center's activities.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Bentley, S. D., G. S. Vernikos, L. A. Snyder, C. Churcher, C. Arrowsmith, T. Chillingworth, A. Cronin, P. H. Davis, N. E. Holroyd, K. Jagels, M. Maddison, S. Moule, E. Rabbinowitsch, S. Sharp, L. Unwin, S. Whitehead, M. A. Quail, M. Achtman, B. Barrell, N. J. Saunders, and J. Parkhill. 2007. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 3:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carson, S. D., B. Stone, M. Beucher, J. Fu, and P. F. Sparling. 2000. Phase variation of the gonococcal siderophore receptor FetA. Mol. Microbiol. 36:585-593. [DOI] [PubMed] [Google Scholar]
- 3.Claus, H., M. C. Maiden, R. Maag, M. Frosch, and U. Vogel. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813-1819. [DOI] [PubMed] [Google Scholar]
- 4.Claus, H., M. C. Maiden, D. J. Wilson, N. D. McCarthy, K. A. Jolley, R. Urwin, F. Hessler, M. Frosch, and U. Vogel. 2005. Genetic analysis of meningococci carried by children and young adults. J. Infect. Dis. 191:1263-1271. [DOI] [PubMed] [Google Scholar]
- 5.Elias, J., D. Harmsen, H. Claus, W. Hellenbrand, M. Frosch, and U. Vogel. 2006. Spatiotemporal analysis of invasive meningococcal disease, Germany. Emerg. Infect. Dis. 12:1689-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elias, J., and U. Vogel. 2007. IS1301 fingerprint analysis of Neisseria meningitidis strains belonging to the ET-15 clone. J. Clin. Microbiol. 45:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Findlow, H., U. Vogel, J. E. Mueller, A. Curry, B. M. Njanpop-Lafourcade, H. Claus, S. J. Gray, S. Yaro, Y. Traore, L. Sangare, P. Nicolas, B. D. Gessner, and R. Borrow. 2007. Three cases of invasive meningococcal disease caused by a capsule null locus strain circulating among healthy carriers in Burkina Faso. J. Infect. Dis. 195:1071-1077. [DOI] [PubMed] [Google Scholar]
- 8.Jolley, K. A., C. Brehony, and M. C. Maiden. 2007. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol. Rev. 31:89-96. [DOI] [PubMed] [Google Scholar]
- 9.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh, J. W., M. M. O'Leary, K. A. Shutt, and L. H. Harrison. 2007. Deletion of fetA gene sequences in serogroup B and C Neisseria meningitidis isolates. J. Clin. Microbiol. 45:1333-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oppermann, H., B. Thriene, H. M. Irmscher, L. Grafe, M. Borrmann, D. Bellstedt, S. Kaynak, W. Hellenbrand, and U. Vogel. 2006. Meningococcal carriers in high school students and possible risk factors. Gesundheitswesen 68:633-637. (In German.) [DOI] [PubMed] [Google Scholar]
- 12.Oster, P., D. Lennon, J. O'Hallahan, K. Mulholland, S. Reid, and D. Martin. 2005. MeNZB: a safe and highly immunogenic tailor-made vaccine against the New Zealand Neisseria meningitidis serogroup B disease epidemic strain. Vaccine 23:2191-2196. [DOI] [PubMed] [Google Scholar]
- 13.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 14.Pettersson, A., A. Maas, D. van Wassenaar, P. van der Ley, and J. Tommassen. 1995. Molecular characterization of FrpB, the 70-kilodalton iron-regulated outer membrane protein of Neisseria meningitidis. Infect. Immun. 63:4181-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, E. A., I. M. Feavers, and M. C. Maiden. 2003. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 149:1849-1858. [DOI] [PubMed] [Google Scholar]
- 17.Urwin, R., J. E. Russell, E. A. Thompson, E. C. Holmes, I. M. Feavers, and M. C. Maiden. 2004. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect. Immun. 72:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Ende, A., C. T. Hopman, and J. Dankert. 1999. Deletion of porA by recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect. Immun. 67:2928-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Ende, A., C. T. Hopman, W. C. Keijzers, L. Spanjaard, E. B. Lodder, P. H. van Keulen, and J. Dankert. 2003. Outbreak of meningococcal disease caused by PorA-deficient meningococci. J. Infect. Dis. 187:869-871. [DOI] [PubMed] [Google Scholar]
- 20.Weber, M. V., H. Claus, M. C. Maiden, M. Frosch, and U. Vogel. 2006. Genetic mechanisms for loss of encapsulation in polysialyltransferase-gene-positive meningococci isolated from healthy carriers. Int. J. Med. Microbiol. 296:475-484. [DOI] [PubMed] [Google Scholar]
- 21.Yazdankhah, S. P., P. Kriz, G. Tzanakaki, J. Kremastinou, J. Kalmusova, M. Musilek, T. Alvestad, K. A. Jolley, D. J. Wilson, N. D. McCarthy, D. A. Caugant, and M. C. Maiden. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42:5146-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]