Abstract
Haemophilus influenzae is an important cause of respiratory infections, including acute otitis media, sinusitis, and chronic bronchitis, which are preceded by asymptomatic H. influenzae colonization of the human pharynx. The aim of this study was to describe the dynamics of pharyngeal colonization by H. influenzae and an intimately related species, Haemophilus haemolyticus, in healthy adults. Throat specimens from four healthy adult carriers were screened for Haemophilus species; 860 isolates were identified as H. influenzae or H. haemolyticus based on the porphyrin test and on dependence on hemin and NAD for growth. Based on tests for hemolysis, for the presence of the 7F3 epitope of the P6 protein, and for the presence of iga in 412 of the isolates, 346 (84%) were H. influenzae, 47 (11%) were H. haemolyticus, 18 (4%) were nonhemolytic H. haemolyticus, and 1 was a variant strain. Carriers A and B were predominantly colonized with nontypeable H. influenzae, carrier C predominantly with b− H. influenzae mutants, and carrier D with H. haemolyticus. A total of 358 H. influenzae and H. haemolyticus isolates were genotyped by pulsed-field gel electrophoresis (PFGE) following SmaI or EagI digestion of their DNA, and the carriers displayed the following: carrier A had 11 unique PFGE genotypes, carrier B had 15, carrier C had 7, and carrier D had 10. Thus, adult H. influenzae and H. haemolyticus carriers are colonized with multiple unique genotypes, the colonizing strains exhibit genetic diversity, and we observed day-to-day and week-to-week variability of the genotypes. These results appear to reflect both evolutionary processes that occur among H. influenzae isolates during asymptomatic pharyngeal carriage and sample-to-sample collection bias from a large, variable population of colonizing bacteria.
Haemophilus species constitute approximately 10% of the total bacterial flora in the human upper respiratory tract (27). Haemophilus influenzae is an opportunistic pathogen in humans that asymptomatically colonizes the pharyngeal mucosa and occasionally the genital mucosa. The rate of carriage of H. influenzae increases from infancy (about 20% in the first year of life) to early childhood (>50% in children 5 to 6 years old), and H. influenzae is recoverable from the upper airways of 20 to 80% of healthy children (2, 15, 22, 32).
Colonization of the human respiratory mucosal surface represents a dynamic process in which bacteria are acquired, replaced, and reacquired many times in a lifetime. Past studies have demonstrated that H. influenzae colonization of the pharynx is characterized by rapid bacterial turnover (9, 12, 50, 58) and carriage of multiple strains at any one time (18, 37, 43, 52, 58). Previous studies have demonstrated a 62% week-to-week turnover rate of H. influenzae isolated from healthy children attending day care; 37 to 43% of throat cultures contained two or more genetically distinct strains (range, zero to five) (14, 56).
In addition to living as a commensal in the respiratory tract, H. influenzae may also cause symptomatic infections. A majority of invasive H. influenzae infections in children, such as bacteremia, meningitis, pneumonia, epiglottitis, and septic arthritis, are caused by the encapsulated H. influenzae type b. Nontypeable (i.e., nonencapsulated) H. influenzae (NTHI) rarely causes invasive disease in healthy hosts but is a significant cause of localized respiratory tract infections, such as otitis media, sinusitis, bronchitis, and conjunctivitis (35). NTHI accounts for 30 to 52% of acute otitis media infections in children (11). NTHI is also the most commonly seen bacterium in acute exacerbations of chronic obstructive pulmonary disease (COPD) (3, 20, 38, 51) and appears to contribute to COPD progression (46). In addition, acute exacerbation has been shown to occur with acquisition of a new strain of H. influenzae (relative risk, 2.96 [95% confidence interval, 2.39 to 3.67]) (51). Strain-specific H. influenzae bactericidal antibodies were seen in 61% of exacerbations associated with new strains compared to 21% associated with preexisting strains (38), further supporting the role of NTHI in COPD exacerbations.
Haemophilus haemolyticus is phylogenetically closely related to H. influenzae and is also found in the pharynx in healthy adults (31). Pathogenicity of H. haemolyticus to humans has not been demonstrated (1), and the organism can be phenotypically differentiated from NTHI in the laboratory by its ability to produce a clear hemolytic zone on horse blood agar. The hemolytic activity of H. haemolyticus, however, may be lost on subculture, resulting in difficulty differentiating it from H. influenzae. Since discriminating H. haemolyticus from H. influenzae is important to fully understand the pathogenesis of H. influenzae disease, in this study, the nonhemolytic isolates were further tested for the presence of a specific epitope on the outer membrane protein P6 and the presence of iga, both of which have been shown to be associated with H. influenzae and not H. haemolyticus (16, 39).
Although H. influenzae pharyngeal colonization in the context of disease has been relatively well studied, its carriage and the dynamics of colonization have not been well characterized in healthy adults. This study used pulsed-field gel electrophoresis (PFGE) to describe the genetic diversity and dynamics of colonization of H. haemolyticus and H. influenzae over 7 months in four healthy adults.
MATERIALS AND METHODS
Study design.
Throat culture samples were obtained from 16 healthy adults after informed consent, as approved by the University of Michigan Institutional Review Board. Screening samples from four participants were positive for NAD- and hemin-dependent Haemophilus species, and further culture samples from these four individuals were obtained five workdays a week for the first month, followed by once a week for the next 6 months.
Bacteriologic methods.
All throat culture samples were collected with a double-tipped swab by the same person to minimize variability in technique. The swabs were streaked on a chocolate agar plate supplemented with 16,500 units/liter bacitracin (BBL, Sparks, MD) and incubated at 37°C with 5% CO2 for 24 h. Up to 30 isolates per throat sample consistent with Haemophilus morphology were selected for further analysis. The porphyrin test and hemolysis of horse erythrocytes (28) were used to differentiate H. influenzae from heme-dependent (i.e., porphyrin-negative) Haemophilus isolates, such as Haemophilus parainfluenzae, and from H. haemolyticus, respectively. The putative H. influenzae and H. haemolyticus isolates were further confirmed by growth on brain heart infusion agar (Difco, Detroit, MI) supplemented with factor X (hemin), V (NAD), or both (Sigma-Aldrich, St. Louis, MO). H. influenzae and H. haemolyticus are both X and V factor dependent. Isolates identified as H. influenzae or H. haemolyticus were stored in sterile skim milk at −80°C.
Serotyping.
All suspected H. influenzae isolates were analyzed by slide agglutination for capsular serotype using polyvalent H. influenzae capsular-serotyping sera (Difco, Detroit, MI). Isolates positive for the capsule were confirmed by agglutination using antisera specific for types a through f. The positive control was H. influenzae type b strain Eagan, and the negative control contained no bacteria. The results from carrier C were ambiguous, and therefore, all isolates from this carrier were further studied by PCR for the presence of capsule genes using the method described by Falla et al. (13). Strains that lacked both the type b-specific gene region and bexA, required for the export of the capsule to the cell surface, were designated nontypeable. Strains that carried the type b-specific gene region but did not have bexA were designated as b− mutants. The positive control in the PCRs was H. influenzae type b strain Eagan, and the negative control contained no DNA.
P6 protein immunodot assay.
The P6 protein of H. influenzae, a 16,000-Da outer membrane protein, demonstrates a high degree of antigenic conservation among H. influenzae strains. Monoclonal antibody 7F3 recognizes an epitope on P6 that is highly specific for H. influenzae strains, as shown by assays of several hundred H. influenzae strains from diverse geographic and clinical origins (42, 45), and this epitope is absent from the P6 of H. haemolyticus. We therefore used the 7F3 antiserum (kindly donated by Timothy Murphy of the Buffalo, NY, Veterans Administration Hospital) to characterize isolates as H. influenzae (possessing the 7F3 epitope) or H. haemolyticus (lacking the 7F3 epitope), based on the method previously described (41). Briefly, 2 μl of bacterial suspension was dotted onto a nitrocellulose membrane and allowed to dry. The membrane was incubated in BLOTTO (5% nonfat dry milk in sterile water) for 1 h at room temperature to block reactive sites. After being washed with phosphate-buffered saline, the membrane was incubated overnight at room temperature in 7F3 antibody diluted 1:1 in BLOTTO. The nitrocellulose was then washed with phosphate-buffered saline and incubated for an hour in goat anti-mouse immunoglobulin G (Sigma A-3562) diluted 1:1,000 in BLOTTO. The immunodots were developed with nitroblue tetrazolium/BCIP (5-bromo-4-chloro-3-indolylphosphate) (Pierce) color developer and visually assessed. H. influenzae type b strain Eagan served as the positive control and H. haemolyticus strain 11P31 (donated by Timothy Murphy of the Buffalo, NY, Veterans Administration Hospital) as the negative control.
DNA isolation and dot blot hybridization analysis for iga.
More than 90% of H. influenzae and less than 10% of H. haemolyticus isolates possess the iga gene, which encodes the enzyme immunoglobulin A1 protease (26). The presence of iga was detected using the dot blot hybridization method previously described (10). Briefly, a DNA probe specific for the conserved β-core region of iga was PCR amplified from H. influenzae type b strain Eagan as previously described (59) using β-core domain primers (BF1, GCAGAATTCAAAGCACAATTTGTTGCA, and BR1, TTATAACGTTAATTCAACAGGCTT) derived from the published sequence (48) of the H. influenzae HK368 iga gene (GenBank accession no. M87492). The probe was fluorescein labeled using the ECF random prime labeling system (Amersham, IL). Crude DNA was isolated from H. influenzae and H. haemolyticus lysates, and the DNA concentrations were standardized by spectrophotometry. DNA samples were then transferred onto nylon membranes using a Bio-Dot Microfiltration Apparatus (Bio-Rad, CA) which generated a fixed 8-by-12 array of DNA dots. The dot blots were hybridized to the fluorescein-labeled DNA probe under stringent conditions using the fluorescein-based ECF detection system (Amersham, IL). The signal intensity of each dot was detected using the Storm system from Molecular Dynamics (Sunnyvale, CA) and reported as a percentage of the positive control after correction for the background signal. Positive controls for the PCR analysis included amplification of 16S rRNA gene sequences (34) and pepN, a peptidase-encoding gene found in all Haemophilus strains studied (10).
Distinguishing H. influenzae and H. haemolyticus isolates.
Based on the H. influenzae characteristics described in Bergey's Manual of Systematic Bacteriology (26) and the conservation of the H. influenzae 7F3 epitope of P6 (42, 45), along with the presence of the iga gene, we developed criteria in our laboratory to distinguish H. influenzae from H. haemolyticus for the purposes of this study (Table 1). H. influenzae isolates were defined as NAD and hemin dependent, nonhemolytic on horse blood agar, and positive for both the 7F3 epitope of P6 (42, 45) and the conserved β-core domain of the iga gene (59). Isolates defined as H. haemolyticus were also NAD and hemin dependent but demonstrated clear hemolysis on horse blood agar (positive control, H. haemolyticus strain 11P31; negative control, H. influenzae type b strain Eagan), did not recognize the 7F3 epitope of P6, and lacked iga. Since H. haemolyticus isolates may lose hemolytic activity during subculture and NAD- and hemin-dependent, nonhemolytic isolates that lack the P6 epitope and iga are phylogenetically more related to H. haemolyticus (16, 39), we designated such isolates nonhemolytic H. haemolyticus. The single nonhemolytic isolate that carried the 7F3 epitope of P6 but not the iga gene was designated a variant strain.
TABLE 1.
Distinguishing H. influenzae and H. haemolyticus isolates
| Species | Porphyrin | Hemolysis | 7F3 epitope of P6 | iga |
|---|---|---|---|---|
| H. influenzae | Negative | Absent | Present | Present |
| H. haemolyticus | Negative | Present | Absent | Absent |
| Nonhemolytic H. haemolyticus | Negative | Absent | Absent | Absent |
| Variant | Negative | Absent | Present | Absent |
USS-PCR.
Uptake signal sequence (USS)-PCR is a repetitive-element genotyping method recently developed in our laboratory that takes advantage of the multiple (1,471 copies in strain Rd) copies of the 9-bp DNA segment (5′-AAGTGCGGT-3′) (54, 55) that promotes double-stranded DNA uptake by the naturally competent Haemophilus bacteria. The PCR consisted of 1 μl of chromosomal DNA, 2 μl 25 mM primer (5′-IIIAAGTGCGGT-3′) (Invitrogen), 2 μl 2.5 mM MgCl2, 2 μl each of a 10 mM concentration of the four deoxynucleoside triphosphates (Invitrogen), 0.5 μl platinum Taq polymerase (Invitrogen), and water to achieve a final volume of 50 μl. The PCR was initiated with a 2-min incubation at 94°C, followed by 30 cycles of 94°C for 30 s, annealing at 45°C for 1 min, and denaturing at 68°C for 5 min. The PCR products were separated by gel electrophoresis using 1% agarose gel for 2 h at 97 V, stained with ethidium bromide, and visualized with a UV transilluminator. Strain differences were defined by a single band difference in the band patterns.
PFGE genotyping.
Genomic DNAs from representative USS-PCR genotypes in each carrier (about one for every two isolates that were identical by USS-PCR) were digested with SmaI and tested by PFGE analysis, using techniques established in our laboratory (47, 56). Following separation of the DNA restriction fragments by gel electrophoresis, the gels were stained with ethidium bromide, visualized under UV light, and photographed for analysis. A previous study from our laboratory showed that restriction patterns from bacterial isolates using SmaI and ApaI restriction enzymes cluster identically (56). Thus, we assumed that those isolates that could not be digested with SmaI after two attempts were genotypically different from SmaI-digestible isolates. Subsequently, digestion with EagI was used to genotype non-SmaI-digestible isolates. Cluster analysis was performed separately for the PFGE band patterns generated by SmaI and EagI using BioNumerics software from Applied Math (Kortrijk, Belgium), and the patterns were confirmed by visual inspection. Band patterns were designated unique on the basis of ≥7 band differences, in accordance with the criteria of Tenover et al. (57) for determining genotypic differences between possible epidemic bacterial strains.
RESULTS
A total of 4,552 Haemophilus isolates from the throat specimens of four healthy adult carriers, collected over 7 months, were analyzed, and the results are shown in Table 2. Briefly, 3,372 (74%) isolates were NAD dependent and hemin independent (i.e., porphyrin positive), namely, H. parainfluenzae and related species, and the prevalences in the four carriers ranged from 46 to 86%. Eight hundred sixty (19%) isolates were NAD- and hemin-dependent (i.e., porphyrin-negative) species, namely, H. influenzae and H. haemolyticus, and the prevalences were 7 to 48%. Four hundred twelve isolates were tested for hemolysis, the presence of the 7F3 epitope of P6, and iga; 346 (84%) were H. influenzae isolates, 47 (11%) were H. haemolyticus, 18 (4%) were nonhemolytic H. haemolyticus, and 1 was a variant strain. Two carriers were predominantly colonized with NTHI, another predominantly with b− H. influenzae mutants, and the remaining carrier with H. haemolyticus.
TABLE 2.
Characteristics of the Haemophilus isolates from the four carriers
| Characteristic | Value in carrier:
|
Total | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| No. of isolates recovered | 1,110 | 1,170 | 1,140 | 1,132 | 4,552 |
| No. (%) porphyrin positive | 889 (80) | 543 (46) | 984 (86) | 956 (84) | 3,372 (74) |
| No. (%) X and V factor dependent | 76 (7) | 559 (48) | 98 (9) | 127 (11) | 860 (19) |
| No. encapsulated/total (%) | 0/74 | 0/542 | 45/65 (69) | 0/126 | 68/840 (8) |
| No. serotype b/total (%) | 0/74 | 0/542 | 45/65 (69) | 0/126 | 68/840 (8) |
| Hemolysis (total no. tested) | 74 | 236 | 74 | 120 | 504 |
| No. (%) positive | 5 (7) | 1 (0.42) | 3 (4) | 102 (85) | 111/504 (22) |
| No. (%) negative | 69 (93) | 235 (99.6) | 71 (96) | 18 (15) | 393/504 (78) |
| P6 epitope (total no. tested) | 74 | 237 | 70 | 118 | 499 |
| No. (%) positive | 63 (85) | 236 (99.6) | 65 (93) | 0 | 364/499 (73) |
| No. (%) negative | 11 (15) | 1 (0.42) | 5 (7) | 118 (100) | 135/499 (27) |
| iga gene (total no. tested) | 57 | 237 | 70 | 48 | 412 |
| No. (%) positive | 45 (79) | 236 (99.6) | 65 (93) | 0 | 346/412 (84) |
| No. (%) negative | 12 (21) | 1 (0.4) | 5 (7) | 48 (100) | 66/412 (16) |
| No. of H. influenzae isolates/total (%) | 45/57 (79) | 236/237 (99.6) | 65/70 (93) | 0/48 (0) | 346/412 (84.0) |
| No. of H. haemophilus isolates/total (%) | 5/57 (9) | 1/237 (0.4) | 3/70 (4) | 38/48 (79) | 47/412 (11.4) |
| No. of nonhemolytic H. haemophilus isolates/total (%) | 6/57 (10) | 0 | 2/70 (3) | 10/48 (21) | 18/412 (4.4) |
| No. of variant isolates (%) | 1/57 (2) | 0 | 0 | 0 | 1/412 (0.2) |
| No. of USS-PCR types | 11 (n = 74) | 16 (n = 541) | 5 (n = 98) | 12 (n = 125) | |
| No. of PFGE genotypes | 11 (n = 51) | 15 (n = 205) | 7 (n = 63) | 10 (n = 39) | |
USS-PCR and PFGE genotypes.
Because the USS-PCR gels exhibited relatively few bands, we chose to use the most stringent criteria for identifying unique USS genotypes, and strain differences were defined by a single band difference in the band patterns. USS-PCR genotyping was thus used as an initial screen to identify strain differences among 838 isolates suspected to be H. influenzae or H. haemolyticus from the four carriers. A subset of 468 isolates representing different USS-PCR genotypes were further analyzed using PFGE.
USS-PCR genotyping of 838 isolates revealed 11, 16, 5, and 12 distinct USS-PCR genotypes in carriers A, B, C, and D, respectively. Four hundred sixty-eight isolates from representative USS-PCR genotypes (about one for every two isolates identical by USS-PCR) were further typed by PFGE, which revealed 11, 15, 7, and 10 distinct PFGE genotypes in carriers A, B, C, and D, respectively (Table 2). One hundred ten isolates could not be digested by either the SmaI or EagI restriction enzyme and were considered to be genotypically different from the rest of the PFGE genotypes identified in each carrier. Based on ≥7 band differences between genotypes, those isolates with similarity indices of >70% clustered into single unique genotypes.
Subsequently, Pearson correlation on the USS-PCR versus PFGE genotypes was done on a subset of isolates from each carrier, and it ranged from 0.5 to 0.7, thus revealing limited correlation between the USS-PCR genotype and the PFGE genotype.
Carrier A.
Throat samples were taken on 37 days over the 7-month study, leading to culture of 1,110 isolates from carrier A. Eight hundred eighty-nine (80%) isolates were heme dependent (porphyrin positive) and therefore belonged to H. parainfluenzae and related species (26). These generally nonpathogenic organisms are oral and pharyngeal commensals (31) and were not analyzed further. Seventy-six (7%) isolates were NAD and hemin dependent and hence belonged to H. influenzae or H. haemolyticus species. Fifty-seven of these 76 isolates were tested for hemolysis, the presence of the 7F3 epitope of P6, and iga; 45 (79%) were H. influenzae, 5 (9%) were H. haemolyticus, 6 (10%) were nonhemolytic H. haemolyticus, and 1 isolate was a variant strain (Table 2). All the H. influenzae isolates were NTHI.
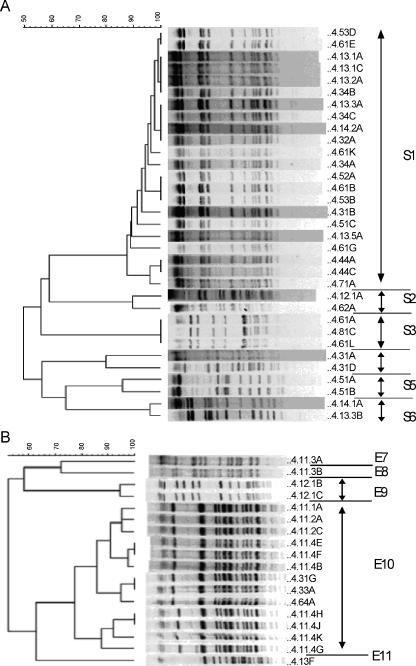
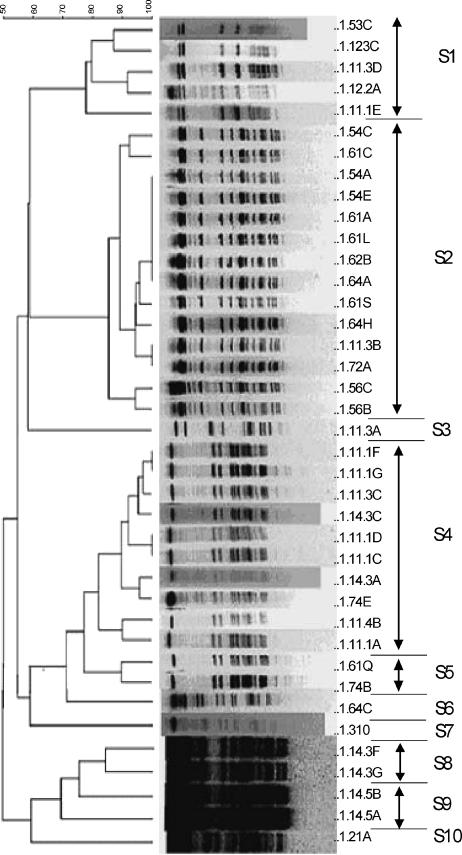
Genotyping of 74 isolates using USS-PCR revealed 11 genotypes. Fifty-one isolates from representative USS-PCR genotypes (about one for every two isolates identical by USS-PCR) were further genotyped by PFGE using the SmaI or EagI restriction enzyme. These 51 isolates consisted of 40 NTHI, 5 H. haemolyticus, and 6 nonhemolytic H. haemolyticus isolates, and they clustered into 11 PFGE genotypes, 6 based on digestion with SmaI and 5 based on lack of digestion with SmaI but successful digestion with EagI (Fig. 1 and Table 3). The results showed simultaneous colonization with both Haemophilus species and with multiple genotypes (Table 3), as well as day-to-day variation in the genotypes and species recovered. The longest duration of carriage of a unique PFGE genotype (S1) was 154 days (Table 3), and this genotype was isolated sporadically on the sampling days during this period.
FIG. 1.
Dendrogram (generated using BioNumerics software) of isolates from carrier A based on the unweighted pair-group method with arithmetic averages analysis of PFGE band patterns following digestion with SmaI (A) or EagI (B). Isolate labels are to the right of each band pattern; the first number on the left is the carrier number, followed in numerical order by the month, week, and day the sample was collected; each isolate from the same day was further designated by an uppercase letter. The PFGE genotypes shown are S1 to S6 in panel A and E7 to E11 in panel B.
TABLE 3.
Distribution of unique PFGE genotypes as defined by digestion with SmaI (S1 to S6) or EagI (E7 to E11) restriction enzyme over time in carrier A
| Day | No. of isolates identified with genotype:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S1a | S2 | S3 | S4 | S5 | S6 | E7b | E8 | E9 | E10 | E11 | |
| 0 | 1c | ||||||||||
| 1 | 2c | ||||||||||
| 2 | 1e | 1e | |||||||||
| 3 | 7c | ||||||||||
| 8 | 1e | 2c | |||||||||
| 14 | 2c | ||||||||||
| 15 | 1c | ||||||||||
| 16 | 1c | 1c | |||||||||
| 17 | 1c | ||||||||||
| 22 | 1c | ||||||||||
| 23 | 1c | ||||||||||
| 60 | 1c | 2d | 1c | 1d | |||||||
| 64 | 1c | ||||||||||
| 71 | 1c | ||||||||||
| 81 | 3c | ||||||||||
| 108 | 2c | ||||||||||
| 116 | 1c | 2d | |||||||||
| 122 | 1c | ||||||||||
| 128 | 2c | ||||||||||
| 141 | 4c | 1c + 2e | |||||||||
| 150 | 1e | ||||||||||
| 161 | 1c | ||||||||||
S genotypes based on digestion of genomic DNA with SmaI.
E genotypes based on failure to be digested with SmaI and successful digestion with EagI.
NTHI.
H. haemolyticus.
Nonhemolytic H. haemolyticus.
One sample (day 141) from carrier A demonstrated three isolates exhibiting the S3 genotype (Table 3); two of these isolates were nonhemolytic H. haemolyticus, and one was H. influenzae. This was confirmed by repeating the PFGE and other tests multiple times on the DNA extracted from the original stocks of these isolates.
In summary, carrier A was colonized predominantly with NTHI, had 11 unique PFGE genotypes, was cocolonized with more than one genotype and more than one species with considerable sample-to-sample variation, and was colonized for 154 days with a single genotype.
Carrier B.
Throat samples were taken on 39 days over the 7-month study, leading to culture of 1,170 isolates from carrier B. Five hundred forty-three (46%) isolates were heme dependent (porphyrin positive) and therefore belonged to H. parainfluenzae and related species (26) and were not analyzed further. Five hundred fifty-nine (48%) isolates were NAD and hemin dependent and hence belonged to H. influenzae or H. haemolyticus species. Two hundred thirty-seven isolates were tested for hemolysis, the presence of the 7F3 epitope of P6, and iga; 236 (99.6%) were H. influenzae, 1 (0.4%) was H. haemolyticus, and none were nonhemolytic H. haemolyticus or variant strains (Table 2). All the H. influenzae isolates were NTHI.
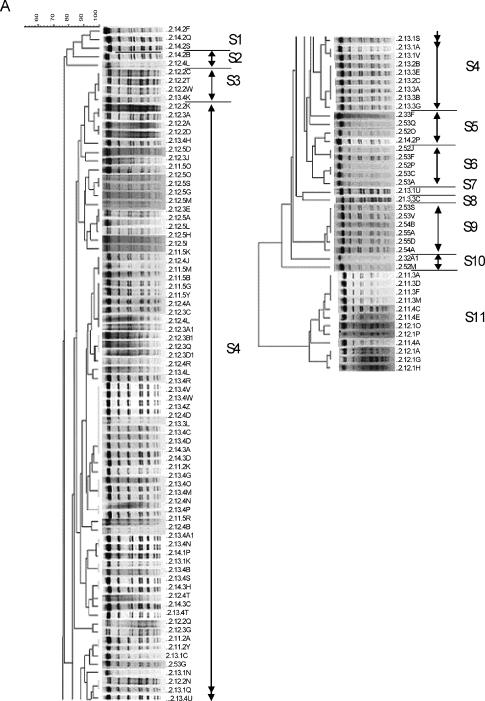
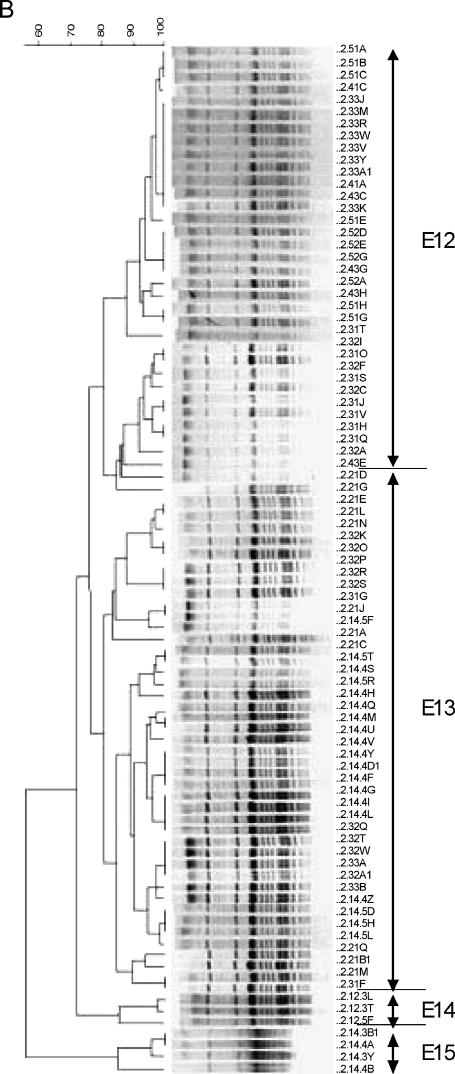
Genotyping using USS-PCR of 540 NTHI isolates and 1 H. haemolyticus isolate revealed 16 genotypes, and 259 NTHI isolates from representative USS-PCR genotypes (about 1 for every 2 isolates identical by USS-PCR) were further genotyped by PFGE using the SmaI or EagI restriction enzyme. Fifty-four (21%) isolates could not be digested by either restriction enzyme after repeated attempts and thus were considered genotypically different from the digestible strains. The remaining 205 NTHI isolates clustered into 15 unique PFGE genotypes (Fig. 2 and Table 4). The results showed simultaneous colonization with multiple genotypes (Table 4), as well as day-to-day variation in the genotypes recovered. The longest duration of carriage of a single unique PFGE genotype (S4) was 129 days, and this genotype was isolated sporadically on the sampling days during this period, with a 106-day interval between the last two isolations.
FIG. 2.
Dendrogram (generated using BioNumerics software) of carrier B isolates digested with SmaI (A) and EagI (B). See the legend to Fig. 1 for an explanation of the isolate labels.
TABLE 4.
Distribution of unique PFGE genotypes as defined by digestion with SmaI (genotypes S1 to S11) or EagI (genotypes E12 to E15) restriction enzyme over time in carrier B
| Day | No. of isolates identified with genotypea:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1b | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | E12c | E13 | E14 | E15 | |
| 1 | 3 | ||||||||||||||
| 2 | 4 | ||||||||||||||
| 3 | 3 | ||||||||||||||
| 4 | 6 | ||||||||||||||
| 7 | 5 | ||||||||||||||
| 8 | 3 | 5 | |||||||||||||
| 9 | 9 | 2 | |||||||||||||
| 10 | 1 | 7 | |||||||||||||
| 11 | 10 | 1 | |||||||||||||
| 15 | 8 | 1 | |||||||||||||
| 16 | 2 | ||||||||||||||
| 17 | 5 | 1 | |||||||||||||
| 18 | 1 | 18 | |||||||||||||
| 21 | 1 | ||||||||||||||
| 22 | 3 | 1 | 1 | ||||||||||||
| 23 | 5 | 2 | |||||||||||||
| 24 | 13 | 2 | |||||||||||||
| 25 | 6 | ||||||||||||||
| 28 | 11 | ||||||||||||||
| 59 | 7 | 2 | |||||||||||||
| 67 | 4 | 9 | |||||||||||||
| 72 | 1 | 8 | 2 | ||||||||||||
| 88 | 2 | ||||||||||||||
| 102 | 4 | ||||||||||||||
| 114 | 6 | ||||||||||||||
| 123 | 1 | 2 | 1 | 4 | |||||||||||
| 129 | 1 | 1 | 3 | 2 | 1 | ||||||||||
| 135 | 2 | ||||||||||||||
| 143 | 2 | ||||||||||||||
All isolates were confirmed to be NTHI.
S genotypes based on digestion of genomic DNA with SmaI.
E genotypes based on failure to be digested with SmaI and successful digestion with EagI.
In summary, carrier B was colonized almost exclusively with NTHI, had 15 unique PFGE genotypes, was cocolonized with more than one genotype on 15 sampling days, and was colonized for 129 days with a single genotype.
Carrier C.
Throat samples were taken on 38 days over the 7-month study, leading to culture of 1,140 isolates from carrier C. Nine hundred eighty-four (86%) isolates were heme dependent (porphyrin positive) and therefore belonged to H. parainfluenzae and related species (26) and were not analyzed further. Ninety-eight (9%) isolates were NAD and hemin dependent and hence belonged to H. influenzae or H. haemolyticus species. Seventy isolates were tested for hemolysis, the presence of the 7F3 epitope of P6, and iga; 65 (93%) were H. influenzae, 3 (4%) H. haemolyticus, and 2 (3%) nonhemolytic H. haemolyticus (Table 2).
Serotyping by slide agglutination revealed mild clumping, so all H. influenzae isolates were serotyped using PCR. Forty-five (69%) isolates were found to be b− mutants. The remaining 20 (31%) isolates failed to generate a PCR product for both bexA and the type-specific region of the cap genes and were thus considered NTHI.
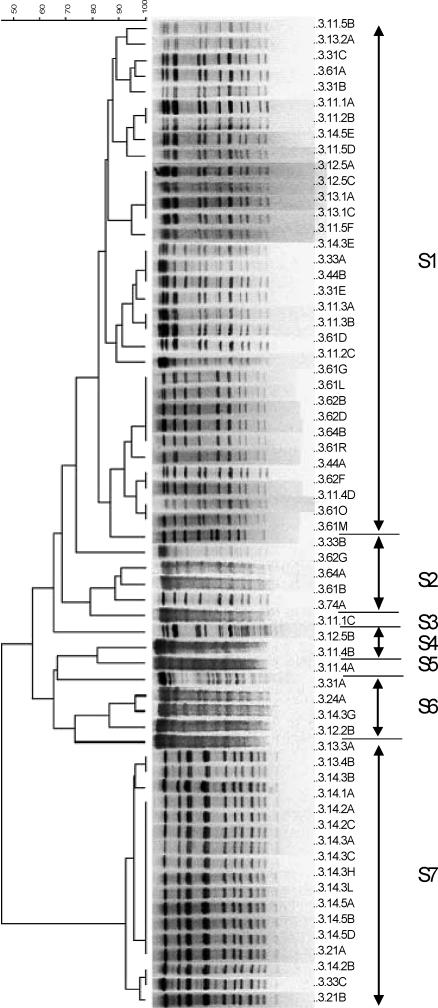
Genotyping of the 98 isolates using USS-PCR revealed five genotypes, and 71 isolates from representative USS-PCR genotypes (about 1 for every 2 isolates identical by USS-PCR) were further genotyped by PFGE using the SmaI or EagI restriction enzyme. These 71 isolates consisted of 66 H. influenzae, 3 H. haemolyticus, and 2 nonhemolytic H. haemolyticus isolates. Eight isolates (four H. influenzae, three H. haemolyticus, and one nonhemolytic H. haemolyticus) could not be digested by either restriction enzyme on repeated attempts and were thus considered to be genotypically different from the digestible strains. The remaining 63 isolates (43 b− H. influenzae mutants, 19 NTHI, and 1 nonhemolytic H. haemolyticus) clustered into seven unique PFGE genotypes (Fig. 3 and Table 5). The results showed simultaneous colonization with both Haemophilus species and with multiple genotypes (Table 5), as well as day-to-day variation in the genotypes recovered. Interestingly, genotype S6 contained both NTHI and b− mutants (Table 5). The longest duration of carriage of a single unique PFGE genotype (S1) was 173 days, and this genotype was isolated sporadically on the sampling days during this period.
FIG. 3.
Dendrogram (generated using BioNumerics software) of carrier C isolates digested with SmaI. See the legend to Fig. 1 for an explanation of the isolate labels.
TABLE 5.
Distribution of unique PFGE genotypes as defined by digestion with SmaI (S1 to S7) restriction enzyme over time in carrier C
| Day | No. of isolates identified with genotype:
|
||||||
|---|---|---|---|---|---|---|---|
| S1a | S2 | S3 | S4 | S5 | S6 | S7 | |
| 0 | 1b | 1b | |||||
| 1 | 2b | ||||||
| 2 | 2b | ||||||
| 3 | 1b | 1b | 1d | ||||
| 4 | 3b | ||||||
| 8 | 1c | ||||||
| 11 | 2b | 1b | |||||
| 15 | 2b | ||||||
| 16 | 1b | . | |||||
| 17 | 1c | ||||||
| 18 | 1c | ||||||
| 21 | 1c | ||||||
| 22 | 3c | ||||||
| 23 | 1b | 1b | 5c | ||||
| 25 | 1b | 3c | |||||
| 31 | 2c | ||||||
| 51 | 1c | ||||||
| 57 | 3b | 1b | |||||
| 71 | 1b | 1b | 1c | ||||
| 106 | 2b | ||||||
| 148 | 7b | 1b | |||||
| 155 | 3b | 1b | |||||
| 172 | 1b | 1b | |||||
| 197 | 1b | ||||||
S genotypes based on digestion of genomic DNA with SmaI.
Capsule-deficient type b mutant.
NTHI.
Nonhemolytic H. haemolyticus.
In summary, carrier C was colonized primarily with b− H. influenzae mutants, as well as NTHI; had seven unique PFGE genotypes; was cocolonized with more than one genotype; and was colonized for 173 days with a single genotype.
Carrier D.
Throat samples were taken on 38 days over the 7-month study, leading to culture of 1,132 isolates from carrier D. Nine hundred fifty-six (84%) isolates were heme dependent (porphyrin positive) and therefore belonged to H. parainfluenzae and related species (26) and were not analyzed further. One hundred twenty-seven (11%) isolates were NAD and hemin dependent and hence belonged to H. influenzae or H. haemolyticus species. Forty-eight isolates were tested for hemolysis, the presence of the 7F3 epitope of P6, and iga; none were H. influenzae, 38 (79%) were H. haemolyticus, and 10 (21%) were nonhemolytic H. haemolyticus, with no variant strain (Table 2).
Genotyping using USS-PCR of 125 isolates revealed 12 genotypes, and 87 isolates from representative USS PCR genotypes (about 1 for every 2 isolates identical by USS-PCR) were further genotyped by PFGE using the SmaI or EagI restriction enzyme. Forty-eight (55%) isolates could not be digested by either restriction enzyme after repeated attempts and thus were considered different from the digestible strains. The remaining 39 isolates (35 H. haemolyticus and 4 nonhemolytic H. haemolyticus) were all digested with SmaI and clustered into 10 unique PFGE genotypes (Fig. 4 and Table 6). The results showed simultaneous colonization with multiple genotypes (Table 6), as well as day-to-day variation in the genotypes recovered. The longest duration of carriage of a single unique PFGE genotype (S4) was 198 days, and this genotype was isolated sporadically on the sampling days during this period, with 174 days between the last two isolations.
FIG. 4.
Dendrogram (generated using BioNumerics software) of carrier D isolates digested with SmaI. See the legend to Fig. 1 for an explanation of the isolate labels.
TABLE 6.
Distribution of unique pulsed field gel electrophoresis (PFGE) genotypes as defined by digestion with SmaI (S1 to S10) restriction enzyme over time in Carrier D
| Day | No. of isolates identified with genotype:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S1a | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | |
| 0 | 1c | 5b | ||||||||
| 2 | 1b | 1b | 1b | 1b | ||||||
| 3 | 1b | |||||||||
| 8 | 1c | |||||||||
| 9 | 1c | |||||||||
| 23 | 2b | 2b | ||||||||
| 25 | 2b | |||||||||
| 28 | 1b | |||||||||
| 56 | 1b | |||||||||
| 129 | 1c | |||||||||
| 137 | 3b | |||||||||
| 144 | 2b | |||||||||
| 148 | 4b | 1b | ||||||||
| 157 | 1b | |||||||||
| 172 | 2b | 1b | ||||||||
| 185 | 1b | |||||||||
| 197 | 1b | 1b | ||||||||
S genotypes based on digestion of genomic DNA with SmaI.
H. haemolyticus.
Nonhemolytic H. haemolyticus.
In summary, carrier D was colonized predominantly with H. haemolyticus, had 10 unique PFGE genotypes, was cocolonized with more than one genotype, and was colonized for 198 days with a single genotype.
DISCUSSION
Pharyngeal colonization is the initial step in the sequence of events leading to H. influenzae infection and disease. Thus, understanding the dynamics of H. influenzae colonization forms the foundation for understanding the natural history of diseases caused by this bacterium. While previous studies have investigated H. influenzae pharyngeal colonization among healthy adults (4) and among individuals at risk for H. influenzae infections, such as children attending day care centers (8, 14, 56), patients with cystic fibrosis (49), and adults with COPD (38, 43), no previous reports have documented colonization with H. haemolyticus in population-based studies of healthy adults. Pharyngeal colonization with nonhemolytic H. haemolyticus has only recently been described (16, 39). The results of the present study describe the dynamics of H. influenzae and H. haemolyticus colonization over a 7-month period studied by culturing throat specimens from four healthy adult carriers at frequent intervals.
Multiple H. influenzae genotypes are frequently present in the upper respiratory tracts of adults with cystic fibrosis (49) and COPD (43) and children in day care centers (14, 56). In our study, cocolonization with two to five H. influenzae or H. haemolyticus strains, as defined by PFGE genotypes, was seen on 37 out of 132 (28%) sampling days. Furthermore, from 7 to 15 genotypes of H. influenzae and H. haemolyticus were cultured from the four carriers over the course of 7 months. Likely explanations for the high yield of positive cultures and for the genetic diversity seen in each individual include the use of selective agar containing bacitracin, which significantly reduces contamination with non-Haemophilus species and facilitates isolation of Haemophilus; vigorous sampling of the throat, which yields H. influenzae isolation rates equal to or greater than sampling the nasopharynx in adults (19, 33); and analysis of multiple isolates with morphology suggestive of Haemophilus species from each sample.
USS-PCR genotyping (a technique that is more rapid and resource conserving than PFGE) was used as an initial screen to identify strains showing identical genotypes that would not require further genotyping. The USS-PCR genotyping procedure resulted in relatively few bands compared to PFGE and thus may be less sensitive in identifying unique genotypes. Our results subsequently showed limited correlation between USS-PCR genotyping and PFGE genotyping, which could be expected, as the two procedures assess very different characteristics of the bacteria (patterns generated by PCR amplification products of DNA fragments flanked by H. influenzae uptake signal sequences and patterns generated by DNA fragments flanked by restriction enzyme sites, respectively). The lack of good correlation between these two techniques underscores the genetic variability between strains and the many mechanisms of genetic variability. Thus, the results reported in this study represent the minimum genetic diversity among strains. If we had performed PFGE on all 838 isolates cultured from the four subjects, we anticipate we would have documented even more diversity.
We chose to employ the criteria of Tenover et al. (57) to define unique genotypes in the PFGE analysis. These criteria were developed for outbreak investigation to identify the genetic relatedness of epidemiologically clustered strains, with the assumption that bacterial isolates from individual disease outbreaks have undergone few evolutionary changes. These criteria stated that strains whose band patterns differed by seven or more restriction fragments exhibited at least three genetic differences between them and were considered genetically unrelated; strains with zero band differences were considered to be genetically identical, strains with two or three band differences (one genetic difference) were considered closely related, and strains with four to six band differences (two genetic differences) were considered possibly related. While repeated throat samples from the same individual could be viewed as representing a closed epidemiologic unit, our data support the concept that pharyngeal colonization is a dynamic process during which bacterial evolution may occur.
Examination of the dendrograms in Fig. 1, 2, 3, and 4 reveals that some isolates from a single individual collected within a few weeks were clearly different genotypes while others were closely related but exhibited several band differences. This observation is consistent with a population of organisms in the pharynx in which both acquisition of new strains and evolution of existing strains was occurring, as has been described for Helicobacter pylori strains isolated several years apart from single individuals (25). These results are consistent with the recent suggestion that H. influenzae exists in the pharynx in the form of a biofilm (17, 40, 60), a structured community of bacterial cells enveloped in a polymeric matrix and adherent to an inert or living surface (6, 7). We anticipate that some of the day-to-day and week-to-week variability of the genotypes and the skips in persistence of unique strains (as also described by Murphy et al. [38]) resulted from sampling bias in that we obtained only a few colonies from a huge, and somewhat diverse, population of bacteria.
H. influenzae is a naturally competent organism and possesses approximately 1,400 copies (54) of the 9-bp uptake signal sequence that allows genetic exchange by transformation and homologous recombination. Intraspecies (28, 29) and interspecies (30) gene transfer has been demonstrated in H. influenzae. High rates of recombination have also been demonstrated in otitis media isolates of NTHI (5), and hypermutability mutants have been associated with high genetic variability among H. influenzae isolates from the airways of patients with cystic fibrosis (49). Thus, H. influenzae is well equipped to survive in its natural environment, the human pharynx, by generating genetic mutants or variants that exhibit various fitness characteristics.
H. haemolyticus, closely related to H. influenzae, is found in the pharynges of healthy humans and often in the bacterial deposits on teeth below the gingival margin. In our study, H. haemolyticus was isolated from carrier D throughout the study period of 7 months and clustered into 10 PFGE genotypes, 9 of which contained only hemolytic isolates while genotype S1 contained one hemolytic and four nonhemolytic isolates. Conceivably, these five isolates represent one classical H. haemolyticus and four H. haemolyticus mutants with either nonfunctional mutations in the hemolysin gene or loss of the gene itself. Further clarification of these strains awaits analysis of their hemolysin genes, which have not yet been defined in H. haemolyticus. Carrier A carried seven genotypes of H. haemolyticus; three genotypes included only nonhemolytic isolates, and three included only hemolytic isolates. On the other hand, genotype S3 from this carrier included two nonhemolytic H. haemolyticus isolates and one H. influenzae isolate. If these three S3-type strains truly represent variants of the same clone, either a parent H. influenzae clone lost both the 7F3 epitope of P6 and iga or a parent nonhemolytic H. haemolyticus gained both, which is less likely. The taxonomy of bacterial strains attempts to group isolates exhibiting identical phenotypic or genotypic features. The failure of a few strains to fit a strict definition of H. influenzae or H. haemolyticus is expected, as bacteria, because of random evolution of individual genes (5, 36), form a continuum of strain characteristics rather than a dichotomy. This blurring of the intersection of two species is similar to that described with Neisseria strains (21).
This study is, to our knowledge, the first to describe the dynamics of H. haemolyticus colonization and to characterize its diversity. Although H. haemolyticus is not a common human pathogen (1, 31), its presence on the respiratory mucosa may serve as a source of genetic material for horizontal gene transfer to H. influenzae and thus enhance the pool of genes available to H. influenzae in sustaining the genetic diversity that fosters its existence in the antibody-laden milieu of the human pharynx.
H. influenzae expressing the type b capsule is found in the pharynx less frequently than nontypeable strains but is responsible for the majority of the cases of H. influenzae invasive infections in nonimmune individuals. Furthermore, type b strains are more clonal than NTHI (44, 53). Capsule-deficient, or b−, H. influenzae mutants carry the b-type cap region but do not have the bexA gene required for exportation of the capsule to the cell surface (13, 24). To our knowledge, long-term colonization with b− mutants has not been documented, although b− mutants have been isolated from the throats of patients with type b infections (23). In our study, carrier C was colonized with b− H. influenzae strains of five different genotypes, one of which (S6) also contained two isolates of NTHI. This observation is difficult to explain; the apparent NTHI strains, which demonstrated absence of cap region genes by PCR on repeated testing, may be Hib variants with complete deletion of the cap gene region and thus could not be distinguished from NTHI by our techniques. Alternatively, this observation may represent a limitation of PFGE in identifying unique clonal differences among strains. In addition, this carrier may have been colonized with the b− mutant over enough time for significant evolutionary events to occur that altered the restriction enzyme genes upon which our typing system depends, leading to unique genotypes.
This study has demonstrated immense genetic diversity of H. influenzae and H. haemolyticus isolated from each of four individuals over 7 months. These results may reflect the extensive recombination and DNA uptake known to occur in H. influenzae and are also consistent with bacterial growth in multimicrobial biofilms (17, 38, 40, 60). Identifying host characteristics that increase the risk of colonization with pathogenic strains, discovering the virulence factors in H. influenzae that cause it to establish infection, and knowledge of environments that are conducive to H. influenzae disease will further our understanding of the pathogenesis of H. influenzae disease and will facilitate the development of preventive strategies for diseases caused by these organisms.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Albritton, W. L., J. L. Brunton, M. Meier, M. N. Bowman, and L. A. Slaney. 1982. Haemophilus influenzae: comparison of respiratory tract isolates with genitourinary tract isolates. J. Clin. Microbiol. 16:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aniansson, G., B. Alm, B. Andersson, P. Larsson, O. Nylen, H. Peterson, P. Rigner, M. Svanborg, and C. Svanborg. 1992. Nasopharyngeal colonization during the first year of life. J. Infect. Dis. 165(Suppl. 1):S38-S42. [DOI] [PubMed] [Google Scholar]
- 3.Cazzola, M., R. Ariano, V. Gioia, V. Mancini, R. Rimoldi, G. Scala, S. Scoccia, and G. Girbino. 1990. Bacterial isolates and cigarette smoking in patients with chronic bronchitis: results from an Italian multicenter survey. Clin. Ther. 12:105-117. [PubMed] [Google Scholar]
- 4.Chi, D. H., J. O. Hendley, P. French, P. Arango, F. G. Hayden, and B. Winther. 2003. Nasopharyngeal reservoir of bacterial otitis media and sinusitis pathogens in adults during wellness and viral respiratory illness. Am. J. Rhinol. 17:209-214. [PubMed] [Google Scholar]
- 5.Cody, A. J., D. Field, E. J. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. W. Hood, and E. R. Moxon. 2003. High rates of recombination on otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 3:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dabernat, H., M. A. Plisson-Saune, C. Delmas, M. Seguy, G. Faucon, R. Pelissier, H. Carsenti, C. Pradier, M. Roussel-Delvallez, J. Leroy, M. J. Dupont, F. De Bels, and P. Dellamonica. 2003. Haemophilus influenzae carriage in children attending French day care centers: a molecular epidemiological study. J. Clin. Microbiol. 41:1664-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhooge, I., M. Vaneechoutte, G. Claeys, G. Verschraegen, and P. Van Cauwengerge. 2000. Turnover of Haemophilus influenzae isolates in otitis-prone children. Int. J. Pediatr. Otorhinol. 54:7-12. [DOI] [PubMed] [Google Scholar]
- 10.Ecevit, I. Z., K. W. McCrea, M. M. Pettigrew, A. Sen, C. F. Marrs, and J. R. Gilsdorf. 2004. Prevalence of the hifBC, hmw1A, hmw2A, hmwC, and hia genes in Haemophilus influenzae isolates. J. Clin. Microbiol. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eskola, J., and T. Kilpi. 2000. Potential of bacterial vaccines in the prevention of acute otitis media. Pediatr. Infect. Dis. J. 19:72-80. [DOI] [PubMed] [Google Scholar]
- 12.Faden, H., L. Duffy, A. Williams, D. A. Krystofik, and J. Wolf. 1995. Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first 2 years of life. J. Infect. Dis. 172:132-135. [DOI] [PubMed] [Google Scholar]
- 13.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. McCoy, C. F. Marrs, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 15.Fontanals, D., R. Bou, I. Pons, I. Sanfeliu, A. Dominguez, V. Pineda, J. Renau, C. Munoz, C. Latorre, and F. Sanches. 2000. Prevalence of Haemophilus influenzae carriers in the Catalan preschool population. Eur. J. Clin. Microbiol. Infect. Dis. 19:301-304. [DOI] [PubMed] [Google Scholar]
- 16.Fung, W. W., C. A. O'Dwyer, S. Sinha, A. L. Brauer, T. F. Murphy, J. S. Kroll, and P. R. Langford. 2006. Presence of copper- and zinc-containing superoxide dismutase in commensal Haemophilus haemolyticus isolates can be used as a marker to discriminate them from nontypeable H. influenzae isolates. J. Clin. Microbiol. 44:4222-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallaher, T. K., S. Wu, P. Webster, and R. Aguilera. 2006. Identification of biofilm proteins in non-typeable Haemophilus influenzae. BMC Microbiol. 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gratten, M., J. Montgomery, G. Gerega, H. Gratten, H. Siwi, A. Poli, and G. Koki. 1989. Multiple colonization of the upper respiratory tract of Papua New Guinea children with Haemophilus influenzae and Streptococcus pneumoniae. Southeast Asian J. Trop. Med. Public Health 20:501-509. [PubMed] [Google Scholar]
- 19.Greenberg, D., A. Broides, I. Blancovich, N. Peled, N. Givon-Lavi, and R. Dagan. 2004. Relative importance of nasopharyngeal versus oropharyngeal sampling for isolation of Streptococcus pneumoniae and Haemophilus influenzae from healthy and sick individuals varies with age. J. Clin. Microbiol. 42:4604-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groeneveld, K., L. van Alphen, P. P. Eijk, G. Visschers, H. M. Jansen, and H. C. Zanen. 1990. Endogenous and exogenous reinfections by Haemophilus influenzae in patients with chronic obstructive pulmonary disease: the effect of antibiotic treatment on persistence. J. Infect. Dis. 161:512-517. [DOI] [PubMed] [Google Scholar]
- 21.Hanage, W. P., C. Fraser, and B. G. Spratt. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harabuchi, Y., H. Faden, N. Yamanaka, L. Duffy, J. Wolf, and D. Krystofik. 1994. Nasopharyngeal colonization with nontypeable Haemophilus influenzae and recurrent otitis media. J. Infect. Dis. 170:862-866. [DOI] [PubMed] [Google Scholar]
- 23.Hoiseth, S. K., and J. R. Gilsdorf. 1988. The relationship between type b and nontypable Haemophilus influenzae isolated from the same patient. J. Infect. Dis. 158:643-645. [DOI] [PubMed] [Google Scholar]
- 24.Hoiseth, S. K., E. R. Moxon, and R. P. Silver. 1986. Genes involved in Haemophilus influenzae type b capsule expression are part of an 18-kilobase tandem duplication. Proc. Natl. Acad. Sci. USA 83:1106-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilian, M. 2005. Genus III. Haemophilus Winslow, Broadhurst, Buchanan, Krumwiede, Rogers and Smith 1917, 561AL, p. 883-904. In G. M. Garrity et al. (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer-Verlag, New York, NY.
- 27.Kilian, M. 1991. Haemophilus, p. 463-470. In A. Balows, K. L. Herrmann, H. D. Isenberg, and J. J. Shadomy (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, DC.
- 28.Kroll, J. S., and E. R. Moxon. 1990. Capsulation in distantly related strains of Haemophilus influenzae type b: genetic drift and gene transfer at the capsulation locus. J. Bacteriol. 172:1374-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroll, J. S., E. R. Moxon, and B. M. Loynds. 1994. Natural genetic transfer of a putative virulence-enhancing mutation to Haemophilus influenzae type a. J. Infect. Dis. 169:676-679. [DOI] [PubMed] [Google Scholar]
- 30.Kroll, J. S., K. E. Wilks, J. L. Farrant, and P. R. Langford. 1998. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA 95:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuklinska, D., and M. Kilian. 1984. Relative proportions of Haemophilus species in the throat of healthy children and adults. Eur. J. Clin. Microbiol. 3:249-252. [DOI] [PubMed] [Google Scholar]
- 32.Leach, A. J., J. B. Boswell, V. Asche, T. G. Nienhuys, and J. D. Mathews. 1994. Bacterial colonization of the nasopharynx predicts very early onset and persistence of otitis media in Australian aboriginal infants. Pediatr. Infect. Dis. J. 13:983-989. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman, D., E. Shleyfer, H. Castel, A. Terry, I. Harman-Boehm, J. Delgado, N. Peled, and D. Lieberman. 2006. Nasopharyngeal versus oropharyngeal sampling for isolation of potential respiratory pathogens in adults. J. Clin. Microbiol. 44:525-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, J. J., C. L. Perng, S. Y. Lee, and C. C. Wan. 2000. Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J. Clin. Microbiol. 38:2076-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marrs, C. F., G. P. Krasan, K. W. McCrea, D. L. Clemans, and J. R. Gilsdorf. 2001. Haemophilus influenzae—human specific bacteria. Front. Biosci. 6:e41-e60. [DOI] [PubMed] [Google Scholar]
- 36.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moller, L. V., A. G. Regelink, H. Grasselier, J. E. Dankert-Roelse, J. Dankert, and L. van Alphen. 1995. Multiple Haemophilus influenzae strains and strain variants coexist in the respiratory tract of patients with cystic fibrosis. J. Infect. Dis. 172:1388-1392. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, T. F., A. L. Brauer, A. T. Schiffmacher, and S. Sethi. 2004. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 170:266-272. [DOI] [PubMed] [Google Scholar]
- 39.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81-89. [DOI] [PubMed] [Google Scholar]
- 40.Murphy, T. F., and C. Kirkham. 2002. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, T. F., C. Kirkham, and D. J. Sikkema. 1992. Neonatal, urogenital isolates of biotype 4 nontypeable Haemophilus influenzae express a variant P6 outer membrane protein molecule. Infect. Immun. 60:2016-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy, T. F., M. B. Nelson, K. C. Dudas, J. M. Mylotte, and M. A. Apicella. 1985. Identification of a specific epitope of Haemophilus influenzae on a 16,600-dalton outer membrane protein. J. Infect. Dis. 152:1300-1307. [DOI] [PubMed] [Google Scholar]
- 43.Murphy, T. F., S. Sethi, K. L. Klingman, A. B. Brueggemann, and G. V. Doern. 1999. Simultaneous respiratory tract colonization by multiple strains of nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J. Infect. Dis. 180:404-409. [DOI] [PubMed] [Google Scholar]
- 44.Musser, J. M., J. S. Kroll, E. R. Moxon, and R. K. Selander. 1988. Clonal population structure of encapsulated Haemophilus influenzae. Infect. Immun. 56:1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson, M. B., R. S. Munson, Jr., M. A. Apicella, D. J. Sikkema, J. P. Molleston, and T. F. Murphy. 1991. Molecular conservation of the P6 outer membrane protein among strains of Haemophilus influenzae: analysis of antigenic determinants, gene sequences, and restriction fragment length polymorphisms. Infect. Immun. 59:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel, I. S., T. A. Seemungal, M. Wilks, S. J. Lloyd-Owen, G. C. Donaldson, and J. A. Wedzicha. 2002. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax 57:759-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettigrew, M. M., B. Foxman, Z. Ecevit, C. F. Marrs, and J. Gilsdorf. 2002. Use of pulsed-field gel electrophoresis, enterobacterial repetitive intergenic consensus typing, and automated ribotyping to assess genomic variability among strains of nontypeable Haemophilus influenzae. J. Clin. Microbiol. 40:660-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulsen, K., J. Reinholdt, and M. Kilian. 1992. A comparative genetic study of serologically distinct Haemophilus influenzae type 1 immunoglobulin A1 proteases. J. Bacteriol. 174:2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roman, F., R. Canton, M. Perez-Vazquez, F. Baquero, and J. Campos. 2004. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 42:1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuelson, A., A. Freijd, J. Jonasson, and A. A. Lindberg. 1995. Turnover of nonencapsulated Haemophilus influenzae in the nasopharynges of otitis-prone children. J. Clin. Microbiol. 33:2027-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 52.Smith-Vaughan, H. C., A. J. Leach, T. M. Shelby-James, K. Kemp, D. J. Kemp, and J. D. Mathews. 1996. Carriage of multiple ribotypes of non-encapsulated Haemophilus influenzae in aboriginal infants with otitis media. Epidemiol. Infect. 116:177-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith-Vaughan, H. C., K. S. Sriprakash, A. J. Leach, J. D. Mathews, and D. J. Kemp. 1998. Low genetic diversity of Haemophilus influenzae type b compared to nonencapsulated H. influenzae in a population in which H. influenzae is highly endemic. Infect. Immun. 66:3403-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, H. O., M. L. Gwinn, and S. L. Salzberg. 1999. DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol. 150:603-616. [DOI] [PubMed] [Google Scholar]
- 55.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 56.St. Sauver, J., C. F. Marrs, B. Foxman, P. Somsel, R. Madera, and J. R. Gilsdorf. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg. Infect. Dis. 6:622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tenover, F. C., R. D. Arbeit, R. V. Goering, A. P. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trottier, S., K. Stenberg, and C. Svanborg-Eden. 1989. Turnover of nontypable Haemophilus influenzae in the nasopharynges of healthy children. J. Clin. Microbiol. 27:2175-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vitovski, S., K. T. Dunkin, A. J. Howard, and J. R. Sayers. 2002. Nontypeable Haemophilus influenzae in carriage and disease. JAMA 287:1699-1705. [DOI] [PubMed] [Google Scholar]
- 60.Webster, P., S. Wu, G. Gomez, M. Apicella, A. G. Plaut, and J. W. St Geme III. 2006. Distribution of bacterial proteins in biofilms formed by non-typeable Haemophilus influenzae. J. Histochem. Cytochem. 54:829-842. [DOI] [PubMed] [Google Scholar]