Abstract
Extraintestinal pathogenic Escherichia coli (ExPEC) strains of human and avian origin show similarities that suggest that the avian strains potentially have zoonotic properties. However, the phylogenetic relationships between avian and human ExPEC strains are poorly documented, so this possibility is difficult to assess. We used PCR-based phylotyping and multilocus sequence typing (MLST) to determine the phylogenetic relationships between 39 avian pathogenic E. coli (APEC) strains of serogroups O1, O2, O18, and O78 and 51 human ExPEC strains. We also compared the virulence genotype and pathogenicity for chickens of APEC strains and human ExPEC strains. Twenty-eight of the 30 APEC strains of serogroups O1, O2, and O18 were classified by MLST into the same subcluster (B2-1) of phylogenetic group B2, whereas the 9 APEC strains of serogroup O78 were in phylogenetic groups D (3 strains) and B1 (6 strains). Human ExPEC strains were closely related to APEC strains in each of these three subclusters. The 28 avian and 25 human strains belonging to phylogenetic subcluster B2-1 all expressed the K1 antigen and presented no significant differences concerning the presence of other virulence factors. Moreover, human strains of this phylogenetic subcluster were highly virulent for chicks, so no host specificity was identified. Thus, APEC strains of serotypes O1:K1, O2:K1, and O18:K1 belong to the same highly pathogenic clonal group as human E. coli strains of the same serotypes isolated from cases of neonatal meningitis, urinary tract infections, and septicemia. These APEC strains constitute a potential zoonotic risk.
A wide range of extraintestinal infections in humans and vertebrate animals are caused by Escherichia coli strains that belong to the large group of extraintestinal pathogenic E. coli (ExPEC) as defined by Russo and Johnson (50). These strains are frequently categorized into pathotypes according to the host's clinical symptoms. The pathotypes include uropathogenic E. coli (UPEC), isolated from humans and animals with urinary tract infections; neonatal-meningitis E. coli (NMEC), isolated from human neonates; septicemic E. coli, isolated from humans and animals with septicemia cases of various origins; and avian pathogenic E. coli (APEC), responsible for colibacillosis in poultry. The most frequent form of avian colibacillosis is a systemic infection that starts in the respiratory tracts of chickens and turkey chicks. It is characterized by fibrinous lesions of internal organs (airsacculitis, pericarditis, perihepatitis) associated with septicemia (4, 15).
Phylogenetic studies based on the E. coli reference collection (ECOR), a set of 72 E. coli strains isolated from a variety of animal hosts and a variety of geographical origins (44), show that there are four main phylogenetic groups of E. coli, designated A, B1, B2, and D (25, 53), and that most ExPEC strains belong to group B2, although some belong to group D (31, 46). However, none of the strains of the ECOR collection is of avian origin, and the place of APEC strains in the phylogenetic tree of E. coli has not been extensively studied.
Molecular epidemiology studies show that many APEC strains can be grouped into a limited number of clones (11, 12, 19, 33, 42). The clonal nature of APEC strains has also been demonstrated by phylogenetic analyses (11, 12, 64-66). Several studies also reveal the prevalence of various serogroups and of particular combinations of virulence-associated genes among APEC strains. These observations suggest that there may be only a limited number of virulence genotypes (5, 19, 41, 47, 48).
Some avian and human ExPEC strains share common characteristics, including various serogroups and various virulence-associated factors such as the iron acquisition systems aerobactin and the iro gene cluster, P and S fimbriae, the K1 capsule, IbeA invasin, the autotransporter Tsh, cytolethal distending toxin (CDT), and hemolysin F (7, 38, 47). Furthermore, some avian strains, especially those harboring the K1 capsule, have been shown to be closely related to the O18:K1 clone in phylogenetic group B2, originally identified by Achtman and colleagues (1, 39). However, the phylogenetic relationships between human and avian strains are not well documented. Despite similarities and genetic relationships between human and avian strains, the extent of the zoonotic risk associated with APEC strains remains unclear (2, 19, 47). A better knowledge of the phylogenetic relationships and virulence factor (VF) patterns of APEC strains would contribute to answering this question.
Here we document the phylogenetic location of APEC strains in the E. coli population and their phylogenetic relationships with human ExPEC strains. We focused on strains belonging to serogroups prevalent among APEC strains (O1, O2, O18, and O78) that are also common among human ExPEC strains. We used the rapid PCR-based phylotyping method of Clermont et al. (8), which allows one to assign E. coli strains to one of the four major phylogenetic groups. The multilocus sequence typing (MLST) method was then used to locate APEC strains in the phylogenetic tree of E. coli (36). MLST is currently one of the best phylogenetic grouping methods for investigating the genetic relationships between clinical pathogenic strains and reference strains. It has been found to be discriminating and reproducible (40) and has the advantage over pulsed-field gel electrophoresis of allowing pertinent phylogenetic analysis as well (60). For MLST analysis, we chose a set of six housekeeping genes previously used by Adiri et al. (2) in a comparison of O78 strains of avian and human origins. To estimate the zoonotic potential of avian E. coli, we compared the virulence genotypes and pathogenicities for chicks of APEC and human ExPEC strains.
MATERIALS AND METHODS
Bacterial strains.
A total of 102 Escherichia coli isolates were used in this study: 51 ExPEC strains of human origin, 39 APEC strains, 4 strains from animals (other than poultry) with septicemia, 2 enterohemorrhagic E. coli (EHEC) strains, and 6 strains from the feces of healthy human (n = 4) or animal (n = 2) hosts.
The 39 APEC strains were recovered previously from the heart blood or livers of chickens or turkeys with clinical signs of colibacillosis and were lethal for 1-day-old chicks following subcutaneous inoculation. Strain 789 AC/1 was kindly provided by E. Ron (2), and the other 38 strains were selected from a collection of 1,601 avian colibacillosis isolates collected in Europe between 1995 and 2000 (57) as belonging to the most frequently isolated serogroups of APEC: O1, O2, O18, and O78.
Human strains used included isolates from the main clinical extraintestinal sources (newborn meningitis, septicemia, and urinary tract infections) as well as strains from the ECOR collection and archetypal ExPEC strains to facilitate comparison with other phylogenetic studies. Twenty of the strains used were isolated from the cerebrospinal fluid of neonates and belonged to serogroup O2, O18, or O78. They included six strains from France (61), six strains from The Netherlands (29), five strains from Hungary (10), two strains from Finland (32, 54), and the NMEC type strain RS218 (1). Sixteen strains were isolated from humans with septicemia: eight from the blood of newborns without meningitis (61), seven from adults with septicemia in France (this study), and one from a septicemic patient in Hungary (10). Fifteen strains were from urinary tract infections, including one strain from Hungary (10), five from Germany (68, 69), six from the ECOR collection (44) (http://foodsafe.msu.edu/whittam/ecor), and the UPEC type strains CFT073, 536, and J96 (22, 43, 62). Four animal ExPEC strains were isolated from cases of septicemia in a sheep (S5), a pig (P72), and calves (BM2-1 and Orne6) (9, 13, 45, 56). Five strains of the ECOR collection (ECOR5, ECOR17, ECOR26, ECOR55, and ECOR70) (44), isolated from feces from healthy humans or animals, and the nonpathogenic K-12 strain MG1655 (6) were also included. Two EHEC strains were also included in the study: EDL933 (ATTC 43895) and SAKAI (RIMD0509952) (24). Escherichia fergusonii (ATCC35469T) was used as an outgroup for phylogenetic analysis. The bacterial strains were routinely grown in Luria-Bertani broth (LB) at 37°C and stored at −70°C in 20% glycerol until use.
Serotyping.
For strains that had not been serotyped previously, slide agglutination with specific antisera (Biovac, Angers, France) was used to test for O1, O2, and O78 antigens. We tested for the O18 antigen by agglutination using the O18 antiserum kindly provided by Jorge Blanco (Laboratorio de Referencia de Escherichia coli, Lugo, Spain). The presence of K1 capsular antigen was detected phenotypically using the Wellcogen Neisseria meningitidis B/E. coli kit (Oxoid), and the neuC gene was detected by PCR as described below.
PCR-based phylotyping.
Strains were classified into the four main phylogenetic groups of the ECOR collection by PCR as described by Clermont et al. (8) using three primer pairs: chuA.1-chuA.2, yjaA.1-yjaA.2, and TspE4C2.1-TspE4C2.2 (Table 1). Strains were assigned to phylogenetic group A, B1, B2, or D according to the amplification of the chuA and yjaA genes and the TspE4C2.1 fragment. Strains MG1655, ECOR26, ECOR62, and ECOR50 were used as controls for phylogenetic groups A, B1, B2, and D, respectively.
TABLE 1.
Primers used for PCR amplificationsa
| Gene | Primer name | Primer sequence (5′ to 3′) | Size of PCR product (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| fimA | fimA1b | CGGCTCTGTCCCTSAGT | 500 | 52 | 39 |
| fimA2b | GTCGCATCCGCATTAGC | ||||
| fimAMT78 | fimA201 | TCTGGCTGATACTACACC | 266 | 52 | 37 |
| fimA215 | ACTTTAGGATGAGTACTG | ||||
| fimH | fimH2 | GATCTTTCGACGCAAATC | 389 | 52 | 3 |
| fimH17 | CGAGCAGAAACATCGCAG | ||||
| sfa/foc | sfa1 | CTCCGGAGAACTGGGTGCATCTTAC | 410 | 64 | 34 |
| sfa2 | CGGAGGAGTAATTACAAACCTGGCA | ||||
| papC | pap1 | GACGGCTGTACTGCAGGGTGTGGCG | 328 | 61 | 34 |
| pap2 | ATATCCTTTCTGCAGGGATGCAATA | ||||
| felA | fel1 | GGTCAASCAGCTAAAAACGGTAAGG | 239 | 61 | 39 |
| fel2 | CCTTCAGAAACAGTACCGCCATTCG | ||||
| stgC | stgC-F | TCTGGTTCACATACACTACG | 208 | 58 | 35 |
| stgC-R | CCAATCATAATCTGGCTTCT | ||||
| neuC | neu1 | AGGTGAAAAGCCTGGTAGTGTG | 676 | 61 | 39 |
| neu2 | GGTGGTACATCCCGGGATGTC | ||||
| iutA | iutA1 | ATGAGCATATCTCCGGACG | 587 | 58 | 39 |
| iutA15 | CAGGTCGAAGAACATCTGG | ||||
| iroD (D11) | 20F | ATGCTGAACATGCAACAACA | 1,230 | 50 | 51 |
| 20R | TCAACCCTGTAGTAAACCAAT | ||||
| ibeA | ibeAF | TGAACGTTTCGGTTGTTTTG | 814 | 55 | 21 |
| ibeAR | TGTTCAAATCCTGGCTGGAA | ||||
| tsh | tsh03 | GGTGGTGCACTGGAGTGG | 640 | 58 | 17 |
| tsh15 | AGTCCAGCGTGATAGTGG | ||||
| cdt | cdt-s1 | GAAAGTAAATGGAATATAAATGTCCG | 466 | 55 | 59 |
| cdt-as1 | AAATCACCAAGAATCATCCAGTTA | ||||
| cdt-s2 | GAAAATAAATGGAACACACATGTCCG | 466 | 55 | 59 | |
| cdt-as2 | AAATCTCCTGCAATCATCCAGTTA | ||||
| hlyF | hlyF1 | TCGTTTAGGGTGCTTACCTTCAAC | 444 | 60 | This study |
| hlyF2 | TTTGGCGGTTTAGGCATTCC | ||||
| A9 | 13F | TTTCGACTGCTGGATGAAC | 934 | 50 | 51 |
| 13R | AATCATGATTGACCGTGC | ||||
| A12 | 16F | ATGCACTCGATAAAAAAAGT | 860 | 50 | 51 |
| 16R | TTAAGAAGGTCGATATACGT | ||||
| D1 | 17F | ATGAATTCACAATTACTGGC | 1,998 | 50 | 51 |
| 17R | TTAGCTGTTCAGTAGCTCAC | ||||
| D7 | cat31 | TCAGTAAGAACGAAAGTGTG | 565 | 50 | This study |
| cat32 | ACAGGAACAATCCCGTGGAT | ||||
| D10 | D10F2 | ATCTTTACCGTCCTCACC | 135 | 50 | 39 |
| D10R2 | CGTACCGCCTTCATTATC | ||||
| chuA | chuA.1 | GACGAACCAACGGTCAGGAT | 279 | 59 | 8 |
| chuA.2 | TGCCGCCAGTACCAAAGACA | ||||
| yjaA | yjaA.1 | TGAAGTGTCAGGAGACGCTG | 211 | 59 | 8 |
| yjaA.2 | ATGGAGAATGCGTTCCTCAAC | ||||
| tspE4.C2 | tspE4C2.1 | GAGTAATGTCGGGGCATTCA | 152 | 59 | 8 |
| tspE4C2.2 | CGCGCCAACAAAGTATTACG | ||||
| adk | adk-F | CGGGCGCGGGGAAAGGGACTC | 595 | 59 | 2 |
| adk-R | GCGCGAACTTCAGCAACCG | ||||
| gcl | gcl-F | GCGTTCTGGTCGTCCGGGTCC | 758 | 58 | 2 |
| gcl-R | GCCGCAGCGATTTGTGACAGACC | ||||
| zwf | gdh-F | TCGGCGTAGGGCGTGCTGAC | 796 | 59 | 2 |
| gdh-R | CTGCTCTTGTTCGCGCCCTCTTC | ||||
| mdh | mdh-F | CCCGGTGTGGCTGTCGATCTGA | 706 | 59 | 2 |
| mdh-R | CGCCGTTTTTACCCAGCAGCAGC | ||||
| metA | metA-F | CGCAACACGCCCGCAGAGC | 601 | 59 | 2 |
| metA-R | GCCAGCTCGCTCGCGGTGTATT | ||||
| ppk | ppk-F | TGCCGCGCTTTGTGAATTTACCG | 758 | 58 | 2 |
| ppk-R | CCCCGGCGCAGAGAAGATAACGT |
VFs searched for by PCR are as follows: fimA and fimH, type 1 fimbriae; fimAMT78, type 1 variant of APEC strain MT78; sfa/foc, S and F1C fimbriae; papC, P fimbriae; felA, F11 variant of P fimbriae; stgC, Stg fimbriae; neuC, K1 antigen; iutA, aerobactin receptor; iroD (D11), siderophore; ibeA, invasin; tsh, autotransporter Tsh; cdt, cytolethal distending toxin; hlyF, putative hemolysin; A9, A12, D1, D7, and D10, genomic fragments putatively associated with ExPEC virulence; chuA, yjaA, and TspE4.C2, sequences used for classification of E. coli strains into phylogenetic groups; adk, gcl, zwf, mdh, metA, and ppk, housekeeping genes used for MLST.
Primers fimA1 and fimA2 were designed from consensus sequences of various fimA genes.
Virulence for chicks.
The virulence of each E. coli isolate was determined using a lethality test for 1-day-old chicks as previously described (14). Groups of five 1-day-old specific-pathogen-free (SPF) chicks were inoculated subcutaneously with 0.5 ml of a 24-h LB culture (in stationary phase, at about 108 CFU), and the mortality was recorded 4 days postinoculation. The 50% lethal dose (LD50) was determined for some strains by inoculating chicks with 10-fold dilutions of the culture and was calculated by the method of Reed and Muench (14).
Experimental infection of SPF chickens via the air sacs.
Thirteen groups of 12 3.5-week-old White Leghorn SPF chickens from the INRA Infectiology Platform were reared in separate cages with food and water available ad libitum. The experiments were conducted as previously described with some modifications (17). Each chicken was inoculated in the right thoracic air sac with 0.1 ml (1 × 107 CFU) of a bacterial inoculum consisting of a 24-h LB stationary-phase culture of E. coli. Blood samples of 50 μl were collected aseptically from each chicken 24 h and 48 h later and were incubated in 2 ml of brain heart infusion (BHI) for qualitative detection of E. coli. Positive growth of E. coli in BHI was confirmed by plating enriched cultures on Drigalski agar (Bio-Rad).
All birds were euthanized by injection of Nesdonal (Rhône-Mérieux, Lyon, France) 48 h postinoculation and underwent necropsy. Macroscopic fibrinous lesions were observed and scored (air sacs, 0 to 4; pericardium, 0 to 2; liver, 0 to 2). The colonization of organs by E. coli was detected as follows: swabs of the left thoracic air sac and of the pericardial fluid were incubated in BHI; 100 μl of heart blood and liver fragments was streaked onto Drigalski agar.
All experiments involving chickens were licensed by the “Préfecture d'Indre et Loire, France,” agreement 006707.
Virulence genotyping.
Virulence genes were detected by PCR amplification in a Perkin-Elmer 9700 temperature cycler (Applied Biosystems). Crude DNA extracts were prepared by a rapid boiling method. Four multiplex PCR assays were designed to detect simultaneously (i) fimA (with consensus primers), a fimA variant (fimAMT78), and fimH, (ii) neuC, felA, and papC, (iii) tsh and iutA, and (iv) the cdt gene. Single PCR assays were used to detect four other genes (sfa/foc, iroD, ibeA, and hlyF) and five genomic fragments (A9, A12, D1, D7, and D10) previously identified in APEC strain BEN2908 and putatively associated with the virulence of ExPEC isolates (51). The corresponding primers are listed in Table 1. DNA fragments were amplified in a 25-μl PCR mixture including 5 μl of DNA crude extract, 1 U of Taq DNA polymerase (Promega), 12.5 pmol each of the forward and reverse primers, and 5 nmol of each deoxynucleoside triphosphate (Promega) in 1× buffer. PCR conditions were as follows: 94°C for 3 min; 30 cycles of 94°C for 1 min, the annealing temperature for 1 min, and 72°C for at least 30 s according to the size of the amplified fragment (1 min/kbp); and a final extension at 72°C for 10 min. The following E. coli strains were used as positive controls in PCR assays: strain BEN2908 (51) for the fimA, fimAMT78, fimH, neuC, iutA, and ibeA genes; strain MT189 (16) for felA and papC; strain χ7122 (17) for tsh, iroD, and stgC; strain KH576 (67) for iutA; strain 536 (23) for sfa/foc; strain E6468/62 (52) for cdt; strain SP15 (29) for hlyF; and strain BEN2908 or CFT073 (51, 62) for genomic fragments A9, A12, D1, D7, and D10. The negative-control strains were E. coli MG1655 and the nonpathogenic E. coli avian strain EC79 (6, 14).
MLST.
The phylogenetic relationships between strains were studied using the MLST method initially described by Maiden et al. (36). Six housekeeping genes were chosen according to the method of Adiri et al. (2): adk (adenylate kinase), gcl (glyoxylate carboligase), zwf (also commonly referred to as gdh) (glucose-6-phosphate dehydrogenase), mdh (malate dehydrogenase), metA (homoserine transsuccinylase), and ppk (polyphosphate kinase). The corresponding primers are listed in Table 1. DNA fragments (600 to 800 bp) were amplified in a Perkin-Elmer 9700 temperature cycler (Applied Biosystems) in a total volume of 50 μl containing 8 μl of DNA crude extract as a template, 0.5 U of Taq DNA polymerase (Promega), 25 pmol of the forward and reverse primers, and 5 nmol of each deoxynucleoside triphosphate (Promega) in 1× buffer. PCR conditions were as follows: 94°C for 5 min; 30 cycles of 94°C for 40 s, 58°C (or 59°C) for 45 s, and 72°C for 45 s; and a final extension at 72°C for 5 min. For strains CFT073, EDL933, and SAKAI, sequences were extracted from the GenBank database. The amplicons were sequenced on both strands by Genome Express (Meylan, France). No gcl fragment could be amplified by PCR from the Escherichia fergusonii strain, and recent sequencing of the whole genome of E. fergusonii (strain ATCC 35469T) confirmed the absence of gcl from this strain (E. Denamur, personal communication).
Sequences were then aligned using Clustal X (58), and a maximal common readable sequence was defined for each gene: adk (451 bp), gcl (534 bp), zwf (530 bp), mdh (468 bp), metA (472 bp), and ppk (545 bp). Sequences of each strain were then concatenated.
Phylogenetic analysis.
The PILEUP program (Genetics Computer Group, Madison, WI) was used for multiple sequence alignments and the phylogenetic inference package PHYLIP for phylogenetic analyses (20). Phylogenetic relationships were inferred using (i) the DNAPARS program, based on the principle of maximum parsimony, and (ii) the DNADist program (for estimation of the distances between pairs of sequences) followed by neighbor-joining programs (building of the trees) based on the principle of phenetics. Bootstrap support percentages were calculated for each branch point of the tree by the Seqboot procedure (100 replicates), and the majority-rule consensus tree was determined by the CONSENSE program. Trees were plotted using DRAWGRAM.
Statistical analysis.
Prevalences of VFs in the various phylogenetic groups were compared using a chi-square test. P values of ≤0.02 were considered significant.
A factorial analysis of correspondences (FCA) was undertaken to describe associations between the presence of virulence genes, the phylogenetic group, and the clinical and avian or human origin of the strains by using SPAD 6.0 software (Decisia, France). Only strains from chickens (39 strains) and humans (55 strains, excluding EHEC strains) were included in this analysis. The informative variables retained and used for calculations included the presence or absence of 16 genes or genomic fragments (neuC, sfa/foc, papC, felA, fimAMT78, ibeA, iutA, tsh, cdt, iroD [D11], hlyF, stgC, A9, D1, D7, and D10). Each strain was defined by its coordinates, these coordinates being the values obtained for each variable for this strain. A modality was defined as a possible value for each variable (e.g., ibeA = 0 or ibeA = 1), and the coordinates of each modality correspond to whether or not it was observed for each of the 94 strains. The coordinates obtained for each strain and each modality were transformed using matrices into new coordinates on several factorial axes. The variance of the distribution of the strains or modalities was calculated for each factorial axis. Factorial axes were then ordered by decreasing level of variance. Results are represented graphically in a plane defined by two axes: F1, which accounts for most of the variance, and F2, which accounts for the largest part of the variance not accounted for by F1. Data concerning the clinical and animal origins of the strains and the phylogenetic groups of the strains were considered supplementary characteristics and projected on the F1/F2 plane.
Nucleotide sequence accession numbers.
The sequences of the housekeeping genes used in this study have been deposited in GenBank and are available under the following accession numbers: DQ999496 to DQ999598 (adk), DQ999291 to DQ999392 (gcl), DQ999188 to DQ999290 (zwf [gdh]), EF011985 to EF012087 (mdh), DQ999085 to DQ999187 (metA), and DQ999393 to DQ999495 (ppk).
RESULTS
Location of APEC strains in the E. coli population.
We used PCR-based phylotyping to classify the 102 E. coli strains studied into the four main phylogenetic groups of E. coli (8). Results for strains of the ECOR collection tested corresponded to their previously reported phylogenetic classifications, with the exception of strain ECOR70, which belongs to phylogenetic group B1 (25) but was identified as A by the PCR-based phylotyping method, as already described by Clermont et al. (8).
Most of the strains studied belonged to phylogenetic group B2 (76 strains), as expected for ExPEC strains (31, 46); some were in phylogenetic group D (7 strains: BEN955, BEN961, BEN1189, BEN3013, BEN3025, ECOR50, and Orne6). PCR-based phylotyping assigned only 2 strains to phylogenetic group B1 (ECOR26 and S5), and 15 strains, including strain ECOR70, showed a “group A profile”: the absence of chuA and of TspE4.C2 and the presence of yjaA.
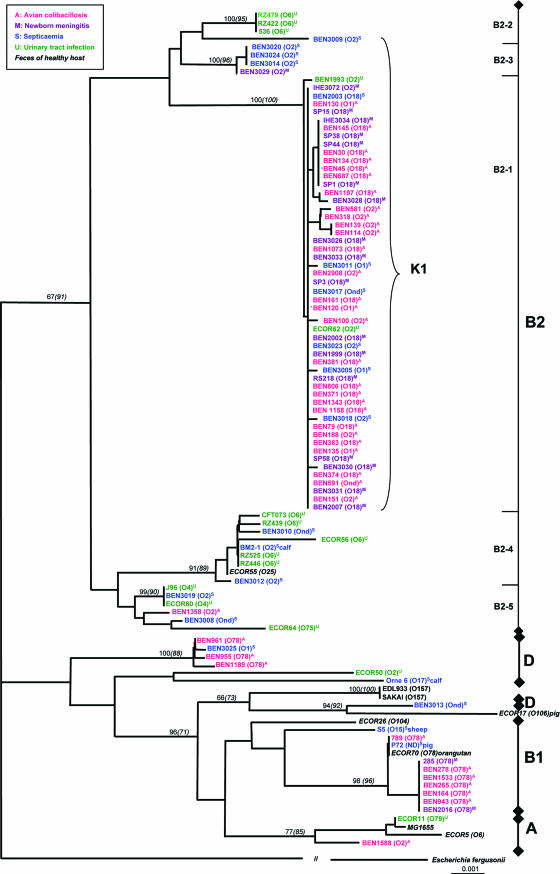
The sequences of six housekeeping genes (adk, gcl, zwf, mdh, metA, and ppk) were determined and used to generate phylogenetic trees by the neighbor-joining method (Fig. 1) and the maximum-parsimony method. Irrespective of the method used, similar main clusters of strains were consistently identified with high bootstrap values. The phylogenetic tree obtained by the neighbor-joining method presented a large cluster of 76 strains (bootstrap value, 67) comprising all strains of phylogenetic group B2 distributed into five subclusters. The main subcluster, referred to as B2-1 (bootstrap value, 100), included 28 avian and 25 human ExPEC strains belonging to serotypes O18:K1, O2:K1, and O1:K1. The human ExPEC strains were from cases of septicemia (6 strains), newborn meningitis (17 strains), or urinary tract infections (2 strains). They include the archetypal NMEC strains RS218 and IHE3034 and the reference strain ECOR62. The phylogenetic distance between these strains is close to zero.
FIG. 1.
Phylogenetic relationships between 102 E. coli isolates originating from human clinical extraintestinal sources (n = 51), avian colibacillosis (n = 39), nonpoultry animal cases of septicemia (n = 4), healthy hosts (n = 6), and hemolytic-uremic syndrome (n = 2). The phylogenetic tree was constructed by neighbor-joining analysis based on the sequence analysis of six housekeeping genes. The phylogenetic groups and subclusters are given on the right. Bootstrap values are given above the nodes where values are higher than 60% (bootstrap values from the maximum-parsimony analysis are in parentheses). The serogroup of each strain (when known) is given in parentheses. The clinical origins of ExPEC strains are coded by colors and letters as follows: A (red), avian colibacillosis; M (purple), neonatal meningitis; S (blue), septicemia; U (green), urinary tract infection; no letter (black), strain from a healthy host. The animals from which the six nonpoultry isolates originated are given. E. coli EDL933 and SAKAI are EHEC strains.
Other subclusters in the B2 phylogenetic group contained only human strains, with the exception of one APEC strain in subcluster B2-5. Subcluster B2-2 contained three human strains (serogroup O6) from urinary tract infections, including the archetypal UPEC strain 536 (bootstrap value, 100). Another subcluster, named B2-3, included four human strains of the O2:K1 serotype, isolated mainly from patients with septicemia or newborn meningitis (bootstrap value, 100). Subclusters B2-4 and B2-5 included human strains isolated from patients with septicemia or urinary tract infections. UPEC archetypal strains CFT073 (serogroup O6) and J96 (serogroup O4) were in B2-4 and B2-5, respectively.
Another subcluster with a high bootstrap value (100) comprised strains belonging to phylogenetic group D; it included three O78 APEC strains (BEN955, BEN961, and BEN1189) and a strain from a human with septicemia of respiratory origin (BEN3025). Thus, the PCR-determined phylogenetic groups B2 and D and the locations of strains in the phylogenetic trees were in good agreement.
However, strains ECOR50 and Orne 6, identified as belonging to phylogenetic group D by the PCR-based phylotyping method, were not classified by any analysis into the same subcluster as other D strains. They may belong to a different subcluster of phylogenetic group D, but no sentinel strain was used that could allow allocating them to one of the D subgroups as recently defined by Johnson et al. (30). EHEC strains EDL933 and SAKAI showed a “phylogenetic group D pattern,” as usually observed when the PCR-based phylotyping method of Clermont et al. (8) is used.
Reference strains of phylogenetic group A (ECOR5, ECOR11, and MG1655) were clustered with the APEC strain BEN1588 (phylogenetic group A) with a bootstrap value of 77. Strain ECOR17, identified as belonging to phylogenetic group A both by multilocus enzyme electrophoresis (53) and by PCR-based phylotyping (this study), was not clustered with other group A reference strains by our MLST method.
An interesting cluster (bootstrap value, 98) contained 10 strains, including strain ECOR70 (serogroup O78), 8 O78 strains of human and avian origins, and a strain from a pig with septicemia (E. coli P72). All these strains were first identified as belonging to phylogenetic group A by PCR-based phylotyping. Misclassification into other phylogenetic groups of some B1 strains, in particular strain ECOR70, has been reported by this phylotyping method (8), so we investigated these strains in more detail. Lymberopoulos et al. (35) have demonstrated that the presence of the stgC gene is significantly associated with phylogenetic groups B1 and D and not with groups A and B2; we therefore tested for the presence of the stgC gene in the 102 E. coli strains of the study. The results confirmed the observations of Lymberopoulos et al. (35): none of the strains of phylogenetic group B2 possessed the stgC gene, whereas strains ECOR26 and ECOR70 (phylogenetic group B1) did. The 10 strains of the “ECOR70 cluster,” initially assigned to phylogenetic group A, all possessed the stgC gene, whereas ECOR5, ECOR11, and MG1655, known to belong to phylogenetic group A, and strain BEN1588 did not. Thus, the 10 strains of the “ECOR70 cluster” were finally assigned to phylogenetic group B1 rather than A (Fig. 1).
APEC strains of serogroups O1, O2, and O18 were clearly differentiated from O78 strains: 28 of the 30 APEC strains of serogroups O1, O2, and O18 belonged to phylogenetic group B2 (subcluster B2-1; bootstrap value, 100), whereas the 9 APEC strains of serogroup O78 were grouped into two subclusters with high bootstrap values, in phylogenetic groups D (3 strains) and B1 (6 strains). Human ExPEC strains appeared to be closely related to APEC strains in each of these three subclusters.
The virulence genotype correlates with the phylogenetic classification of the strains.
We tested for the presence of 13 VF genes commonly found in ExPEC strains and of 5 genomic fragments putatively associated with the virulence of ExPEC isolates (51). Differences in the incidence of each individual VF gene between the main phylogenetic group in our study (B2) and other phylogenetic groups (A, B1, and D) were tested using the chi-square test: the presence of fimAMT78, sfa/foc, neuC, ibeA, cdt, A9, D1, D7, D10, and iroD (D11) was significantly more frequent (P ≤ 0.02) in B2 strains than in strains belonging to phylogenetic groups A, B1, and D (Table 2). A theoretical virulence index (VI) was calculated for each group by scoring the total number of VF genes present (fimA, sfa/foc, papC, neuC, iutA, iroD, ibeA, tsh, cdt, hlyF, A9, A12, D1, D7, D10) and then dividing by the number of strains in the group. The VI was higher for phylogenetic group B2 (11.3/15) than for other groups (5.6/15).
TABLE 2.
Distribution of VF genes in the phylogenetic groups for 100 E. coli isolates of different originsa
| VF gene | No. (%) of strains positive for the indicated VF gene
|
|||
|---|---|---|---|---|
| Phylogenetic groups
|
B2 phylogenetic subclusters
|
|||
| B2 | A, B1, or D | B2-1 | Other B2 subclusters | |
| fimA | 76 (100) | 23 (96) | 53 (100) | 23 (100) |
| fimAMT78 | 46 (61)b | 2 (8) | 44 (83)b | 2 (9) |
| fimH | 76 (100) | 24 (100) | 53 (100) | 23 (100) |
| sfa/foc | 47 (62)b | 3 (13) | 30 (57) | 17 (74) |
| papC | 34 (45) | 6 (25) | 18 (34)b | 16 (70) |
| felA | 17 (22) | 4 (17) | 17 (32) | 0 |
| neuC | 62 (82)b | 0 | 53 (100)b | 9 (39) |
| iutA | 60 (79) | 20 (83) | 50 (94)b | 10 (43) |
| iroD (D11) | 72 (95)b | 18 (75) | 52 (98) | 20 (87) |
| ibeA | 40 (53)b | 0 | 37 (70)b | 3 (13) |
| tsh | 24 (32) | 8 (33) | 22 (42)b | 2 (9) |
| cdt | 27 (36)b | 2 (8) | 24 (45)b | 3 (13) |
| hlyF | 52 (68) | 16 (67) | 50 (94)b | 2 (9) |
| A9 | 64 (84)b | 1 (4) | 50 (94) | 14 (61) |
| A12 | 76 (100) | 20 (83) | 53 (100) | 23 (100) |
| D1 | 73 (96)b | 5 (21) | 50 (94) | 23 (100) |
| D7 | 75 (99)b | 6 (25) | 52 (98) | 23 (100) |
| D10 | 76 (100)b | 7 (29) | 53 (100) | 23 (100) |
Isolates were from human clinical extraintestinal sources (n = 51), avian colibacillosis (n = 39), nonpoultry animal cases of septicemia (n = 4), and healthy hosts (n = 6). A VI was calculated for each phylogenetic subcluster by adding the number of strains positive for fimA, sfa/foc, papC, neuC, iutA, iroD (D11), ibeA, tsh, cdt, hlyF, A9, A12, D1, D7, and D10, and dividing by the total number of strains in the group. The VIs were 11.3 for phylogenetic group B2; 5.6 for A, B1, or D; 12.2 for the B2-1 subcluster; and 9.2 for other B2 subclusters.
The incidence of the corresponding gene was significantly different (P ≤ 0.02) between phylogenetic groups or subclusters as assessed by the chi-square test.
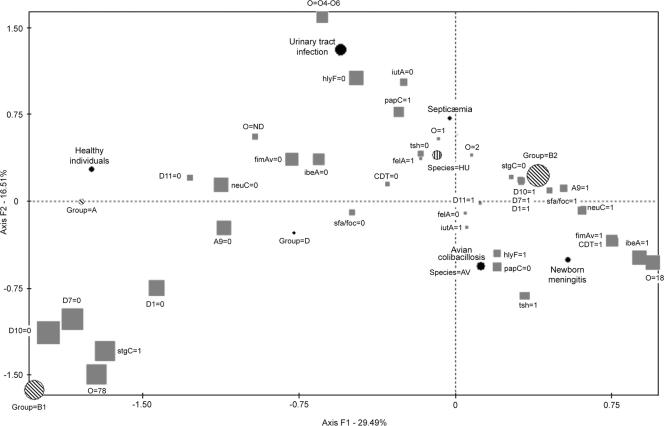
FCA was also used to search for associations of multiple combinations of VF genes with the phylogenetic status of strains and with their origin (human or avian). FCA confirmed that strains of phylogenetic group B2 were clearly differentiated from strains of the other groups and were tightly associated with the presence of VF genes sfa/foc, neuC, A9, D1, D7, and D10 and the absence of stgC (Fig. 2). Among human and avian isolates of phylogenetic group B2, those associated with serogroup O18 were also associated with the same VF genes. By contrast, human isolates from urinary tract infections that were associated with serogroups O4 and O6 showed characteristics different from those of avian colibacillosis isolates (Fig. 2). FCA confirmed that APEC strains belonging to phylogenetic group B2 were associated with VF genes different from those of APEC strains of phylogenetic group B1 (Fig. 2).
FIG. 2.
Association of VFs with phylogenetic groups in E. coli strains originating from human clinical extraintestinal sources (n = 51), feces of healthy humans (n = 4), or avian colibacillosis (n = 39). In this schematic representation of the FCA, the distributions of VFs and major serotypes are represented on a factorial plane defined by the most discriminating axes: F1 and F2. Factorial axis F1 accounts for 29.49% of the variance, and factorial axis F2 accounts for 16.51% of the remaining variance. The strength of the associations is indicated by the distance between the symbols and the sizes of the symbols (the larger the size of the symbol, the stronger the association). The species of the host (avian or human), phylogenetic group (A, B1, B2, or D), clinical origin (avian colibacillosis, neonatal meningitis, septicemia, urinary tract infection, or healthy individual), O serogroup, and VFs are indicated. The presence or absence of a virulence factor is indicated by “1” or “0,” respectively.
Avian and human strains in subcluster B2-1 could not be differentiated by their virulence genotypes.
The major subcluster in the phylogenetic tree (B2-1, with 53 strains) appeared to be a homogeneous group of human and avian strains with very low, sometimes even undetectable phylogenetic distances between strains (Fig. 1). Thus, we investigated if the B2-1 subcluster was associated with a particular genotype, compared with other B2 subclusters, and if avian and human strains could be differentiated by their virulence genotypes. The frequencies of various VF genes in the B2-1 subcluster—fimAMT78 (83%), neuC (100%), iutA(94%), ibeA (70%), tsh (42%), cdt (45%), and hlyF (94%)—were higher than their frequencies in other B2 subclusters. Other VF genes were significantly less frequent in subcluster B2-1 than in other B2 subclusters (for example, papC [34%]) (Table 2). When present on strains of the B2-1 subcluster, P fimbriae were of the F11 variant type, as shown by the presence of the felA gene. The B2-1 subcluster included avian (n = 28) and human (n = 25) strains that did not differ significantly in the incidence of any of the VF genes studied (data not shown). These results were confirmed by FCA, which demonstrated that among human and avian isolates of phylogenetic group B2, those associated with serogroup O18 (which belonged to the B2-1 subcluster) were also associated with the same VF genes. Conversely, human isolates from urinary tract infections that were associated with serogroups O4 and O6 (which belonged to subclusters B2-2, B2-4, and B2-5) showed characteristics different from those of avian colibacillosis isolates (Fig. 2).
Other VF genes commonly assessed in studies of human-source ExPEC, such as hlyA and cnf, are infrequently present on E. coli isolates from neonatal meningitis and avian colibacillosis cases (29, 39, 61) and thus were not systematically tested in the present study. However, in phylogenetic subcluster B2-1, all 28 avian isolates were hlyA negative and 27 were cnf negative while 15 of 17 human isolates tested were negative for both genes (data not shown).
Human ExPEC strains of subcluster B2-1 were highly virulent for chickens.
We tested the hypothesis of host specificity of human B2 strains by determining their virulence for chickens in several models of infection.
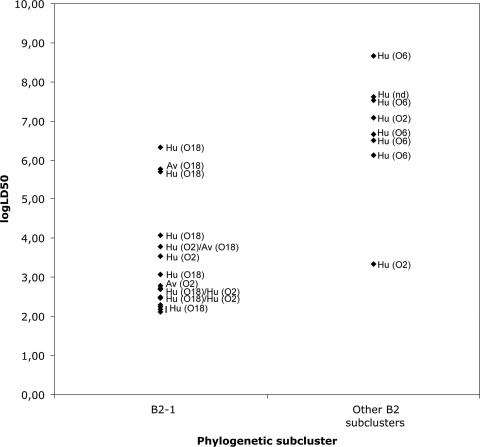
All ExPEC strains of phylogenetic group B2 were lethal when used to inoculate 1-day-old chicks subcutaneously: 72 of the 76 B2 strains killed 5/5 chicks, 2 strains killed 4/5 chicks (ECOR55 and CFT073), and 2 strains killed 1 or 2 chicks (RZ525 and ECOR56). The LD50s for 1-day-old chicks were also calculated for 25 strains of phylogenetic group B2: two groups of strains could be differentiated on the basis of the LD50 (Fig. 3). Strains of phylogenetic subcluster B2-1 were more virulent than strains of other B2 subclusters (Fig. 3). Of 17 strains belonging to phylogenetic subcluster B2-1, 14 were highly virulent (LD50, ≤1.20E + 04 CFU). All these strains belonged to serotype O18:K1 or O2:K1 and were isolated in cases of meningitis, septicemia, urinary tract infection, or avian colibacillosis. Three strains showed lower virulence levels: RS218, IHE3034, and BEN79, with LD50s between 5.10E + 05 and 2.10E + 06 CFU. Of the eight strains of other B2 phylogenetic subgroups that had been isolated from humans with septicemia or urinary tract infections, all but one (BEN3012) were less virulent (LD50, >1.0E + 06 CFU) than strains of phylogenetic subcluster B2-1. Most belonged to serogroup O6, which is infrequently isolated in cases of avian colibacillosis.
FIG. 3.
LD50s for chicks of human (n = 22) and avian (n = 3) ExPEC strains of phylogenetic group B2. Decimal dilutions of bacterial cultures of ExPEC strains of human and avian origins were inoculated into 1-day-old chicks, and the lethality was recorded. The virulence of the E. coli strains was expressed as the LD50 for chicks; the log LD50 is represented. The O serogroups and origins (Hu, human; Av, avian) of the strains are given.
Because the lethality test on 1-day-old chicks bypasses the first steps of natural infection in chickens (which usually starts in the respiratory tract), we tested the pathogenicities of nine human strains by using bacterial cultures to inoculate the thoracic air sacs of 3.5-week-old SPF chickens. Three APEC strains and one nonpathogenic strain of avian origin were also used for comparison. Human isolates of phylogenetic subcluster B2-1 caused a typical avian colibacillosis similar to that caused by avian strains, and the criteria used to assess avian colibacillosis (bacterial contamination of air sacs, blood, pericardial fluid, liver, and typical fibrinous lesions) were fulfilled (Table 3). In contrast, only a few chickens developed the disease when inoculated with the archetypal human strains RS218 and IHE3034, showing few lesions and no contamination of pericardial fluid or the liver. This result may be due to the fact that strains RS218 and IHE3034 have undergone numerous subcultures, possibly resulting in genetic modifications and partial loss of virulence properties, as is sometimes observed for intensively used reference strains.
TABLE 3.
Pathogenicities of ExPEC strains of different clinical origins for SPF chickens following intra-air sac inoculationa
| E. coli strain | Phylogenetic subcluster | Clinical originb | Serotype | % of chickens with contamination in:
|
Lesion score/8 (SD) | Death (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| Air sacs | Blood | Pericardial fluid | Liver | ||||||
| SP15 | B2-1 | Hu M | O18:K1 | 100 | 100 | 100 | 100 | 7.0 (0.6) | 41.6 |
| BEN3031 | B2-1 | Hu M | O18:K1 | 100 | 100 | 100 | 100 | 5.8 (1.2) | 0 |
| BEN3033 | B2-1 | Hu M | O18:K1 | 100 | 100 | 100 | 91.7 | 5.1 (0.8) | 33.3 |
| SP3 | B2-1 | Hu M | O18:K1 | 75 | 50 | 58.3 | 75 | 3.6 (3.1) | 33.3 |
| IHE3034 | B2-1 | Hu M | O18:K1 | 50 | 33.3 | 33.3 | 16.7 | 2.6 (1.8) | 0 |
| RS218 | B2-1 | Hu M | O18:K1 | 11.1 | 5.5 | 5.5 | 5.5 | 1.6 (1.2) | 0 |
| BEN79 | B2-1 | Av C | O18:K1 | 91.7 | 83.3 | 83.3 | 100 | 4.9 (0.8) | 25 |
| BEN374 | B2-1 | Av C | O18:K1 | 100 | 100 | 100 | 100 | 7.1 (0.9) | 30 |
| IHE3072 | B2-1 | Hu M | O2:K1 | 100 | 100 | 100 | 100 | 6.2 (1.0) | 25 |
| BEN2908 | B2-1 | Av C | O2:K1 | 100 | 100 | 100 | 100 | 5.0 (1.6) | 37.5 |
| BEN3009 | B2-2 | Hu Sc | O2:K1 | 25 | 0 | 0 | 0 | 1.5 (1.2) | 0 |
| BEN3012 | B2-4 | Hu Sd | O2:K1 | 100 | 58.3 | 100 | 100 | 4.8 (0.8) | 0 |
| EC79e | A | Av healthy | O2 | 6 | 0 | 0 | 0 | 0.5 | 0 |
Groups of 12 chickens were inoculated in the air sacs with bacterial cultures of E. coli strains. Bacterial colonization of internal organs, fibrinous lesions, and deaths were recorded.
Strains were from newborn meningitis (n = 7), human septicemia (n = 2), and avian colibacillosis (n = 3). Hu, human origin; Av, avian origin; M, newborn meningitis; S, septicemia; C, avian colibacillosis.
Isolated from an adult with a urinary tract infection.
Isolated from a newborn with septicemia.
Nonpathogenic E. coli strain of avian origin used as a negative control (14).
Neither pathogenicity for chicks nor MLST and virulence genotyping data allowed us to distinguish APEC strains from human ExPEC strains belonging to the highly pathogenic subcluster B2-1; thus, we could find no evidence of host specificity for strains belonging to this subcluster.
DISCUSSION
In this study we used two approaches, PCR-based phylotyping and MLST, to establish the phylogenetic positions of 39 APEC strains belonging to the prevalent serogroups O1, O2, O18, and O78 in relation to 63 E. coli strains of human and other mammalian origins. The positions of the 102 strains studied in the phylogenetic trees reconstructed from MLST analysis were consistent with results obtained by PCR-based phylotyping, with a few exceptions. Strain ECOR17, identified as belonging to phylogenetic group A both by multilocus enzyme electrophoresis (44) and by PCR-based phylotyping (this study), was not clustered with other A strains by use of our MLST method. As discussed by Johnson et al. (30), this could be a consequence of recombined genes or gene fragments at the loci we used for MLST. Strain ECOR70, originally identified as belonging to phylogenetic group B1 (44), and the other O78 strains located in the same cluster by MLST analysis were first identified as belonging to group A by the PCR-based phylotyping method. As discussed by Clermont et al. (8), the PCR method may fail to identify some B1 strains, so we used the association between the presence of the stgC gene, serogroup O78, and phylogenetic group B1 (35) to demonstrate that this cluster of strains was more likely to belong to phylogenetic group B1. Moreover, this result is in agreement with the O78 clade described by Johnson et al. (30), which includes strain ECOR70 and an O78 E. coli strain from a case of human bacteremia. Thus, testing for the stgC gene could be a useful complement to the PCR-based phylotyping method of Clermont et al. (8) for the discrimination of strains belonging to phylogenetic groups B1 and A.
Comparison of the phylogenetic trees we obtained with those published in previous studies revealed similar subclusters of strains according to various sentinel strains, especially in the B2 phylogenetic group. The four important clades described by Johnson et al. (30) in group B2 were detected as B2-1 (strains RS218, IHE3034, and ECOR62), B2-2 (strain 536), B2-4 (strains CFT073 and ECOR56), and B2-5 (strains J96 and ECOR60). Similar clades in the B2 group were also differentiated by Escobar-Páramo et al. and Hommais et al. (18, 26).
Most of the APEC strains in this study (29/39) belong to phylogenetic group B2, and fewer belong to the B1, D, and A phylogenetic groups (6 strains, 3 strains, and 1 strain, respectively). This distribution does not necessarily reflect the incidence of the four main phylogenetic groups in the APEC population, because common serogroups (O1, O2, and O18) of avian and human ExPEC strains were overrepresented in our sample, and serogroup O78, which is prevalent among avian colibacillosis isolates in numerous countries (19, 42, 47), was underrepresented.
Only three major phylogenetic clusters could be identified among APEC strains of the predominant serogroups included in our study: B1, B2, and D. This finding is consistent with previous observations of a few clone complexes of APEC isolates (39, 63, 65). We demonstrated an association between the phylogenetic clusters of APEC strains and their serogroups. Although the O1, O2, and O18 APEC strains originated from various geographical locations (France, Spain, and Belgium) and had been isolated over a 10-year period (and can thus be considered representative of APEC strains), they were very closely related: all but one were clustered in the same phylogenetic subcluster (B2-1). Also, APEC strains of serogroup O78 all fell into phylogenetic groups B1 and D. This agrees with the observation of two phylogenetic clusters of O78 APEC strains by Adiri et al., using MLST analysis (2); it also clearly demonstrates that O78 strains are phylogenetically different from O1, O2, and O18 APEC strains. A similar pattern of differences was also observed by virulence genotyping: the three clusters of APEC strains were associated with different sets of virulence genes. As already reported by Mokady et al. and by Ron, O2 and O78 APEC strains differ with respect to several VFs (38, 49).
Phylogenetic trees reconstructed from MLST analysis demonstrated close relationships between human and avian ExPEC strains belonging to the same phylogenetic group, whether B1, B2, or D. The few strains in groups B1 and D did not allow useful analysis, and further investigations are currently under way with a larger sample of O78 E. coli strains to analyze the relationships between human and avian strains in these groups.
However, phylogenetic group B2 included 28 APEC strains and 25 human ExPEC strains, all in a single, highly homogeneous subcluster designated B2-1: human and avian ExPEC strains belonging to this subcluster were highly virulent for chickens and possessed the same major VFs (fimAMT78, neuC, iutA, ibeA, tsh, cdt, and hlyF). These results suggest that some particular subclusters of strains within the ExPEC population have little or no host specificity. As already suggested by Johnson et al. (28), this highly pathogenic clonal group including O1:K1, O2:K1, and O18:K1 E. coli strains isolated in cases of neonatal meningitis, septicemia, or urinary tract infections, as well as avian colibacillosis, contradicts the classical definition of pathotypes as host specific in the case of ExPEC isolates.
Whereas we could demonstrate the virulence of human strains for chickens, it is difficult to assess the virulence of avian strains for humans. However, previous studies have shown that APEC isolates are lethal for mice when inoculated intraperitoneally (10). Moreover, experimental models of mammalian infections, such as those developed by Skyberg et al. and Johnson et al., indicate that APEC strains are able to express pathogenicity for mammals (27, 55).
According to our results, avian and human strains belonging to subcluster B2-1 can be indistinguishable in terms of phylogenetic location and the presence of VFs. Moreover, no host specificity could be shown: avian and human strains of this phylogenetic subcluster were highly and similarly virulent for chickens. These results are in favor of the possibility that E. coli strains of avian origin are virulent for humans and consequently constitute a zoonotic risk.
Acknowledgments
We are grateful to Philippe Gilot for critical reading of the manuscript. Eliora Ron, Roland Quentin, Jörg Hacker, Erick Denamur, Eric Oswald, and Jorge Blanco are fully acknowledged for the kind gift of strains and for helpful advice. We especially thank Thomas Whittam for providing E. coli strains from the ECOR collection. We thank Michèle Pelloile and Laila Bakri for skillful participation in this work and Nguessan Yao, Nathalie Lallier, and Patrice Cousin for valuable technical help at various stages of the work.
This work was supported by a grant from the European Commission (“Colirisk project,” QLK2-CT-2002-00944).
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Achtman, M., A. Mercer, B. Kusecek, A. Pohl, M. Heuzenroeder, W. Aaronson, A. Sutton, and R. P. Silver. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adiri, R. S., U. Gophna, and E. Z. Ron. 2003. Multilocus sequence typing (MLST) of Escherichia coli O78 strains. FEMS Microbiol. Lett. 222:199-203. [DOI] [PubMed] [Google Scholar]
- 3.Arné, P., D. Marc, A. Brée, C. Schouler, and M. Dho-Moulin. 2000. Increased tracheal colonization in chickens without impairing pathogenic properties of avian pathogenic Escherichia coli MT78 with a fimH deletion. Avian Dis. 44:343-355. [PubMed] [Google Scholar]
- 4.Barnes, H. J., J.-P. Vaillancourt, and W. B. Gross. 2003. Colibacillosis, p. 631-652. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames, IA.
- 5.Blanco, J. E., M. Blanco, A. Mora, W. H. Jansen, V. Garcia, M. L. Vazquez, and J. Blanco. 1998. Serotypes of Escherichia coli isolated from septicaemic chickens in Galicia (northwest Spain). Vet. Microbiol. 61:229-235. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Chérifi, A., M. Contrepois, B. Picard, P. Goullet, I. Orskov, and F. Orskov. 1994. Clonal relationships among Escherichia coli serogroup O78 isolates from human and animal infections. J. Clin. Microbiol. 32:1197-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contrepois, M., Y. Bertin, J. P. Girardeau, B. Picard, and P. Goullet. 1993. Clonal relationships among bovine pathogenic Escherichia coli producing surface antigen CS31A. FEMS Microbiol. Lett. 106:217-222. [DOI] [PubMed] [Google Scholar]
- 10.Czirók, E., M. Dho, M. Herpay, I. Gado, and H. Milch. 1990. Association of virulence markers with animal pathogenicity of Escherichia coli in different models. Acta Microbiol. Hung. 37:207-217. [PubMed] [Google Scholar]
- 11.da Silveira, W. D., A. Ferreira, M. Lancellotti, I. A. Barbosa, D. S. Leite, A. F. de Castro, and M. Brocchi. 2002. Clonal relationships among avian Escherichia coli isolates determined by enterobacterial repetitive intergenic consensus (ERIC)-PCR. Vet. Microbiol. 89:323-328. [DOI] [PubMed] [Google Scholar]
- 12.da Silveira, W. D., M. Lancellotti, A. Ferreira, V. N. Solferini, A. F. de Castro, E. G. Stehling, and M. Brocchi. 2003. Determination of the clonal structure of avian Escherichia coli strains by isoenzyme and ribotyping analysis. J Vet. Med. B 50:63-69. [DOI] [PubMed] [Google Scholar]
- 13.De Rycke, J., L. Phan-Thanh, and S. Bernard. 1989. Immunochemical identification and biological characterization of cytotoxic necrotizing factor from Escherichia coli. J. Clin. Microbiol. 27:983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dho, M., and J. P. Lafont. 1984. Adhesive properties and iron uptake ability in Escherichia coli lethal and nonlethal for chicks. Avian Dis. 28:1016-1025. [PubMed] [Google Scholar]
- 15.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 16.Dho-Moulin, M., J. F. van den Bosch, J. P. Girardeau, A. Brée, T. Barat, and J. P. Lafont. 1990. Surface antigens from Escherichia coli O2 and O78 strains of avian origin. Infect. Immun. 58:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dozois, C. M., M. Dho-Moulin, A. Brée, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escobar-Páramo, P., O. Clermont, A. B. Blanc-Potard, H. Bui, C. Le Bouguénec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 19.Ewers, C., T. Janssen, S. Kiessling, H. C. Philipp, and L. H. Wieler. 2004. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet. Microbiol. 104:91-101. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46:101-111. [DOI] [PubMed] [Google Scholar]
- 21.Germon, P., Y. H. Chen, L. He, J. E. Blanco, A. Brée, C. Schouler, S. H. Huang, and M. Moulin-Schouleur. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179-1186. [DOI] [PubMed] [Google Scholar]
- 22.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 23.Hacker, J., S. Knapp, and W. Goebel. 1983. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 154:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hommais, F., S. Pereira, C. Acquaviva, P. Escobar-Paramo, and E. Denamur. 2005. Single-nucleotide polymorphism phylotyping of Escherichia coli. Appl. Environ Microbiol. 71:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, J. R., O. Clermont, M. Menard, M. A. Kuskowski, B. Picard, and E. Denamur. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141-1150. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, J. R., P. Delavari, A. L. Stell, T. S. Whittam, U. Carlino, and T. A. Russo. 2001. Molecular comparison of extraintestinal Escherichia coli isolates of the same electrophoretic lineages from humans and domestic animals. J. Infect. Dis. 183:154-159. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, J. R., K. L. Owens, C. R. Clabots, S. J. Weissman, and S. B. Cannon. 2006. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 8:1702-1713. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 32.Korhonen, T. K., M. V. Valtonen, J. Parkkinen, V. Vaisanen-Rhen, J. Finne, F. Orskov, I. Orskov, S. B. Svenson, and P. H. Makela. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La Ragione, R. M., and M. J. Woodward. 2002. Virulence factors of Escherichia coli serotypes associated with avian colisepticaemia. Res. Vet. Sci. 73:27-35. [DOI] [PubMed] [Google Scholar]
- 34.Le Bouguénec, C., M. Archambaud, and A. Labigne. 1992. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 30:1189-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lymberopoulos, M. H., S. Houle, F. Daigle, S. Léveillé, A. Brée, M. Moulin-Schouleur, J. R. Johnson, and C. M. Dozois. 2006. Characterization of Stg fimbriae from an avian pathogenic Escherichia coli O78:K80 strain and assessment of their contribution to colonization of the chicken respiratory tract. J. Bacteriol. 188:6449-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marc, D., and M. Dho-Moulin. 1996. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J. Med. Microbiol. 44:444-452. [DOI] [PubMed] [Google Scholar]
- 38.Mokady, D., U. Gophna, and E. Z. Ron. 2005. Extensive gene diversity in septicemic Escherichia coli strains. J. Clin. Microbiol. 43:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moulin-Schouleur, M., C. Schouler, P. Tailliez, M. R. Kao, A. Brée, P. Germon, E. Oswald, J. Mainil, M. Blanco, and J. Blanco. 2006. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 44:3484-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemoy, L. L., M. Kotetishvili, J. Tigno, A. Keefer-Norris, A. D. Harris, E. N. Perencevich, J. A. Johnson, D. Torpey, A. Sulakvelidze, J. G. Morris, Jr., and O. C. Stine. 2005. Multilocus sequence typing versus pulsed-field gel electrophoresis for characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 43:1776-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ngeleka, M., L. Brereton, G. Brown, and J. M. Fairbrother. 2002. Pathotypes of avian Escherichia coli as related to tsh-, pap-, pil-, and iuc-DNA sequences, and antibiotic sensitivity of isolates from internal tissues and the cloacae of broilers. Avian Dis. 46:143-152. [DOI] [PubMed] [Google Scholar]
- 42.Ngeleka, M., J. K. Kwaga, D. G. White, T. S. Whittam, C. Riddell, R. Goodhope, A. A. Potter, and B. Allan. 1996. Escherichia coli cellulitis in broiler chickens: clonal relationships among strains and analysis of virulence-associated factors of isolates from diseased birds. Infect. Immun. 64:3118-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Normark, S., D. Lark, R. Hull, M. Norgren, M. Baga, P. O'Hanley, G. Schoolnik, and S. Falkow. 1983. Genetics of digalactoside-binding adhesin from a uropathogenic Escherichia coli strain. Infect. Immun. 41:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oswald, E., J. De Rycke, J. F. Guillot, and R. Boivin. 1989. Cytotoxic effect of multinucleation in HeLa cell cultures associated with the presence of Vir plasmid in Escherichia coli strains. FEMS Microbiol. Lett. 49:95-99. [DOI] [PubMed] [Google Scholar]
- 46.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097-2110. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36:241-256. [DOI] [PubMed] [Google Scholar]
- 49.Ron, E. Z. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 9:28-32. [DOI] [PubMed] [Google Scholar]
- 50.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 51.Schouler, C., F. Koffmann, C. Amory, S. Leroy-Setrin, and M. Moulin-Schouleur. 2004. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology 150:2973-2984. [DOI] [PubMed] [Google Scholar]
- 52.Scott, D. A., and J. B. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selander, R. K., D. Caugant, and T. S. Whittam. 1987. Genetic structure and variation in natural populations of Escherichia coli, p. 1625-1648. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. I. American Society for Microbiology, Washington, DC. [Google Scholar]
- 54.Selander, R. K., T. K. Korhonen, V. Vaisanen-Rhen, P. H. Williams, P. E. Pattison, and D. A. Caugant. 1986. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect. Immun. 52:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skyberg, J. A., T. J. Johnson, J. R. Johnson, C. Clabots, C. M. Logue, and L. K. Nolan. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chick embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74:6287-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, H. W. 1974. A search for transmissible pathogenic characters in invasive strains of Escherichia coli: the discovery of a plasmid-controlled toxin and a plasmid-controlled lethal character closely associated, or identical, with colicine V. J. Gen. Microbiol. 83:95-111. [DOI] [PubMed] [Google Scholar]
- 57.Stordeur, P., D. Marlier, J. Blanco, E. Oswald, F. Biet, M. Dho-Moulin, and J. Mainil. 2002. Examination of Escherichia coli from poultry for selected adhesin genes important in disease caused by mammalian pathogenic E. coli. Vet. Microbiol. 84:231-241. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence aligment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tóth, I., F. Herault, L. Beutin, and E. Oswald. 2003. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). J. Clin. Microbiol. 41:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urwin, R., and M. C. J. Maiden. 2003. Multilocus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 61.Watt, S., P. Lanotte, L. Mereghetti, M. Moulin-Schouleur, B. Picard, and R. Quentin. 2003. Escherichia coli strains from pregnant women and neonates: intraspecies genetic distribution and prevalence of virulence factors. J. Clin. Microbiol. 41:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White, D. G., M. Dho-Moulin, R. A. Wilson, and T. S. Whittam. 1993. Clonal relationships and variation in virulence among Escherichia coli strains of avian origin. Microb. Pathog. 14:399-409. [DOI] [PubMed] [Google Scholar]
- 64.White, D. G., R. A. Wilson, D. A. Emery, K. V. Nagaraja, and T. S. Whittam. 1993. Clonal diversity among strains of Escherichia coli incriminated in turkey colisepticemia. Vet. Microbiol. 34:19-34. [DOI] [PubMed] [Google Scholar]
- 65.White, D. G., R. A. Wilson, A. S. Gabriel, M. Saco, and T. S. Whittam. 1990. Genetic relationships among strains of avian Escherichia coli associated with swollen-head syndrome. Infect. Immun. 58:3613-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whittam, T. S., and R. A. Wilson. 1988. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect. Immun. 56:2458-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams, P. H. 1979. Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. Infect. Immun. 26:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zingler, G., G. Blum, U. Falkenhagen, I. Orskov, F. Orskov, J. Hacker, and M. Ott. 1993. Clonal differentiation of uropathogenic Escherichia coli isolates of serotype O6:K5 by fimbrial antigen typing and DNA long-range mapping techniques. Med. Microbiol. Immunol. (Berlin) 182:13-24. [DOI] [PubMed] [Google Scholar]
- 69.Zingler, G., M. Ott, G. Blum, U. Falkenhagen, G. Naumann, W. Sokolowska-Kohler, and J. Hacker. 1992. Clonal analysis of Escherichia coli serotype O6 strains from urinary tract infections. Microb. Pathog. 12:299-310. [DOI] [PubMed] [Google Scholar]