Abstract
We report a novel sequence of the serotype II capsular locus of group B streptococcus that resolves inconsistencies among the results of various groups and the sequence in GenBank. This locus was found in diverse lineages and presents genes consistent with the complete synthesis of the type II polysaccharide.
Streptococcus agalactiae (group B streptococcus [GBS]) is an important cause of bacterial sepsis and meningitis in neonates (13) and is increasingly associated with invasive infections in adults (6). The surface-exposed capsular polysaccharide (CPS) is essential for virulence of GBS, and its serotype-specific antibodies are protective, justifying its choice as the target of experimental GBS vaccines. Epidemiological studies have relied on the diversity of CPSs, allowing the differentiation of nine serotypes (Ia, Ib, and II to VIII) (13) to distinguish GBS isolates. Even with the advent of molecular typing methods such as pulsed-field gel electrophoresis (7) and multilocus sequence typing (MLST) (9), serotyping remains an important methodology in epidemiological studies. However, there is a trend to replace the traditional serotyping methods with genotypic methods (4, 10, 16) with increases in accuracy and typeability. Recently, it has been suggested that, similarly to Streptococcus pneumoniae (12), GBS undergoes capsular transformation (9), raising the prospect that the efficacy of any future vaccine may be hampered by capsular switches. The recent identification of serotype V and the genetic structure of its capsular locus imply that this serotype may have emerged recently, indicating that the diversification of GBS CPSs may be ongoing (5). Knowledge of the genetic determinants of existing serotypes is essential for understanding the mechanisms by which antigenic diversity arises, for monitoring putative capsular transformation events, and for the development of genetic methodologies for the determination of serotype.
Previous publications detailing the development of genetic serotyping techniques (10, 16) reported inconsistencies between the results obtained and the sequence of the type II locus deposited in GenBank (accession no. AY375362). The authors attribute these inconsistencies to differences in the GBS clones analyzed (10) or to mistakes in the sequence of the cpsHII gene (16). The original publication reporting the cps type II locus had already noted that its genetic makeup was not congruent with the determined CPS and suggested that the latter could have been in error or that some of the functions necessary for its synthesis could be located outside the cps locus (5).
To examine the capsular locus of GBS strains, we developed a molecular serotyping method based on PCR-based restriction fragment length polymorphism (RFLP) similar to a previously published method (10). Our technique relies on the amplification and double digestion with the MspI and XbaI endonucleases of the cpsG-neuA region of the capsular locus. The expected pattern was inferred from the cps locus sequences deposited in GenBank for serotypes Ia, Ib, II to V, and VII (5), determined using Vector NTI 10 software (Invitrogen, Carlsbad, CA).
Briefly, total bacterial DNA was isolated from bacterial cultures grown overnight in Todd-Hewitt broth (Oxoid, Hampshire, England) at 37°C by mutanolysin, RNase, and pronase treatment; phenol-chloroform-isoamyl alcohol extraction; and ethanol precipitation (1). The cpsG-neuA region of the cps cluster was amplified by PCR with an Expand long-template PCR system (Roche, Manheim, Germany) according to the manufacturer's instructions. The oligonucleotide primers used were loEFrev, previously described (14), and cpsG-UP (GAAGCTGAGATTGTTATCACACATGGCGG). PCR products were purified by using a high pure PCR product purification kit (Roche, Manheim, Germany), simultaneously digested with 10 U of MspI and 10 U of XbaI in Tango buffer (all from Fermentas, Vilnius, Lithuania) and separated in a 2% agarose gel.
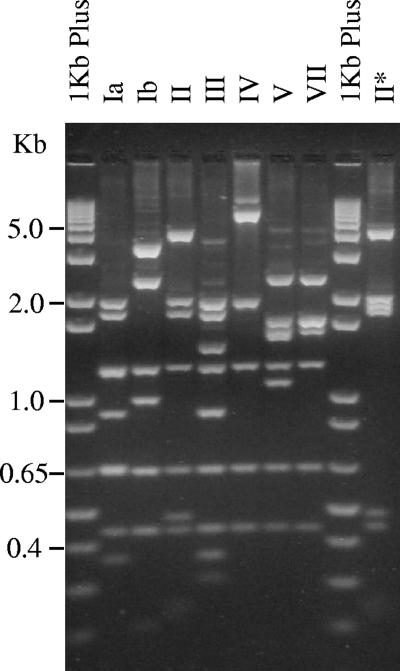
The RFLP patterns of three isolates of each serotype, representing distinct pulsed-field gel electrophoresis clusters, revealed the expected digestion profile discriminating the various serotypes, with the exception of isolates presenting serotype II (Fig. 1), in which both the size of the cpsG-neuA region and the digestion pattern differed from those predicted. In isolates of serotype II, the PCR product was 10.5 kb long and the MspI/XbaI digestion yielded nine fragments (Fig. 1). In contrast, the analysis of the type II sequence available in GenBank (accession no. AY375362) revealed that the amplified fragment was expected to be 8.7 kb long. Furthermore, the in silico digestion of this region resulted in seven fragments, of 293, 428, 626, 1,183, 1,361, 1,715, and 3,080 bp, which was not compatible with the pattern observed (Fig. 1). To exclude the possibility that this discrepancy was due to particular clones, we typed an additional 15 isolates of serotype II from our collection, presenting distinct MLST sequence types (ST) differing in up to five alleles: ST10, ST12, ST28, ST267, and ST292. The MspI/XbaI digestion patterns obtained were identical to one another and to those of the three isolates analyzed initially, with one exception (Fig. 1). Sequencing of the region expected to contain the absent recognition sequence of MspI of the single isolate presenting a different pattern (Fig. 1) indicated that this was due to a single point mutation (G/A at position 12022 of GenBank accession no. EF990364). This mutation was particular to that isolate and was not a characteristic of the genetic lineage, since other isolates sharing ST12 presented the pattern characteristic of serotype II.
FIG. 1.
RFLP-specific band patterns of the capsular loci of serotypes Ia, Ib, II to V, and VII. A fragment encompassing the cpsG-neuA region was digested simultaneously with MspI and XbaI. “1Kb Plus” denotes the 1-kb Plus DNA ladder (Invitrogen, Carlsbad, CA). Numbers on the left indicate the sizes of the fragments. The rightmost lane is a different pattern presented by a single isolate of serotype II that is compatible with a missing recognition sequence of the MspI endonuclease.
Considering these findings and previous observations (10, 16), we decided to sequence the capsule polysaccharide locus of two serotype II isolates: one representing a frequent ST in our collection (strain 318905, ST28) and DSM2134, a reference strain from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). MLST analysis of DSM2134 revealed a larger-than-expected PCR fragment targeting the glcK gene, and sequencing revealed the presence of a mobile genetic element, related to group II introns. A similar finding was reported previously for eight isolates of bovine origin (3), suggesting that this may be a characteristic of a particular lineage; however, no sequence data are available, which prevents the comparison of the data reported here with the previous findings. The sequence of the glcK allele was deposited in GenBank, and for the purpose of MLST analysis, the nucleotide sequence of the mobile genetic element was ignored and ST61 was assigned to the strain.
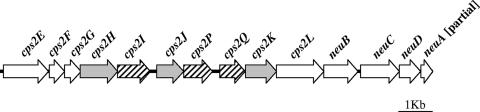
The sequences of the GBS cps2 loci of DSM2134 and 318905 were mostly identical, although these isolates were recovered in different geographic locations years apart, were isolated from cow's milk or responsible for a human infection, and represented two extremely different GBS lineages differing in six of the seven loci used in MLST. As expected, the cps2 locus is constituted by a central “serotype-specific” region flanked by genes that are conserved in all cps loci of GBS (Table 1 and Fig. 2). The central region is larger than the previously reported sequence (5) and presented two additional open reading frames (ORFs), cps2P and cps2Q, that are predicted to encode glycosyl transferases that did not group with any other previously described glycosyl transferases of GBS (Table 1). The remaining “serotype-specific” ORFs were designated cps2I and cps2J since their putative products grouped with high bootstrap values with the corresponding proteins of other GBS cps loci (data not shown). This allowed us to propose a function for every protein encoded in the cps locus such that all of the glycosyl transferases necessary for the synthesis of the type II CPS (8) now reside in this locus, as is common to most streptococcal capsular loci (2). Only 18 substitutions in the protein-coding regions were noted between the two serotype II cps loci, and most of those were synonymous.
TABLE 1.
Characteristics of the putative products of the ORFs associated with S. agalactiae serotype II
| Putative protein | Highest-scoring homologue (GenBank accession no.)a | E-value (TBLASTX) | Organism (serotype)b | Proposed function |
|---|---|---|---|---|
| Cps2E | CpsE (AAL56285) | 0 | S. agalactiae (III) | Glycosyl transferase |
| Cps2F | CpsF (NP_735687) | 8 × 10−83 | S. agalactiae (III) | β-1,4-Galactosyl transferase enhancer |
| Cps2G | CpsG (AAK43608) | 2 × 10−89 | S. agalactiae (IV) | β-1,4-Galactosyl transferase |
| Cps2H | CpsH (NP_688177) | 7 × 10−47 | S. agalactiae (V) | CPS polymerase |
| Cps2Ic | CpsI (AAF18944) | 2 × 10−72 | Streptococcus suis | β-1,3-N-acetylglucosaminyltransferase |
| Cps2J | CpsJ (AAK43612) | 1 × 10−34 | S. agalactiae (IV) | Putative glycosyl transferase |
| Cps2P | BT_2870 (NP_811782) | 4 × 10−21 | Bacteroides thetaiotaomicron | Putative glycosyl transferase |
| Cps2Q | Eps3O (AAL23738) | 3 × 10−29 | Streptococcus thermophilus | Putative glycosyl transferase |
| Cps2K | CpsK (AAK11668) | 2 × 10−52 | S. agalactiae (VI) | α-2,3-N-Acetylneuraminyl transferase |
| Cps2L | CpsL (AAK11669) | 0 | S. agalactiae (VI) | CPS repeat unit transporter |
| NeuB | NeuB (NP_688170) | 0 | S. agalactiae (V) | N-Acetylneuraminic acid synthetase |
| NeuC | NeuC (AAR25957) | 0 | S. agalactiae (VII) | N-Acetylglucosamine-2-epimerase |
| NeuD | NeuD (NP_688168) | 1 × 10−116 | S. agalactiae (V) | Putative acetyltransferase |
| NeuAd | NeuA (AAK43618) | 6 × 10−62 | S. agalactiae (IV) | CMP-N-acetylneuraminic acid synthetase |
Serotype II sequences were excluded from the analysis due to the inconsistencies described in the text.
Whenever the highest-scoring homologue was an S. agalactiae protein, the serotype of the strain is also shown.
Although the BLAST highest-scoring homologue is an S. suis protein, this putative protein groups with a high bootstrap value with the CpsI proteins from other GBS serotypes.
The region sequenced does not include the complete gene.
FIG. 2.
Serotype II CPS synthesis locus of S. agalactiae. ORFs are represented as arrows. The length of each arrow reflects the relative size of the gene. White arrows represent ORFs conserved across all GBS serotypes, gray arrows indicate ORFs with the highest BLAST scoring homolog among ORFs of other GBS serotypes, and hatched arrows indicate ORFs with the highest BLAST scoring homolog among species other than S. agalactiae.
Taken together the data presented above strongly argue that the sequence reported here represents the true cps2 capsular locus. In fact, when searching the available databases for similarities, we identified partial sequences of the cps loci of GBS isolates that share a very high identity to the sequence reported here. One, from an isolate expressing serotype II from yet a different genetic lineage (ST19), was obtained during a genomic survey (15), and the other, also from a type II isolate, became available in GenBank while the manuscript was in preparation (accession no. AM498296) (11). Both of these sequences present only minor changes relative to the sequences reported here.
The RFLP method proposed is able to discriminate accurately among the various GBS cps loci and may be a valuable tool in identifying putative capsular transformation events or clarifying ambiguous serotyping results. Our approach improved on a previously published procedure (10) by producing patterns more easily distinguishable by agarose electrophoresis and by generating a single profile for each serotype. The reappraisal of the cps2 locus identified a serotype-specific region unlike that previously reported. This novel genetic arrangement was found in different GBS genetic lineages expressing serotype II, is compatible with the CPS associated with this serotype (8), and explains previous inconsistencies reported in the literature (10, 16), arguing that it represents the true cps2 locus.
Nucleotide sequence accession numbers.
Nucleotide sequences were deposited in GenBank under accession numbers EF990364 to EF990366.
Acknowledgments
This work was partly supported by Fundação para a Ciência e Tecnologia (POCI/SAU-ESP/57646/2004) and by a grant from Fundação Calouste Gulbenkian.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith (ed.). 1999. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 2.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brito, D. A., M. Ramirez, and H. de Lencastre. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 41:2378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens. 2005. Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 7.Figueira-Coelho, J., M. Ramirez, M. J. Salgado, and J. Melo-Cristino. 2004. Streptococcus agalactiae in a large Portuguese teaching hospital: antimicrobial susceptibility, serotype distribution, and clonal analysis of macrolide-resistant isolates. Microb. Drug Resist. 10:31-36. [DOI] [PubMed] [Google Scholar]
- 8.Jennings, H. J., K. G. Rosell, E. Katzenellenbogen, and D. L. Kasper. 1983. Structural determination of the capsular polysaccharide antigen of type II group B streptococcus. J. Biol. Chem. 258:1793-1798. [PubMed] [Google Scholar]
- 9.Luan, S. L., M. Granlund, M. Sellin, T. Lagergard, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manning, S. D., D. W. Lacher, H. D. Davies, B. Foxman, and T. S. Whittam. 2005. DNA polymorphism and molecular subtyping of the capsular gene cluster of group B streptococcus. J. Clin. Microbiol. 43:6113-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poyart, C., A. Tazi, H. Reglier-Poupet, A. Billoet, N. Tavares, J. Raymond, and P. Trieu-Cuot. 2007. Multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. J. Clin. Microbiol. 45:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez, M., and A. Tomasz. 1999. Acquisition of new capsular genes among clinical isolates of antibiotic-resistant Streptococcus pneumoniae. Microb. Drug Resist. 5:241-246. [DOI] [PubMed] [Google Scholar]
- 13.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 14.Sellin, M., C. Olofsson, S. Hakansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen, L., Q. Wang, Y. Li, F. Kong, G. L. Gilbert, B. Cao, L. Wang, and L. Feng. 2006. Use of a serotype-specific DNA microarray for identification of group B streptococcus (Streptococcus agalactiae). J. Clin. Microbiol. 44:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]