Abstract
Clinical isolates of Bordetella pertussis collected during the year 2004 (n = 153) in eight European countries, Denmark, Finland, France, Germany, The Netherlands, Poland, Sweden, and United Kingdom, were analyzed by pulsed-field gel electrophoresis (PFGE), and their PFGE profiles were compared with those of isolates collected in 1999 (n = 102). The 255 isolates produced 59 distinct PFGE profiles. Among the 153 isolates from 2004, 36 profiles were found, while within the 102 isolates from 1999, 33 profiles were detected. One PFGE profile, BpSR11, was dominant (30% to 50%) in all countries except Denmark (10%) and Poland (0%). In comparison with 1999, there was an increase in BpSR11 prevalence in Finland in 2004 from 5% to 40%, coinciding with a major incidence peak. Some other PFGE profiles seemed to be associated with limited dissemination. Poland was the only country in which the most common actual European PFGE profiles were not found. In a dendrogram analysis, all common PFGE profiles were identified within PFGE group IV, and BpSR11 clustered together with PFGE subgroup IVβ. Compared to the 1999 isolates, PFGE group V representative for pertactin variant prn3 strains had disappeared, and a new cluster was seen. In conclusion, some PFGE profiles, such as BpSR11, evidently have a higher capacity to spread, suggesting increased fitness to the present immunological environment. It is therefore of major interest to continue with surveillance programs of B. pertussis isolates, as both waning vaccine-derived immunity and strain variation may play a role in the persistence of pertussis.
Whooping cough or pertussis is still a significant disease with regular outbreaks despite the introduction of mass vaccination and good coverage of the programs. The resurgence of pertussis has been observed in the United States, Europe, Canada, Asia, and Australia (8, 13, 15, 19, 26, 27). Insight into the polymorphism of Bordetella pertussis, the causative agent of pertussis, and its capacity to adapt to population immunity is important to understand pertussis epidemiology (17).
To investigate B. pertussis strains circulating in the European countries with different vaccination programs and to elucidate possible emergence of bacterial variants with increased fitness, a European research program for strain characterization and surveillance, EUpertstrain, was established. The EUpertstrain I project was initiated in 2001 and supported by the European Commission. Initially, the members participating were the pertussis reference groups from Finland, France, Germany, The Netherlands, and Sweden. The EUpertstrain II project was a continuation of EUpertstrain I and was supported by GlaxoSmithKline (Rixensart, Belgium) and Sanofi Pasteur and Sanofi Pasteur MSD (Lyon, France). In addition to the above-mentioned five countries, Denmark, Poland, and United Kingdom joined the project. All eight participating countries use different vaccination schedules and use at least partly different vaccines (Table 1).
TABLE 1.
Vaccination programs up to 2004 in eight countries participating in the two EUpertstrain projects
| Country | Yrs of vaccination program | Vaccination schedulea | Booster schedulea |
|---|---|---|---|
| Finlandb | 1952 | 3, 4, and 5 mo DTwP | 20 to 24 mo DTwP |
| The Netherlands | 1953 | 2, 3, 4, and 11 mo DTwP-IPV-Hib | |
| France | 1959 | 3, 4, and 5 mo DTwP-IPV | 16 to 8 mo DTwP-IPV |
| 1995 | 2, 3, and 4 mo DTwP-IPV-Hib | 16 to 18 mo DTwP-IPV-Hib | |
| 1998 | 2, 3, and 4 mo DTwP-IPV-Hib | 16 to 18 mo DTaP-IPV-Hib, 11 to 13 yr DtaP-IPV | |
| Germany | 1995 | 2, 3, and 4 mo DTaP-IPV-Hib-HB | 11 to 14 mo DTaP-IPV-Hib-HB |
| 2000 | 2, 3, and 4 mo DTaP-IPV-Hib-HB | 11 to 14 mo DTaP-IPV-Hib-HB, 9 to 17 yr dTaP or dTaP-IPV | |
| Sweden | 1953 | 3, 5, and 12 mo DTwP | |
| 1979-1996 | No vaccination | ||
| 1996 | 3, 5, and 12 mo DTPa | ||
| 1998 | 3, 5, and 12 mo DTaP-IPV-Hib | ||
| Denmark | 1961 | 5, 6, 7, and 15 mo DTwP | |
| 1969 | 5 and 9 wk and 10 mo wP (nonadsorbed) | ||
| 1997 | 3, 5, and 12 mo DTaP-IPV | ||
| 2002 | 3, 5, and 12 mo DTaP-IPV/HIB | ||
| 2003 | 5 yr TdaP | ||
| 2004 | 5 yr Tdap-IPV | ||
| Poland | 1960 | 2, 3 to 4, 5, and 16 to 17 mo DTwP | |
| 2004 | 6 yr DtaP | ||
| United Kingdom | 1950 | 3, 5, and 10 mo DTwP | |
| 1990 | 2, 3, and 4 mo DTwP | ||
| 2000/2001 | DtaP-Hibc | ||
| October 2001 | aP added to preschool DT |
The age of the children (in weeks, months, or years) when the vaccine or booster was given and the specific vaccine are shown in the vaccination and booster schedules. Vaccine abbreviations: DTwP, diphtheria-tetanus-whole-cell pertussis; IPV, inactivated poliovirus; Hib, Haemophilus influenzae type b; DTaP, diphtheria-tetanus-acellular pertussis; HB, hepatitis B; wP, whole-cell pertussis; aP, acellular pertussis.
For Finland, acellular pertussis vaccine was given to children at 6 years of age starting January 2003.
In the United Kingdom, three-component aP was used temporarily in the primary DTaP-Hib vaccine from 2000 to 2001.
For epidemiological characterization of B. pertussis isolates, various DNA-based techniques are available, including pulsed-field gel electrophoresis (PFGE) (2, 4, 12, 16), IS1002-based fingerprinting (18, 23), multilocus sequence typing (24), multilocus variable-number tandem repeat analysis (MLVA) (20, 22), and recently whole-genome DNA microarray (6, 9). Multilocus sequence typing has been used successfully to assess variation in a number of B. pertussis surface protein-encoding genes, including genes encoding pertussis toxin (Ptx), pertactin (Prn), tracheal colonization factor (TcfA), and serotype 3 fimbriae (Fim3) (1, 16, 21).
For epidemiological studies, PFGE has the best discriminatory power, and a standard protocol was chosen as the reference method by a group of experts meeting in Paris in 1999 (16). Reference strains were defined and made available. These strains represented five major PFGE groups (I to V) as well as three subgroups in PFGE group IV (7, 25).
The methods to be chosen for epidemiological typing depend on the objective of the study. In a previous study by Caro et al. (7) the PFGE patterns of EUpertstrain I culture collection strains were studied by means of cluster analysis, adequate for establishing the genetic relationships between the strains.
In this paper we used the discriminatory power of PFGE to identify and trace PFGE profiles represented in material collected from the eight countries participating in EUpertstrain II project. For comparison, EUpertstrain I culture collection strains were also included.
MATERIALS AND METHODS
Collection of strains. (i) EUpertstrain I.
In the EUpertstrain I project, which is also referred to as the 1999 period in this paper, sets of approximately 20 isolates from each country, altogether 102 strains, were collected from children less than 5 years of age in Finland, France, Germany, The Netherlands, and Sweden during 1999 to 2001.
(ii) EUpertstrain II.
In the EUpertstrain II project, which is also referred to as the 2004 period in this paper, a collection of 153 isolates from children in the five countries participating in EUpertstrain I project plus Denmark, Poland (13 isolates only), and United Kingdom during the year 2004 was established. The selection criteria were the same as for EUpertstrain I. All isolates in EUpertstrain I and II were lyophilized at the Swedish Institute for Infectious Disease Control (SMI) and redistributed to each partner.
PFGE methodology.
PFGE was performed at the Swedish Institute for Infectious Disease Control (SMI) for all isolates according to standardized recommendations for typing of B. pertussis (16), with the modifications described previously (2). Due to the stability and high resolution of the PFGE method, it was possible to identify separate profiles. The profiles were analyzed by using BioNumerics software version 4.61 (Applied Maths, NV, Belgium) and defined as distinct DNA band patterns that differed by at least one band and designated BpSR1, BpSR2, BpSR3, and so on for those strains first detected in Sweden. If the strain was first identified in a country other than Sweden, such as Poland, it was designated BpPLR1 and so on. A cluster analysis was also performed using BioNumerics software with the unweighted-pair group method using arithmetic average algorithm with 2% band tolerance and 1.5% optimization settings. For the purpose of comparability, the same band tolerance and optimization settings used in the previous EUpertstrain paper (7) were used. Reference strains 18323 (PFGE group I), Tohama I (PFGE group II), Bp134 (PFGE group III), B902 (PFGE group IVα), FR743 (PFGE group IVβ), FIN12 (PFGE group IVγ), and FR287 (PFGE group V) were included in the dendrogram for traceability.
RESULTS
The 255 isolates of the two EUpertstrain collections produced 59 PFGE profiles (Fig. 1). A total of 33 profiles were detected in the 102 isolates belonging to the EUpertstrain collection of the 1999 period, while 36 profiles were found in the 153 isolates from the 2004 period. Ten PFGE profiles were common in the two periods of study.
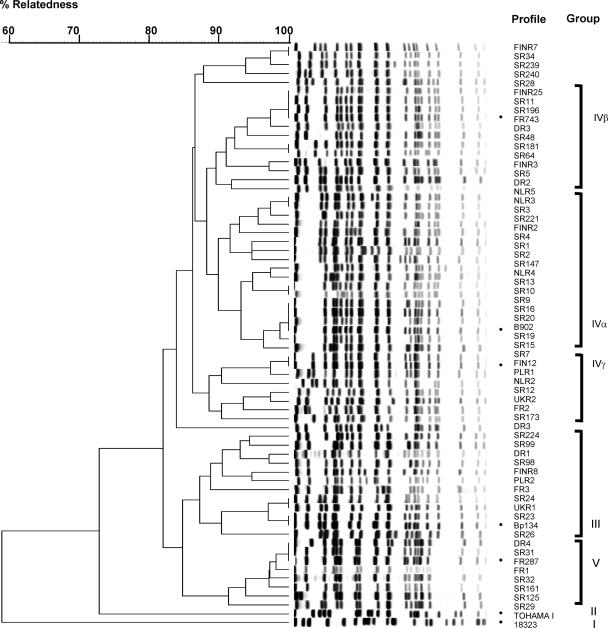
FIG. 1.
Dendrogram of 59 PFGE profiles of B. pertussis circulating in eight European countries in 1999 to 2004. Reference strains for different PFGE groups (7, 25) are indicated (•). The PFGE groups were identified to have an overall relatedness of approximately 83%.
Despite this apparent heterogeneity, some of the profiles appeared to predominate. Table 2 shows the 11 most common profiles found in five or more isolates, covering 53% to 90% of the materials from each country with the exception of Poland. Moreover, 70% of all isolates studied were found to belong to these 11 common PFGE profiles. None of the profiles common in other European countries was found in Poland. In particular, one profile, BpSR11, was predominant in this study. This profile was found in 5% to 45% of the isolates in the collection from the 1999 period and 10% to 50% of the isolates in the collection of the 2004 period, excluding Poland. In comparison with the 1999 period, there was an increase of BpSR11 in Finland from 5% to 40%. BpSR11 was characterized by the following allele combination: ptxA1, ptxC2, prn2, tcfA2, and fim3B (1).
TABLE 2.
Proportions of 11 predominant PFGE profiles identified in eight European countries in the period from 1999 to 2004
| PFGE profile | Cluster/group | Proportion (%) of PFGE profile in the following country and yr:
|
Total no. of isolates | Prevalence (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Finland
|
France
|
Germany
|
The Netherlands
|
Sweden
|
Denmark, 2004 | Poland, 2004 | United Kingdom, 2004 | |||||||||
| 1999 | 2004 | 1999 | 2004 | 1999 | 2004 | 1999 | 2004 | 1999 | 2004 | |||||||
| BpSR11 | IVβ | 5 | 40 | 45 | 50 | 35 | 17 | 30 | 30 | 20 | 35 | 10 | 0 | 45 | 73 | 29 |
| BpSR10 | IVα | 5 | 5 | 10 | 20 | 6 | 6 | 25 | 17 | 0 | 10 | 20 | 0 | 0 | 25 | 10 |
| BpSR5 | IVβ | 5 | 0 | 0 | 0 | 6 | 22 | 15 | 4 | 4 | 5 | 15 | 0 | 15 | 18 | 7 |
| BpSR12 | IVγ | 0 | 0 | 10 | 5 | 6 | 0 | 0 | 4 | 4 | 5 | 25 | 0 | 15 | 15 | 6 |
| BpSR3 | IVα | 0 | 0 | 0 | 10 | 0 | 11 | 0 | 26 | 0 | 0 | 10 | 0 | 0 | 12 | 5 |
| BpSR13 | IVα | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 10 | 0 | 0 | 7 | 3 |
| BpSR16 | IVα | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 6 | 2 |
| BpSR19 | IVα | 0 | 5 | 0 | 0 | 0 | 0 | 20 | 0 | 4 | 0 | 0 | 0 | 0 | 6 | 2 |
| BpSR7 | IVγ | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 6 | 2 |
| BpSR147 | IVα | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 2 |
| BpSR173 | IVγ | 0 | 0 | 5 | 0 | 0 | 17 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 5 | 2 |
| Total | 55 | 60 | 70 | 85 | 53 | 73 | 90 | 85 | 60 | 80 | 90 | 0 | 75 | |||
In both collections, the profiles BpSR10, BpSR5, and BpSR12 were found in six of the eight participating countries at frequencies of 5 to 25%, also indicating a capacity to spread to other countries. The BpSR13 profile was found only in Sweden and Denmark (frequencies of 25% and 10%, respectively), and BpSR16 and BpSR7 were found only in Sweden and Finland (frequencies of 10% to 16%). BpSR173 and BpSR3 were found in France, The Netherlands, and Germany, indicating a limited dissemination of these PFGE types (frequencies of 4% to 26%).
The BpSR7 and BpSR147 profiles were found only during the 1999 period (frequencies of 12% to 15%), and other profiles, like BpSR3 (frequencies of 10% to 26%) and BpSR13 (frequencies of 10% to 25%), were found only during the 2004 period.
There were seven different profiles identified among the Polish isolates. The profiles BpPLR1 and BpPLR2, together representing four isolates, were found exclusively in Polish isolates in the present study. The profile BpSR98, representing three isolates, was also found in Germany. The other Polish profiles, BpSR64 and BpSR98, were found frequently in previous Swedish and French strain collections, while BpSR34, BpSR239, and BpSR240 were found sporadically.
To investigate the genetic relationship between the strains, a cluster analysis was performed (Fig. 1) on strains, including the reference strains for groups I to V, and the genetic relationships between the strains were identified as described elsewhere (16, 25).
The BpSR11 profile clustered with BpSR5 and the group IVβ reference strain FR743. BpSR3, BpSR10, and BpSR13 clustered with the reference strain for group IVα, B902. BpSR12 and BpSR173 clustered with the reference strain FIN12 for group IVγ.
A new cluster was discovered; this cluster included profiles mainly from Poland (BpSR34, BpSR239, and BpSR240) and Finland (BPFINR7) as well as one isolate from Denmark (BpSR28). The remaining Polish isolates clustered with the reference strains for group III (six isolates), group IVβ (three isolates), and group IVγ (one isolate). Group IVβ strains, which were the most common in the other European countries, were represented in Poland by BpSR64. In Finland, strains belonging to groups IVα and IVγ were common during the 1999 period; however, they seemed to be replaced by group IVβ strains more recently (5).
Isolates that clustered with the group V reference strain FR287 were seen sporadically during the 1999 period but not found in the 2004 period.
Differences in the vaccination programs (Table 1) did not seem to have a direct influence on the distribution of profiles. A possible exception is Poland with its very unique pattern of PFGE profiles and the use of a national whole-cell pertussis (Pw) vaccine.
DISCUSSION
In this present work, we studied separate PFGE profiles, as it was shown that these were reproducible and stable, thereby taking advantage of the great discriminatory power of PFGE (2). This type of analysis provides detailed data useful for epidemiology and for the selection of certain predominant profiles, which are of interest for further investigation.
Despite the limited number of isolates from each country, analysis of the EUpertstrain culture collections reveals a gradual expansion of certain PFGE profiles within the B. pertussis population of the participating European countries. BpSR11 represents an example of this, expanding to 30% to 50% in most of the participating countries (Table 2). Strain FR743, the reference strain for group IVβ, was also typed as BpSR11. It was first isolated in France around 1996. Isolates sharing the profile of group IVβ increased in frequency in 1996 to 1997 in the country and has since become the most frequent group in France (7, 25). Some profiles, such as such those in group V, also seem to have disappeared.
A more detailed analysis of temporal changes in the PFGE profiles of Swedish B. pertussis isolates revealed that the BpSR11 profile was first isolated in Uddevalla on the western coast of Sweden in 1997. From 1999, coinciding with a major incidence peak, it was widely spread in Sweden and has been predominant since then (1). The same profile was observed in Finland for the first time in 1999 and was then the most prevalent profile, up to 56% in the incidence peak of 2003 (10). Interestingly, in a Swedish follow-up study, it was shown that BpSR11 was statistically more frequent among pertussis cases with long duration of hospitalization (3).
Weber et al. (25) used a cluster analysis based on the same algorithm for the investigation of relationships between strains. Their French isolates could be classified in five groups. The historical isolates belonged to clusters I and II and were not represented in the circulating strains at all. The other three clusters represented the circulating isolates with a clear trend of shifts from groups III and V to group IV. Grouping is a convenient tool for investigating the lineage of B. pertussis strains over time. In the paper by Caro et al. (7), it was concluded that the isolates belonging to the collection of the 1999 period were found to be very similar and fell into the same major PFGE groups, with a predominance of groups IVβ (44.6%) and IVα (22.8%).
A strong association between PFGE profiles/clusters with prn type and to a lesser extent with fimbrial serotype has been demonstrated in the previous studies (11, 14, 25). Furthermore, a strong association between PFGE profiles and different combinations of the alleles for the ptxA, ptxC, prn, tcfA, and fim3 genes was found in a recent study on the B. pertussis population in Sweden during an acellular pertussis vaccine period between 1997 and 2004, although this is not reflected in PFGE groups (1). Isolates with the allele combinations 1/2/2/2/B (BpSR11, BpSR5, and BpSR12) and 1/2/2/2/A1 (BpSR3, BpSR10, and BpSR13) for the ptxA, ptxC, prn, tcfA, and fim3 genes, respectively, replaced profiles with allele combinations 1/1/2/2/A1 (BpSR16) and 1/1/3/3/A1 (BpSR31) during a period of 7 years after the introduction of acellular pertussis vaccines (1). There seems to be a similar trend in the EUpertstrain results.
Interestingly, Poland was the only country in this study in which the most common European PFGE profiles were not found. This may be partially explained by the very limited number of strains (n = 13) included in this study. Further study with a large number of isolates is needed to ensure that they are representative. It was noted that several changes of vaccine strains had occurred in the nationally produced DTwP (diphtheria-tetanus-whole-cell pertussis) vaccine. It is also important to make these strains available for extended analyses of properties.
In conclusion, some PFGE profiles, such as BpSR11, evidently have a higher capacity to spread, suggesting they show increased fitness in the present immunological environment. Some other profiles are seen mainly in local outbreaks and seem to persist for shorter periods of time. Isolates expressing the genetic marker prn1 were associated with PFGE group III, and isolates expressing prn3 were associated with group V, a group that probably represents “older” types. In this context, it is interesting to notice the fact that both the whole-cell and acellular vaccines currently used in Europe today most often are derived from strains producing Prn1. From a vaccination point of view, it is important to study the background and possible consequences of B. pertussis polymorphism and the mechanisms that might influence the bacterial adaptation to population immunity. It is therefore of major interest to continue with surveillance programs of B. pertussis isolates, as both waning vaccine-derived immunity and strain variation may play a role in the persistence of pertussis.
Acknowledgments
This work was supported by GlaxoSmithKline (Rixensart, Belgium), Sanofi Pasteur, and Sanofi Pasteur MSD (Lyon, France).
Footnotes
Published ahead of print on 15 August 2007.
REFERENCES
- 1.Advani, A., D. Donnelly, L. Gustafsson, and H. O. Hallander. 2007. Changes of the Swedish Bordetella pertussis population in incidence peaks during an acellular pertussis vaccine period between 1997 and 2004. APMIS 115:299-310. [DOI] [PubMed] [Google Scholar]
- 2.Advani, A., D. Donnelly, and H. Hallander. 2004. Reference system for characterization of Bordetella pertussis pulsed-field gel electrophoresis profiles. J. Clin. Microbiol. 42:2890-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advani, A., L. Gustafsson, R.-M. Carlsson, D. Donnelly, and H. Hallander. 2007. Clinical outcome of pertussis in Sweden: association with pulsed-field gel electrophoresis profiles and serotype. APMIS 115:736-742. [DOI] [PubMed] [Google Scholar]
- 4.Bisgard, K. M., C. D. Christie, S. F. Reising, G. N. Sanden, P. K. Cassiday, C. Gomersall, W. A. Wattigney, N. E. Roberts, and P. M. Strebel. 2001. Molecular epidemiology of Bordetella pertussis by pulsed-field gel electrophoresis profile: Cincinnati, 1989-1996. J. Infect. Dis. 183:1360-1367. [DOI] [PubMed] [Google Scholar]
- 5.Caro, V., A. Elomaa, D. Brun, J. Mertsola, Q. He, and N. Guiso. 2006. Bordetella pertussis, Finland and France. Emerg. Infect. Dis. 12:987-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caro, V., D. Hot, G. Guigon, C. Hubans, M. Arrive, G. Soubigou, G. Renauld-Mongenie, R. Antoine, C. Locht, Y. Lemoine, and N. Guiso. 2006. Temporal analysis of French Bordetella pertussis isolates by comparative whole-genome hybridization. Microbes Infect. 8:2228-2235. [DOI] [PubMed] [Google Scholar]
- 7.Caro, V., E. Njamkepo, S. C. Van Amersfoorth, F. R. Mooi, A. Advani, H. O. Hallander, Q. He, J. Mertsola, M. Riffelmann, C. Vahrenholz, C. H. Von Konig, and N. Guiso. 2005. Pulsed-field gel electrophoresis analysis of Bordetella pertussis populations in various European countries with different vaccine policies. Microbes Infect. 7:976-982. [DOI] [PubMed] [Google Scholar]
- 8.Celentano, L. P., M. Massari, D. Paramatti, S. Salmaso, and A. E. Tozzi. 2005. Resurgence of pertussis in Europe. Pediatr. Infect. Dis. J. 24:761-765. [DOI] [PubMed] [Google Scholar]
- 9.Cummings, C. A., M. M. Brinig, P. W. Lepp, S. van de Pas, and D. A. Relman. 2004. Bordetella species are distinguished by patterns of substantial gene loss and host adaptation. J. Bacteriol. 186:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elomaa, A., A. Advani, D. Donnelly, M. Antila, J. Mertsola, Q. He, and H. Hallander. 2007. Population dynamics of Bordetella pertussis in Finland and Sweden, neighbouring countries with different vaccination histories. Vaccine 25:918-926. [DOI] [PubMed] [Google Scholar]
- 11.Hallander, H. O., A. Advani, D. Donnelly, L. Gustafsson, and R.-M. Carlsson. 2005. Shifts of Bordetella pertussis variants in Sweden from 1970 to 2003, during three periods marked by different vaccination programs. J. Clin. Microbiol. 43:2856-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodama, A., K. Kamachi, Y. Horiuchi, T. Konda, and Y. Arakawa. 2004. Antigenic divergence suggested by correlation between antigenic variation and pulsed-field gel electrophoresis profiles of Bordetella pertussis isolates in Japan. J. Clin. Microbiol. 42:5453-5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, Y. S., C. Y. Yang, C. H. Lu, and Y. H. Tseng. 2003. Molecular epidemiology of Bordetella pertussis isolated in Taiwan, 1992-1997. Microbiol. Immunol. 47:903-909. [DOI] [PubMed] [Google Scholar]
- 14.Lin, Y.-C., S.-M. Yao, J.-J. Yan, Y.-Y. Chen, M.-J. Hsiao, C.-Y. Chou, H.-P. Su, H.-S. Wu, and S.-Y. Li. 2006. Molecular epidemiology of Bordetella pertussis in Taiwan, 1993-2004: suggests one possible explanation for the outbreak of pertussis in 1997. Microbes Infect. 8:2082-2087. [DOI] [PubMed] [Google Scholar]
- 15.McIntyre, P., H. Gidding, R. Gilmour, G. Lawrence, B. Hull, P. Horby, H. Wang, R. Andrews, M. Burgess, and National Centre for Immunisation Research and Surveillance of Vaccine Preventable Diseases. 2002. Vaccine preventable diseases and vaccination coverage in Australia, 1999 to 2000. Commonwealth Dept. of Health and Ageing, Canberra, Australia. [DOI] [PubMed]
- 16.Mooi, F. R., H. Hallander, C. H. Wirsing von Konig, B. Hoet, and N. Guiso. 2000. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur. J. Clin. Microbiol. Infect. Dis. 19:174-181. [DOI] [PubMed] [Google Scholar]
- 17.Mooi, F. R., I. H. van Loo, and A. J. King. 2001. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg. Infect. Dis. 7:526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. van der Heide, W. Gaastra, and R. J. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ntezayabo, B., G. De Serres, and B. Duval. 2003. Pertussis resurgence in Canada largely caused by a cohort effect. Pediatr. Infect. Dis. J. 22:22-27. [DOI] [PubMed] [Google Scholar]
- 20.Schouls, L. M., H. G. J. van der Heide, L. Vauterin, P. Vauterin, and F. R. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsang, R. S. W., A. K. H. Lau, M. L. Sill, S. A. Halperin, P. Van Caeseele, F. Jamieson, and I. E. Martin. 2004. Polymorphisms of the fimbria fim3 gene of Bordetella pertussis strains isolated in Canada. J. Clin. Microbiol. 42:5364-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Amersfoorth, S. C. M., L. M. Schouls, H. G. J. van der Heide, A. Advani, H. O. Hallander, K. Bondeson, C. H. W. von Konig, M. Riffelmann, C. Vahrenholz, N. Guiso, V. Caro, E. Njamkepo, Q. He, J. Mertsola, and F. R. Mooi. 2005. Analysis of Bordetella pertussis populations in European countries with different vaccination policies. J. Clin. Microbiol. 43:2837-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Zee, A., S. Vernooij, M. Peeters, J. van Embden, and F. R. Mooi. 1996. Dynamics of the population structure of Bordetella pertussis as measured by IS1002-associated RFLP: comparison of pre- and post-vaccination strains and global distribution. Microbiology 142:3479-3485. [DOI] [PubMed] [Google Scholar]
- 24.van Loo, I. H., K. J. Heuvelman, A. J. King, and F. R. Mooi. 2002. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J. Clin. Microbiol. 40:1994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber, C., C. Boursaux-Eude, G. Coralie, V. Caro, and N. Guiso. 2001. Polymorphism of Bordetella pertussis isolates circulating for the last 10 years in France, where a single effective whole-cell vaccine has been used for more than 30 years. J. Clin. Microbiol. 39:4396-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. 2001. Pertussis surveillance: a global meeting, Geneva, 16-18 October 2000. World Health Organization document no. WHO/V&B/01.19. Department of Vaccines and Biologicals, World Health Organization, Geneva, Switzerland. http://www.who.int/vaccines-documents/DocsPDF01/www605.pdf.
- 27.Zanardi, L., F. Pascual, K. Bisgard, T. Murphy, and M. Wharton. 2002. Pertussis—United States,1997-2000. Morb. Mortal. Wkly. Rep. 51:73-76. [PubMed] [Google Scholar]