Abstract
We report the development and evaluation of a Salmonella O-group-specific Bio-Plex assay to detect the six most common serogroups in the United States (B, C1, C2, D, E, and O13) plus serotype Paratyphi A. The assay is based on rfb gene targets directly involved in O-antigen biosynthesis; it can be completed 45 min post-PCR amplification. The assay correctly and specifically identified 362 of 384 (94.3%) isolates tested in comparison to traditional serotyping. Seventeen isolates (4.4%) produced results consistent with what is known about the molecular basis for serotypes but different from the results of traditional serotyping, and five isolates (1.3%) generated false-negative results. Molecular determination of the serogroup for rough isolates was consistent with a common serotype in most instances, indicating that this approach has the potential to provide O-group information for isolates that do not express an O antigen. We also report the sequence of the O-antigen-encoding rfb gene cluster from Salmonella enterica serotype Poona (serogroup O13). Compared with other, previously characterized rfb regions, the O13 rfb gene cluster was most closely related to Escherichia coli O127 and O86. The O-group Bio-Plex assay described here provides an easy-to-use, high-throughput system for rapid detection of common Salmonella serogroups.
Serotyping of salmonellae is a valuable phenotypic subtyping tool for understanding the epidemiology of this important food-borne pathogen. Salmonella isolates are serotyped using the Kauffmann-White scheme according to their O, H, and Vi antigens (4, 40). The O antigen contains multiple repeats of an oligosaccharide unit (O unit), which, together with lipid A and core oligosaccharides, form the lipopolysaccharide present in the outer membranes of gram-negative bacteria (7). Many of the genes required for O-antigen biosynthesis are organized in a large regulon termed the rfb gene cluster (47). rfb gene clusters have been characterized from a growing number of gram-negative bacteria; this operon is located between galF and gnd in Salmonella enterica and Escherichia coli (49). In general, rfb genes have a low G+C content (usually less than 40%); the deviation in G+C content from that of typical S. enterica genes (51%) suggests that rfb DNA originated in species other than S. enterica and was captured by lateral gene transfer (46, 56). Typically, three classes of genes are found in rfb clusters: (i) genes for synthesis of nucleotide sugars specific to the respective O antigen, (ii) sugar transferase genes to build the O subunit, and (iii) the O-antigen polymerase (wzy) and transport protein (wzx) genes for assembly of the O subunit into the O antigen (49).
There are 46 O serogroups described in the Kauffmann-White scheme; serogroups were originally designated by alphabetic letters, and then it was necessary to continue with numbers 51 to 67. For consistency in the scheme, all serogroups were given a number designation; however, the most common serogroups (A to E) are commonly designated by letter.
The antigenic differences among the 46 Salmonella O serogroups are due mainly to genetic variation in their respective rfb gene clusters (58). The rfb gene clusters from 15 Salmonella serogroups have been characterized or sequenced: A (32), B (24), C1 (28), C2 (5), D1 (32), D2 (9), D3 (9), E (54), O6,14 (18), O17 (19), O18 (19), O35 (56), O30, O50 (50), and O54 (26). The O antigens of serogroups A, B, C2, D1, D2, D3, and E have related structures, and their rfb regions are related. The Salmonella enterica serotype Paratyphi A rfb region has been shown to be a minor modification of a serogroup D1 rfb region (32); it has a frameshift mutation that inactivates tyv, a sugar biosynthesis gene required for the biosynthesis of tyvelose. Serogroup D1 strains have tyvelose as their O-antigen side chain sugar, whereas serotype Paratyphi A has paratose, the substrate for tyvelose, as its side chain sugar. Thus, a small genetic change is responsible for a substantial O-antigen difference. Serogroup E1 originally was divided into three serogroups (E1, E2, and E3); later, the Kauffmann-White scheme classified E2 (O3,15) and E3 (O3,15,34) strains with O3,10 strains as belonging to serogroup E1 (42), since the antigenic differences detected between the three O groups are thought to be due to the presence of lysogenic phage. The rfb gene cluster from a serogroup E1 strain has been sequenced; hybridization data from Xiang et al. (58) suggest that serogroup E2, E3, and E4 isolates have rfb gene clusters very similar to that of E1. Serogroup O54 is a heterogeneous group, with 14 serotypes currently reported; this O antigen is encoded on a plasmid (26, 41), and strains harboring this plasmid still have a genomic rfb region. The rare serogroup O67 is a known variant of serogroup B (31). Three Salmonella O-antigen structures are known to be shared with other enteric organisms: Salmonella serogroup O35 and E. coli O111 have the same antigen structure, as do Salmonella serogroup O50 and E. coli O55, and Salmonella serogroup O30, E. coli O157, and Citrobacter freundii F90. The respective rfb gene clusters from Salmonella, E. coli, and C. freundii with identical O antigens are, in turn, also highly related to one another (50, 56).
Despite the utility of serotyping, problems associated with antiserum production and isolates for which the serotype antigen cannot be detected have led many to take advantage of molecular approaches to serotyping. The sequences of sugar transferase genes and of the transport (wzx) and O-antigen polymerase (wzy) genes tend to be specific to a particular O antigen. PCR assays based on these O-antigen-specific genes have been proposed for molecular typing of a growing number of pathogens, including Salmonella, E. coli, and Shigella spp. (12, 16, 18, 20, 34, 55, 57). These assays are an attractive alternative to traditional approaches to O-antigen determination, but they do not adapt easily to a multiplex format. In addition, these gene targets can be highly diverse at the DNA sequence level, even among serogroups of the same species, and they are not always in the same gene order in each rfb cluster. Thus, it can be difficult to target and sequence these genes. It is often necessary to sequence the entire rfb gene cluster in order to identify the relevant O-group-specific genes to target for molecular typing.
We report here the development of a sevenplex, bead-based suspension array to detect the six most common serogroups in the United States (B, C1, C2, D, E, and O13), plus serotype Paratyphi A, using Luminex technology (13). This approach combines PCR with a multiplexed bead-based suspension array detection system. O-antigen rfb gene cluster DNA sequences, on which to base PCR primer and probe selection, were publicly available for six of the seven serogroups targeted. No rfb DNA sequence was available for Salmonella serogroup O13, so we also report here the characterization of the rfb gene cluster from Salmonella serogroup O13. The O-antigen gene clusters of Salmonella O13, E. coli O127, and E. coli O86 were shown to be closely related.
MATERIALS AND METHODS
Strains.
All isolates were from the Salmonella Reference Collection in the Foodborne and Diarrheal Diseases Branch, CDC. Isolates were serotyped according to the method of Brenner and McWhorter-Murlin (4). Serotypes were designated according to the Kauffmann-White scheme (38). Salmonella enterica stock strain 99-0277 (Salmonella enterica serotype Poona, 13,22:z:1,6) was selected for rfb sequence analysis. The O-group sevenplex assay was evaluated in parallel with traditional serotyping methods on a panel of 368 isolates that represented all subspecies/serogroup combinations in the current Kauffmann-White scheme.
DNA sequencing of the Salmonella O13 rfb gene cluster.
The Salmonella serogroup O13 rfb region was amplified by PCR using the Expand Long PCR kit (Roche) according to the manufacturer's instructions and oligonucleotide primers corresponding to the middle of the JUMPstart sequence and the 5′ end of gnd, as previously described (31). A combination of subcloning and primer walking was used to sequence the rfb region; all sequencing was performed on a Perkin-Elmer Applied Biosystems 377 automated DNA sequencer using the BigDye Terminator cycle sequencing ready reaction mix according to the manufacturer's instructions (Perkin-Elmer Applied Biosystems, Foster City, CA). DNA sequence data were assembled and analyzed using Lasergene 99 (DNASTAR) software. The National Center for Biotechnology Information (NCBI) BLAST network server was used to search sequence databases for sequences with homology to the open reading frames (ORFs) found in the O13 rfb sequence. The TMHMM program was used for predicting the numbers of transmembrane helices in the Wzx and Wzy proteins (27). The ClustalW and T-coffee programs (39) were used for performing sequence alignments, and the Pfam protein family database (2) was searched for protein domain similarity.
O-group Bio-Plex assay. (i) PCR amplification.
PCR primers corresponding to specific rfb genes were used to generate amplicons of 90 to 417 bp depending on the O group (Table 1). The forward primer was biotinylated at the 5′ end to allow binding of the reporter dye, streptavidin-R-phycoerythrin, and subsequent detection on the Bio-Plex platform. The target sequences for all O groups were amplified in a single multiplex PCR. PCR amplification was performed in 25-μl volumes using a Hot Start PCR kit (Promega, Madison, WI) according to the manufacturer's instructions. PCR amplification was carried out in a thermal cycler (MJ Research, MA) with the following parameters: initial denaturation at 95°C for 15 min; then 30 cycles of 94°C for 30 s (denaturation), 48°C for 60 s (annealing), and 72°C for 90 s (extension); and 72°C for 10 min (final extension). PCR amplicons were then used directly with the coupled beads in the hybridization reaction described below, and the O-group targets were detected in a single Bio-Plex assay.
TABLE 1.
Group primers and capture probe information
| Serogroup | Gene target | Forward PCR primer | Reverse PCR primer | O-group-specific capture probe |
|---|---|---|---|---|
| Serotype Paratyphi A/D | prt | GA GTT TAT ATG CAT ATA CTA A | TAC TGG TAA ACT TAT C | GC GGC GGC GAA CTC ATT Ta/GC GGC GGC GAG TTC ATT Tb |
| TGC TGG TAA ATT TAT Cc | GT GGC TGC GAG TTC ATT Tc | |||
| B | abe | GC ATA AAA CAT CAT CTC TCA TAA A | CGC ATA TGT TGA TAA TTA AAA TCT TT | TT TAT TGC CAA ATC AAG AAG C TT TAA TGG TTT TA |
| C1 | wzy | GC TTT AAT CCA ATA TAA AA | TAA ATA TAT GAT AGT TCC AAA TAA | AT GAA ATG TCG ACG CAC ATA GAA TAA GAA TAA GC |
| C2 | abe | CA GAC GTT TGT TTT ATA A | GAT CGT ACA ATC AAT ATC | TC CCT CTC ACC GTA AAC ATG TTC TAA ACG TAA ATT |
| E | wzx | CT GAT TTA ACC GGG TAT TAT TTA T | CTT TTC TAT GCA GTG GTT TAA | AG GAT ATA TGT TAA GGC TGT ACT TAT ACC TAG CT |
| O13 | wzy | CT CTT GAT GAA TGT TAT TA | GTT AAC CCC TCC TAA TA | AT ATA TAG ATG TTT ATT ATA TTT GCA TTC CCC A |
Serogroup A probe.
Serogroup D probe.
Reverse primer and probe sequence for subspecies II serogroup D.
(ii) Coupling of probes to microspheres (beads).
Oligonucleotide probes based on rfb genes specific for serotype Paratyphi A and serogroups B, C1, C2, D, and O13 (Table 1) were synthesized with an amino-modified 6-carbon spacer at the 5′ end of each probe. Carbodiimide coupling (13) was then used to covalently attach the probes to carboxylated beads. In brief, 200 pmol of each oligonucleotide probe, 2.5 × 106 microspheres (Luminex Corp., Austin, TX), and 25 μg of freshly prepared N-(3-dimethylaminodipropyl)-N′-ethylcarbodiimide (EDC; Pierce Chemicals, IL) were combined in 25 μl of 100 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 4.5. The reaction mixtures were incubated in the dark at room temperature for 30 min with constant shaking, and the EDC treatment and subsequent incubation step were repeated once. After coupling, the microspheres were washed with 1.0 ml of 0.02% Tween 20 (Sigma, St. Louis, MO), followed by a second wash with 0.5 ml of 0.1% sodium dodecyl sulfate (Sigma). Coupled microspheres were then suspended in 50 μl of TE buffer (0.01 M Tris-EDTA, pH 8.0) and stored at 4°C in the dark. All seven of the O-group-specific bead sets were mixed by combining equal volumes (3 μl) of each, and the mixture was stored in the dark at 4°C until use. The hybridization capture probes are listed in Table 1.
(iii) Hybridization of bead-coupled probes to PCR amplicons.
Immediately before use in the hybridization reaction, the O-group bead mixture was brought to a final volume of 1 ml in 1.5× TMAC buffer (1.5 M tetramethylammonium chloride, 75 mM Tris, 6 mM EDTA, and 0.15% Sarkosyl [pH 8.0]). For the O-group Bio-Plex assay, 33 μl of bead mix was added to 5 μl of O-group PCR product and 12 μl of TE buffer in a single well of a 96-well microtiter plate (Thermowell; VWR International, West Chester, PA). The reaction mixture was incubated first for 5 min at 94°C and then for 30 min at 52°C to denature the DNA and allow hybridization of the probes to the PCR amplicons.
(iv) Detection of hybridized amplicons.
The microspheres were then suspended in 75 μl of detection buffer (R-phycoerythrin-conjugated streptavidin [Molecular Probes, Eugene, OR] diluted to 4 μg/ml in 1× TMAC hybridization buffer). Samples were incubated for an additional 10 min at 52°C and then analyzed on the Bio-Plex (Bio-Rad, Hercules, CA). The median fluorescence intensity (MFI) for each bead set was calculated automatically by the Bio-Plex software. A positive signal was defined as an MFI giving 6 times the background fluorescence intensity for each bead-probe set.
PFGE analysis.
Preparation of Salmonella DNA, macrorestriction analysis using the restriction enzymes XbaI and BlnI, and pulsed-field gel electrophoresis (PFGE) were performed according to the standardized PulseNet Salmonella protocol (48). Thiourea was added to the PFGE running buffer to prevent degradation of the DNA from Salmonella enterica serotype Koessen and Salmonella enterica serotype Panama (45). Gel analysis was performed using BioNumerics (version 4.0; Applied Maths, Sint-Martens-Latem, Belgium). Cluster analysis with BioNumerics was determined with the Dice coefficient using the unweighted-pair group method with arithmetic averages. Isolates were assigned to the same PFGE pattern when they clustered at 100% similarity (when they were indistinguishable by the similarity matrix and visually with settings of 1.50 for both optimization and position tolerance with both enzymes).
Nucleotide sequence accession number.
The sequence of the Salmonella serogroup O13 rfb gene cluster has been deposited in GenBank and assigned accession number EF204526.
RESULTS
Salmonella serogroup O13 rfb cluster.
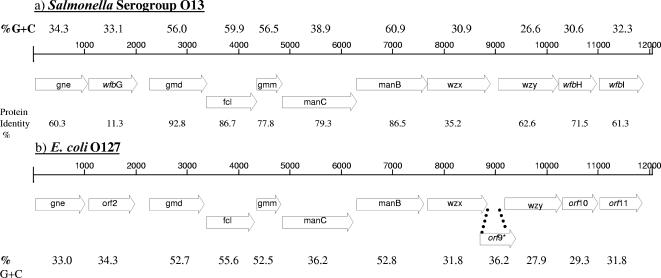
Analysis of the DNA sequence from the Salmonella enterica serogroup O13 rfb gene cluster revealed 12.0 kb flanked by JUMPstart and the gnd gene and containing 11 ORFs (Fig. 1). The sequences were analyzed using BLAST, and the genes were named in accordance with bacterial polysaccharide gene database (BPGD) nomenclature (46). Seven of the 11 genes from the serogroup O13 gene cluster had G+C contents ranging from 26.6 to 38.9% (Fig. 1), a feature characteristic of other rfb gene clusters. The other four genes, gmd, fcl, gmm, and manB, had G+C contents ranging from 56.0 to 60.9%, respectively. All of the genes from serogroup O13 were transcribed in the same direction. Further details on the sequence analysis of the Salmonella serogroup O13 rfb gene cluster can be found in the supplemental material.
FIG. 1.
Comparison of organization and G+C contents of rfb gene clusters from Salmonella O13 (a) (this study) and E. coli O127 (b) (17). The percentage of G+C was calculated and plotted for each 100 bases. Putative ORFs are represented by open arrows in which the assigned gene names are given. *, Salmonella O13 glycosyltransferases had 3.2% to 11.7% amino acid identity to E. coli O127 orf9.
The rfb gene cluster from Salmonella serogroup O13 had high amino acid identities to those of the two closely related E. coli O antigens, O127 and O86 (17). Nine of 11 Salmonella O13 genes shared 61% to 93% amino acid identity with the corresponding genes in E. coli O127 (Fig. 1). This included 63% identity between the respective O-antigen polymerase genes from the two serogroups; this gene is usually very divergent even between Salmonella serogroups. The O-antigen polymerase forms an α-(1→2) linkage between O units in E. coli O127; the same linkage may be formed between Salmonella O13 subunits. The O-antigen transport gene, wzx, had 35.2% protein identity between the two serogroups. One glycosyltransferase gene (wfbG) from Salmonella O13 had low protein homologies to orf2 in E. coli O127 and wcmA in E. coli O86, genes that also encode glycosyltransferases. The E. coli O127 and O86 transferases are responsible for an α-d-N-acetylgalactosamine (GalNAc)-(1→3)-β-d-GalNAc linkage, which suggests that this linkage may not be present in the Salmonella O13 antigen structure. One gene each present in the E. coli O127 (orf9) and O86 (wcmB) rfb gene clusters, encoding a glycosyltransferase for the α-d-Gal-(1→3)-β-d-Gal linkage in E. coli O86 and an O-acetyltransferase in O127, is not seen in the Salmonella O13 rfb cluster, again suggesting that this particular linkage and an O-acetyl group are not present in the Salmonella O13 rfb cluster.
O-group Bio-Plex primer and probe design.
Visual OMP (version 4.2) software (DNA Software Inc., Ann Arbor, MI) was used to design and simulate the performance of the O-group PCR primers and capture probes. PCR primers and probes were based on gene targets from previously described PCR assays for serogroups A/D, B, and C2 and from new PCR assays described in this study for serogroups C1, E, and O13 (Table 1). Luk et al. (34) described three PCR assays for identification of Salmonella serogroups A/D, B, and C2 based on sequences within the rfb genes encoding abequose (abe) and paratose synthase (prt). We targeted the same rfb genes for the development of primers and probes specific for serogroups B, C2, and D and serotype Paratyphi A (Table 1), except that the sizes of the PCR amplicons were adjusted to optimize positive-signal intensity on the Bio-Plex platform; the primers were designed to make smaller amplicons of 122 bp to 150 bp (Table 1). The rfb gene cluster of serogroup A is highly related to that of serogroup D (32); the PCR assay for serogroup A/D described by Luk et al. (34) did not differentiate between these two serogroups. Alignment of the rfb gene clusters from the genome sequences of Salmonella enterica serotype Enteritidis (PT4), Salmonella enterica serotype Typhi (TY2 and CT18), and Salmonella serotype Paratyphi A (ATCC 9150) (www.salmonella.org) showed a 2-bp difference between the serogroup D rfb DNA sequences and that of Salmonella serotype Paratyphi A in the PCR target region. We targeted this 2-bp difference for design of the serogroup D- and serotype Paratyphi A-specific capture probes. The rfb gene clusters from serogroups C1 and E have been sequenced previously (28, 54). We targeted wzy (O-antigen polymerase gene) and wzx (O-antigen transport gene) to generate a 417-bp and a 117-bp PCR amplicon, respectively. O13-specific PCR primers were derived from a sequence within the O-antigen polymerase gene, wzy (Fig. 1), to generate a 90-bp PCR amplicon. Initially, capture probes of 18 to 21 bp were designed for each O group. By lengthening the size of the probes to 33 to 35 bp for serogroups B, C1, C2, and O13, positive-signal intensity was increased 10-fold. For serogroup D and serotype Paratyphi A, where the DNA sequence difference between the two target regions is 2 bp, a probe length of 18 bp gave optimum positive-signal intensity. Probe hybridization was evaluated at different temperatures (37°C, 52°C, 53°C), and the fluorescence signal intensity and stringency of hybridization were found to be optimal at a hybridization temperature of 52°C.
Data output and analysis.
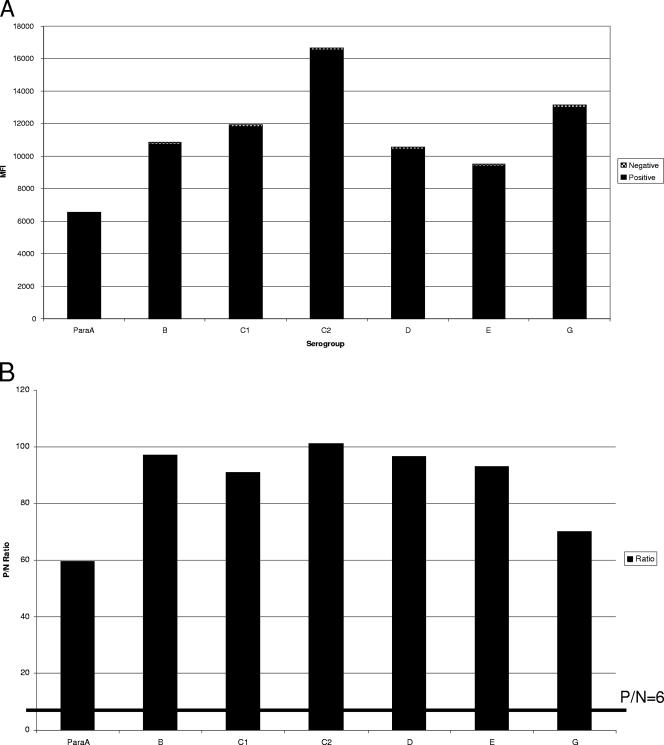
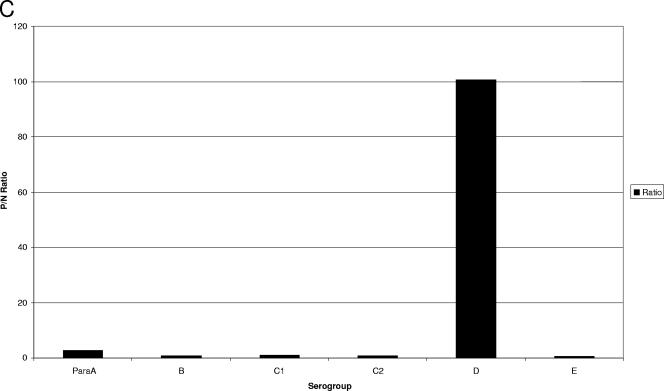
For the O-group Bio-Plex assay, the Bio-Plex instrument is set to measure the reporter fluorescence for a minimum of 100 microspheres of each of the seven microsphere sets present in a given reaction well. The output, or “raw data,” is reported as the MFI measured for each of the seven O-group-specific microsphere sets. Bio-Plex software can also generate a “ratio” for each microsphere set in a given reaction well. This is generated by dividing the MFI of each isolate for a given microsphere set by the MFI obtained for a negative control (consisting of all reaction components except DNA, which is replaced by water). An arbitrary cutoff ratio of 6 times the background was established to score a particular isolate as positive or negative with each O-group-specific capture probe microsphere set. Examples of raw and ratio data averaged for five isolates for each O-group microsphere bead set are shown in Fig. 2. To give an example of the ratio data for all seven bead sets for a given sample, Fig. 2C shows the ratio values for a Salmonella enterica serotype Dublin isolate, which belongs to serogroup D. The isolate had a positive ratio of 100.15 for serogroup D, while it was negative with the other six bead sets, with ratios ranging from 0.58 to 2.7.
FIG. 2.
Examples of raw MFIs (A) and positive/negative (P/N) ratios (B) for each O-group-specific bead set averaged for five positive samples each. (C) P/N ratios for each O-group-specific bead set for a Salmonella serovar Dublin isolate.
Comparison of O-group determination by traditional versus molecular serotyping.
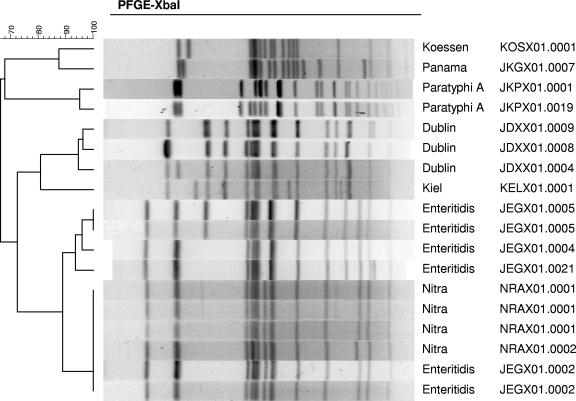
The O-group Bio-Plex assay was validated on a panel of 393 strains in parallel with traditional methods. A comparison of the serogroup data generated by the O-group Bio-Plex assay and traditional serotyping is shown in Table 2. For 384 isolates, an O group could be determined by traditional methods, and the traditional and molecular methods identified the same O group for 362 of the 384 (94%). Seventeen isolates produced different results by the two methods, but these were consistent with what is known about the phenotypic and genetic bases for serotypes. An isolate of Salmonella enterica serotype Crossness, the only serotype belonging to serogroup O67, was positive for serogroup B by the Bio-Plex assay, but O67 is a minor variant of serogroup B (31). Serogroup O54 is encoded on a plasmid, and serogroup O54 strains also have a chromosomally encoded rfb region; 10 of 14 serogroup O54 serotypes were positive for one of the serogroups in the Bio-Plex assay (Table 3). Serogroup A consists of four serotypes including serotype Paratyphi A. Isolates of the three serogroup A serotypes Salmonella enterica serotype Nitra (4), Salmonella enterica serotype Kiel (1), and Salmonella enterica serotype Koessen (1) were positive with the serogroup D probe and negative with the serotype Paratyphi A probe. These six serogroup A isolates were subtyped with a representative isolate from the corresponding serotype in serogroup D by PFGE, together with a representative of serotype Paratyphi A. Cluster analysis demonstrated similarities from 81% to 100% between the corresponding pairs of serogroup A and D isolates (Fig. 3). The serotype Paratyphi A isolates were less similar (68%).
TABLE 2.
Comparison of O-group determination by traditional serotyping and the Salmonella Bio-Plex assay
| Result by: | No. in the following subspecies:
|
Grand total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Bio-Plex assay | Traditional serotyping | I | II | IIIa | IIIb | IV | V | VI | |
| A | A | 5 | 5 | ||||||
| B | 54 | 2 | 2 | ||||||
| 67 | 1 | 1 | |||||||
| B | 29 | 3 | 32 | ||||||
| Rough | 3 | 3 | |||||||
| C1 | 54 | 2 | 2 | ||||||
| C1 | 28 | 4 | 1 | 3 | 2 | 38 | |||
| C2 | 54 | 3 | 3 | ||||||
| C2 | 31 | 1 | 32 | ||||||
| Rough | 2 | 2 | |||||||
| D | A | 6 | 6 | ||||||
| D1 | 20 | 5 | 25 | ||||||
| D2 | 3 | 1 | 4 | ||||||
| D3 | 3 | 3 | |||||||
| E | 54 | 3 | 3 | ||||||
| E1 | 17 | 3 | 20 | ||||||
| E4 | 1 | 1 | |||||||
| Negative | 11 | 2 | 1 | 1 | 1 | 1 | 1 | 7 | |
| 16 | 4 | 1 | 1 | 1 | 7 | ||||
| 17 | 2 | 1 | 1 | 1 | 2 | 7 | |||
| 18 | 2 | 1 | 1 | 1 | 1 | 6 | |||
| 21 | 2 | 1 | 2 | 1 | 1 | 7 | |||
| 28 | 2 | 1 | 1 | 4 | |||||
| 30 | 2 | 1 | 3 | ||||||
| 35 | 1 | 1 | 1 | 1 | 4 | ||||
| 38 | 2 | 1 | 1 | 2 | 1 | 7 | |||
| 39 | 1 | 1 | 2 | ||||||
| 40 | 2 | 1 | 2 | 1 | 1 | 2 | 9 | ||
| 41 | 1 | 1 | 1 | 1 | 2 | 6 | |||
| 42 | 1 | 1 | 1 | 1 | 1 | 5 | |||
| 43 | 1 | 1 | 1 | 1 | 1 | 5 | |||
| 44 | 1 | 1 | 1 | 1 | 2 | 1 | 7 | ||
| 45 | 1 | 1 | 1 | 1 | 1 | 5 | |||
| 47 | 1 | 2 | 1 | 1 | 1 | 6 | |||
| 48 | 1 | 2 | 4 | 4 | 1 | 2 | 1 | 15 | |
| 50 | 2 | 1 | 1 | 1 | 1 | 1 | 7 | ||
| 51 | 1 | 1 | 1 | 1 | 1 | 5 | |||
| 52 | 2 | 1 | 1 | 4 | |||||
| 53 | 1 | 1 | 1 | 1 | 1 | 5 | |||
| 54 | 4 | 4 | |||||||
| 55 | 1 | 1 | |||||||
| 56 | 1 | 1 | 2 | ||||||
| 57 | 2 | 1 | 2 | 1 | 6 | ||||
| 58 | 2 | 1 | 3 | ||||||
| 59 | 1 | 2 | 1 | 4 | |||||
| 60 | 1 | 1 | 1 | 3 | |||||
| 61 | 1 | 1 | 2 | ||||||
| 62 | 1 | 1 | |||||||
| 63 | 1 | 1 | |||||||
| 65 | 1 | 1 | 2 | ||||||
| 66 | 1 | 1 | |||||||
| Rough | 2 | 1 | 3 | ||||||
| 6, 14 | 5 | 1 | 1 | 1 | 1 | 1 | 10 | ||
| O13 | 2 | 3 | 5 | ||||||
| O13 | Rough | 1 | 1 | ||||||
| O13 | 26 | 1 | 2 | 29 | |||||
| Grand total | 231 | 54 | 29 | 34 | 26 | 12 | 7 | 393 | |
TABLE 3.
Properties of serogroup O54 serotypes
| Serotype | Antigenic formula | O54-negative variant | Salmonella serotype with an indistinguishable antigenic formula | Bio-Plex O group determination |
|---|---|---|---|---|
| Tonev | 21,54:b:e,n,x | 21:b:e,n,xa | Minnesotab | Negative |
| Winnipeg | 54:e,h:1,5 | 8:e,h:1,5c | Ferruchb | C2 |
| Rossleben | 3,54:e,h:1,6 | Unknown | E | |
| Borreze | 54:f,g,s:− | 4,12:f,g,s:-d | Agona | B |
| Uccle | 3,54:g,s,t:− | 3..:g,s,t:−e | E | |
| Poeseldorf | 8,20,54:i:z6 | 8,20:i:z6a | Kentuckyb | C2 |
| Ochsenwerder | 6,7,54:k:1,5 | 6,7:k:1,5e | Thompsonb | C1 |
| Newholland | 4,12,54:m,t:− | 4,12:m,t:−c | Bananab | B |
| Czernyring | 54:r:1,5 | Unknown | Negative | |
| Steinwerder | 3,15,54:y:1,5 | 3,15:y:1,5a | Orionb | E |
| Canton | 54:z10:e,n,x | 6,8:z10:e,n,xc | Hadarb | C2 |
| Barry | 54:z10:e,n,z15 | 6,7:z10:e,n,z15c | Mbandakab | C1 |
| Yerba | 54:z4,z23:− | Unknown | Negative | |
| Mundubbera | 54:z29:− | Unknown | Negative |
FIG. 3.
PFGE cluster analysis of Salmonella serogroup A and D isolates.
Nine isolates were designated “rough” by traditional serotyping, indicating that they did not express O antigen; six of the nine isolates gave positive results for one of the serogroups covered in the assay (Table 2). By combining the molecular result for the O group with the H-antigen result by traditional serotyping, we were able to designate a serotype for all six isolates (Table 4).
TABLE 4.
Molecular characterization of rough isolates
| Subspecies | Result by traditional serotyping | O group by Bio-Plex assay | H antigen | Possible Salmonella serotype |
|---|---|---|---|---|
| I | I rough:i:1,2 | B | i:1,2 | Typhimurium |
| I | I rough:r:1,2 | B | r:1,2 | Heidelberg |
| IV | IV rough:z4,z23:− | Negative | z4,z23:− | |
| I | I rough:z10:e,n,z15 | Negative | z10:e,n,z15 | |
| I | I rough:z:1,6 | G | z:1,6 | Poona |
| I | I rough:r:1,2 | B | r:1,2 | Heidelberg |
| I | I rough:i:z6 | C2 | i:z6 | Kentucky |
| I | I rough:d:1,2 | C2 | d:1,2 | Virginia |
| I | I rough:k:1,5 | Negative | k:1,5 |
False-negative results were initially obtained for 14/383 isolates: all subspecies II serogroup D1 (n = 5), D2 (n = 1), and D3 (n = 3) isolates were negative for serogroup D, while 2 subspecies IIIb and 3 Salmonella bongori serogroup O13 isolates were negative for serogroup O13, in the O-group Bio-Plex assay. We determined there was no amplification of the target DNA for the subspecies II serogroup D isolates. Sequencing of the 137-bp gene target used in the assay design showed that there was a 2-bp difference in both the reverse primer and the probe region for all nine of the isolates. Design of an additional reverse primer and probe incorporating this sequence variation between subspecies II serogroup D isolates and the other subspecies of serogroup D, and their addition to the O-group Bio-Plex assay, resulted in a positive serogroup D result for the subspecies II serogroup D isolates in the assay. Similarly, for the subspecies IIIb and S. bongori serogroup O13 isolates tested, we determined there was no PCR amplification of the target DNA.
DISCUSSION
Salmonella surveillance activities depend on the accuracy of serotype identification. More than 35,000 Salmonella isolates are serotyped by public health laboratories in the United States each year; this information is used to monitor local, regional, and national trends in human salmonellosis, to detect potential outbreaks, and to monitor control efforts. However, in practical terms, serotyping does have limitations. The technique itself is simple, but it is laborious to produce and maintain the more than 250 O and H grouping, typing, and single-factor reagents that are needed to identify all serotypes. More than 350 stock strains and antigens are needed to evaluate and perform quality control for antisera. In addition, expression of a serotype antigen can be lost; rough strains do not express O antigen and thus do not react with any O antisera; mucoid strains produce a capsule that blocks detection of the O antigen; and nonmotile strains do not express flagellar antigen.
Alternative methods, such as PCR-based and enzyme-linked immunosorbent assays, have been described for the identification of specific Salmonella serotypes (22, 52, 53) as well as specific Salmonella O groups (21, 23, 34), but these platforms do not adapt easily to a multiplex format. A growing number of fingerprinting methods have also attempted to identify Salmonella to the serotype level, including randomly amplified polymorphic DNA analysis (51), enterobacterial repetitive intergenic consensus PCR (6), automated ribotyping (38), and PFGE (15). Use of these methods for the identification of Salmonella serotypes, which are based on the establishment and comparison of banding patterns, relies not only on a large and diverse population of samples that reflect the true diversity of all Salmonella serotypes but also on the maintenance and analysis of these patterns in a database, which is not practical. More recently, microarray technology including O- and H-antigen-encoding genes has been used to characterize Salmonella isolates (35). While this platform can be sensitive and specific, the construction of microarray chips requires significant design expertise; the cost of custom-made microarray chips can be prohibitive; and microarray analyses are often restricted to one sample per slide, making them less cost-effective (10).
The Bio-Plex bead-based liquid suspension array has the benefits of ease of use, lower cost, more flexibility in array preparation, and automatic analysis of data, making it a more attractive platform for widespread application. We describe here a molecular method for the determination of common Salmonella serogroups that combines direct hybridization of a biotinylated PCR-amplified target DNA to bead sets coupled with oligonucleotide capture probes specific for each sequence and detection on a Bio-Plex platform. This assay uses Luminex technology, which has the potential to do multiplex analysis combined with high throughput, to provide a rapid means of O-group detection.
The rfb gene cluster sequence was not publicly available for Salmonella serogroup O13 at the time of our Bio-Plex assay design. The O unit of Salmonella serogroup O13 consists of five sugar residues: two of GalNAc, one of galactose, one of glucose, and one of fucose (Fuc). However, the manner in which all of the oligosaccharides are linked is not known (33). Only genes involved in the synthesis of UDP-GalNAc and GDP-Fuc were expected in the O13-antigen gene cluster; genes for the synthesis of UDP-Gal and UDP-Glc, which are involved in other cell functions, are located elsewhere on the chromosome (49). Comparative sequence analysis of the serogroup O13 rfb gene cluster revealed Fuc and GalNAc biosynthesis genes, three potential glycosyltransferases, an O-antigen polymerase gene, and an O-antigen transport gene. The Salmonella O13 rfb gene cluster was closely related to those of E. coli O127 and O86.
Based on the Salmonella rfb gene clusters that have been sequenced to date, the level of sequence similarity for the three different functional types of genes in the rfb region ranges from high for the sugar biosynthesis genes to little or no similarity for the genes involved in processing and assembly functions, such as wzx and wzy. We chose the latter types of genes for design of the serogroup C1-, E-, and O13-specific primers and probes. A PCR-based enzyme-linked immunosorbent assay for identification of serogroups C1 and E based on rfb genes has been reported (21). wzx was targeted for the serogroup C1 assay and the mannosyltransferase gene wbaO for the serogroup E1-specific assay. However, additional sequencing of other Salmonella rfb gene clusters has shown that the Wzx transport protein from Salmonella serogroup C1 is at least 50% similar to the corresponding Wzx protein in Salmonella serogroup O11 (unpublished data). In addition, wbaO is also present in serogroup D2, so one might expect cross-reactivity between serogroups E and D2 with this gene target. Therefore, we targeted wzy and wzx for design of serogroup C1 and E primers and probes, respectively.
Serogroup-specific PCR primers and oligonucleotide capture probes were designed and optimized for use in the Bio-Plex assay in a TMAC-containing hybridization buffer. TMAC stabilizes AT base pairs, minimizing the effect of base composition on hybridization and therefore allowing different sets of probes with different characteristics to be used under the same hybridization conditions (14). To optimize assay conditions, we assessed the effects of temperature on hybridization efficiency. Optimum hybridization occurred in our study at 52°C; we found lower signal strength at lower hybridization temperatures, as reported by other studies (8, 10, 11). PCR amplification primers are typically designed to amplify 100- to 400-bp regions of the target DNA, to help minimize the potential for steric hindrance to affect hybridization efficiency; all PCR amplicons were kept within this size range for our O-group Bio-Plex assay. The target region for most serogroups in the assay differed by whole genes; therefore, probe lengths of 33 to 35 nucleotides provided the optimum signal intensity. The effect of a mismatch has been shown to be greater with increasing temperature and decreasing probe length (1, 14). The difference between serotype Paratyphi A and serogroup D was 2 bp; a decreased probe length of 18 bp had a significant impact on signal intensity at an increased hybridization temperature of 52°C.
This Bio-Plex assay was based on the genes responsible for O-antigen biosynthesis in Salmonella, and thus, in most cases it provided a serogroup designation comparable to traditional methods. All 189 Salmonella isolates that were subtyped as serotype Paratyphi A or serogroup B, C1, C2, D, E, or O13 by traditional serotyping were correctly identified by the Bio-7-Plex assay, except five group 13 isolates belonging to subspecies IIIb or V. Isolates belonging to both E1 and E4 were positive with the serogroup E bead set. One hundred seventy-three isolates belonging to 35 of the serogroups not represented in the assay were negative by the O-group Bio-Plex assay.
Our Bio-Plex assay proved to be more specific than traditional molecular assay formats, since we were more easily able to differentiate between serotype Paratyphi A and serogroup D isolates. Serotype Paratyphi A is the only common serotype among the four currently described serogroup A serotypes. By alignment of the rfb gene cluster DNA sequences from the genome sequences of Salmonella serotype Paratyphi A and three serogroup D sequences, we targeted a 2-bp mismatch within the CDP-paratose synthase (prt) gene, for molecular differentiation of serotype Paratyphi A and serogroup D. All five serotype Paratyphi A isolates tested were positive with the serogroup A probe and negative with the serogroup D probe. The assay did not detect any of the six isolates belonging to the other three serogroup A serotypes; they were positive by the serogroup D assay. These other three serotypes belonging to serogroup A are extremely rare and are closely related to more-common serogroup D serotypes at the molecular level. Molecular characterization of the six non-serotype Paratyphi A O-group serogroup A isolates by PFGE cluster analysis was in agreement with the molecular determination of the serogroup and showed them to be most closely related to and sometimes indistinguishable from their corresponding serotypes in serogroup D. This finding is in agreement with the results of a previous study looking at variation of rfb gene clusters in S. enterica, where colony hybridization was used to show that serogroup A isolates are variants of serogroup D (58), as well as with multilocus enzyme electrophoresis (3) and microarray (36, 44) data.
Based on the genetic basis for serotypes, the Bio-Plex assay also generated different but appropriate O-group designations for serogroups O67 and O54. Both of these serogroups, like serogroup A, are rare and related to more common serogroups; O67 is a variant of serogroup B (31), and the O54 serotypes have chromosomally encoded O antigens (see Table 3). Using our Bio-7-Plex assay, we were able to identify chromosomally encoded O antigens for 10 of the 14 serotypes in serogroup O54. By traditional serotyping, besides factor O54, seven of the serotypes are known to express additional O factors (Table 3). Studies on pWQ799, a 6.9-kb plasmid from Salmonella enterica serotype Borreze, demonstrated that the transfer and mobilization of this plasmid provide a mechanism for lateral transfer of O-antigen biosynthesis genes in S. enterica. This explains the coexpression of the O54 factor with other O factors in different Salmonella serotypes that are encoded by chromosomal genes (25). Previous work where the plasmid was cured from 10 of the O54 serotypes resulted in derivatives that failed to produce factor O54 but expressed other antigenic specificities (29, 30, 41, 43) (Table 3). We tested all 14 of the O54 serotypes in our sevenplex O-group assay to examine the chromosomally based rfb O-antigen gene cluster in these strains. For the 10 serotypes where O54-negative variants have been described, the PCR results correlated with the serological data; the derivatives that serologically expressed serogroup B, C1, C2, and E factors were positive by the corresponding serogroup B, C1, C2, and E Bio-Plex assays, respectively. Popoff and Le Minor (41) were unable to isolate O54-negative variants for Salmonella enterica serotype Rossleben, Salmonella enterica serotype Czernyring, Salmonella enterica serotype Mundubbera, and Salmonella enterica serotype Yerba, so whether additional chromosomally based antigenic factors were present was not determined. One of these four O54 serotypes was positive by the Bio-Plex assay; Salmonella serotype Rossleben was positive for serogroup B. Interestingly, 4 of the 10 serotypes that produced O54-negative variants did not coexpress antigenic factor O54 with any other factor, so production of O54 can completely inhibit or hide expression of other antigenic factors. Plans are ongoing to design primers and probes specific for serogroup O54 and to add them to the Salmonella O-group Bio-Plex assay so that we can continue to correlate the molecular and traditional determinations of O groups. Serogroup O54 has been provisionally kept in the Kauffmann-White scheme (42), with 8 of 14 serotypes considered O54+ variants of other serotypes (see Table 3). The molecular determination for classifying serogroups certainly supports this and suggests that other O54 serotypes, as well as serogroups A and O67, should be considered minor variants of more common serotypes from serogroups B, C1, C2, and E (for O54), D (for A), and B (for O67).
False-negative results were observed for subspecies IIIb and V serogroup O13 isolates. We are in the process of sequencing the 90-bp target region from subspecies IIIb and V serogroup G isolates to look for sequence variation around the primer/probe sites. Once the variation is identified, additional PCR primers/probes will be incorporated into the O-group Bio-Plex assay.
Molecular O-group determination by PCR has previously been reported for rough Salmonella isolates (23). Our data provide further evidence of the advantage of a molecular approach to serotyping, because it has the potential to provide serotype information for rough isolates. The Bio-Plex assay identified an O group consistent with a common serotype for six of nine rough isolates tested.
The work described in this study forms the basis for the development of an O-group-specific Bio-Plex assay for detection of common Salmonella serogroups based on target sequences directly involved in the formation of the O antigens. Determination of Salmonella serogroups based on DNA markers appears to provide a viable alternative to traditional serotyping methods. The assay has a 96-well format and can be completed 45 min post-PCR amplification, therefore providing a high-throughput assay that should be amenable to the characterization of large numbers of isolates with minimal hands-on time. We have been investigating the O-antigen gene clusters from additional serogroups for which no sequence information is currently available (18, 19) and have identified serogroup-specific DNA targets that we are currently incorporating into the Salmonella O-group Bio-Plex assay. We also intend to identify additional rfb targets specific for other Salmonella serogroups and to add these to the Salmonella O-group assay. Our overall goal is to combine this serogroup identification system with DNA targets specific for H antigens (37) in order to establish a comprehensive DNA-based scheme for identification of the major Salmonella serotypes (P. I. Fields, C. Fitzgerald, J. R. McQuiston, M. L. Collins, L. T. Bryant, and L. L. Gheesling, presented at the International Symposium on Salmonella and Salmonellosis, Saint Malo, France, 2006). The multiplex capabilities of the Bio-Plex platform support the detection of both O and H antigens in a single assay format, and by targeting the genes responsible for serotype antigens, it provides a correlation between serotypes determined by traditional and molecular methods. Further studies are under way to validate the O-group Bio-7-Plex assay together with a Bio-Plex assay for detection of Salmonella H antigens on a panel of 500 isolates submitted to the National Salmonella Reference Laboratory in parallel with traditional serotyping methods and to standardize the procedure so as to allow for direct detection straight from a culture instead of from extracted DNA.
Supplementary Material
Acknowledgments
We thank Linda Gheesling and Lonnie Bryant for assistance with traditional serotyping of the Salmonella isolates in this study, and we appreciate the technical advice provided by Kathryn Kellar on the Bio-Plex platform and by Astrid Tuin (DNA Software Inc.) on the DNA software application. We also thank Bernard Henrissat (University of Marseille, France) for classifying the glycosyltransferases from Salmonella serogroup O13 and Peter Reeves and Kate Wilson (University of Sydney) for assignment of BPGD gene names to the Salmonella serogroup O13 rfb gene cluster.
Use of trade names is for identification only and does not imply endorsement by the Public Health Service or by the U.S. Department of Health and Human Services.
Footnotes
Published ahead of print on 18 July 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Armstrong, B., M. Stewart, and A. Mazumder. 2000. Suspension arrays for high throughput, multiplexed single nucleotide polymorphism genotyping. Cytometry 40:102-108. [PubMed] [Google Scholar]
- 2.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139:1125-1132. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, F. W., and A. C. McWhorter-Murlin. 1998. Identification and serotyping of Salmonella. Centers for Disease Control and Prevention, Atlanta, GA.
- 5.Brown, P. K., L. K. Romana, and P. R. Reeves. 1992. Molecular analysis of the rfb gene cluster of Salmonella serovar Muenchen (strain M67): the genetic basis of the polymorphism between groups C2 and B. Mol. Microbiol. 6:1385-1394. [DOI] [PubMed] [Google Scholar]
- 6.Burr, M. D., K. L. Josephson, and I. L. Pepper. 1998. An evaluation of ERIC PCR and AP PCR fingerprinting for discriminating Salmonella serotypes. Lett. Appl. Microbiol. 27:24-30. [DOI] [PubMed] [Google Scholar]
- 7.Caroff, M., and D. Karibian. 2003. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 338:2431-2447. [DOI] [PubMed] [Google Scholar]
- 8.Cowan, L. S., L. Diem, M. C. Brake, and J. T. Crawford. 2004. Transfer of a Mycobacterium tuberculosis genotyping method, spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. J. Clin. Microbiol. 42:474-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curd, H., D. Liu, and P. R. Reeves. 1998. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J. Bacteriol. 180:1002-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das, S., T. M. Brown, K. Kellar, B. P. Holloway, and C. J. Morrison. 2006. DNA probes for the rapid identification of medically important Candida species using a multianalyte profiling system. FEMS Immunol. Med. Microbiol. 46:244-250. [DOI] [PubMed] [Google Scholar]
- 11.Diaz, M. R., and J. W. Fell. 2004. High-throughput detection of pathogenic yeasts of the genus Trichosporon. J. Clin. Microbiol. 42:3696-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Souza, J. M., L. Wang, and P. Reeves. 2002. Sequence of the Escherichia coli O26 O antigen gene cluster and identification of O26 specific genes. Gene 297:123-127. [DOI] [PubMed] [Google Scholar]
- 13.Dunbar, S. A., C. A. Vander Zee, K. G. Oliver, K. L. Karem, and J. W. Jacobson. 2003. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J. Microbiol. Methods 53:245-252. [DOI] [PubMed] [Google Scholar]
- 14.Dunbar, S. A. 2006. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta 363:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebner, P. D., and A. G. Mathew. 2001. Three molecular methods to identify Salmonella enterica serotype Typhimurium DT104: PCR fingerprinting, multiplex PCR and rapid PFGE. FEMS Microbiol. Lett. 205:25-29. [DOI] [PubMed] [Google Scholar]
- 16.Feng, L., J. Tao, H. Guo, J. Xu, Y. Li, F. Rezwan, P. Reeves, and L. Wang. 2004. Structure of the Shigella dysenteriae 7 O antigen gene cluster and identification of its antigen specific genes. Microb. Pathog. 36:109-115. [DOI] [PubMed] [Google Scholar]
- 17.Feng, L., H. Weiqing, Q. Wang, D. Bastin, and L. Wang. 2005. Characterization of Escherichia coli O86 O-antigen gene cluster and identification of O86-specific genes. Vet. Microbiol. 106:241-248. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald, C., R. Sherwood, L. L. Gheesling, F. W. Brenner, and P. I. Fields. 2003. Molecular analysis of the rfb O antigen gene cluster of Salmonella enterica serogroup O:6,14 and development of a serogroup-specific PCR assay. Appl. Environ. Microbiol. 69:6099-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzgerald, C., L. Gheesling, M. Collins, and P. I. Fields. 2006. Sequence analysis of the rfb loci, encoding proteins involved in the biosynthesis of the Salmonella enterica O17 and O18 antigens: serogroup-specific identification by PCR. Appl. Environ. Microbiol. 72:7949-7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fratamico, P. M., C. E. Briggs, D. Needle, C. Y. Chen, and C. DebRoy. 2003. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J. Clin. Microbiol. 41:3379-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillespie, B. E., A. G. Mathew, F. A. Draughon, B. M. Jayarao, and S. P. Oliveri. 2003. Detection of Salmonella enterica somatic groups C1 and E1 by PCR-enzyme-linked immunosorbent assay. J. Food Prot. 66:2367-2370. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto, Y., Y. Itho, Y. Fujinaga, A. Q. Khan, F. Sultana, M. Miyake, K. Hirose, H. Yamamoto, and T. Ezaki. 1995. Development of nested PCR based on the ViaB sequence to detect Salmonella typhi. J. Clin. Microbiol. 33:775-777. (Erratum, 33:3082.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoorfar, J., D. L. Baggesen, and P. H. Porting. 1999. A PCR-based strategy for simple and rapid identification of rough presumptive Salmonella isolates. J. Microbiol. Methods 35:77-84. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, X. M., B. Neal, F. Santiago, S. J. Lee, L. K. Romana, and P. R. Reeves. 1991. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol. Microbiol. 5:695-713. [DOI] [PubMed] [Google Scholar]
- 25.Keenleyside, W. J., and C. Whitfield. 1995. Lateral transfer of rfb genes: a mobilizable ColE1-type plasmid carries the rfbO:54 (O:54 antigen biosynthesis) gene cluster from Salmonella enterica serovar Borreze. J. Bacteriol. 177:5247-5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keenleyside, W. J., M. Perry, L. Maclean, C. Poppe, and C. Whitfield. 1994. A plasmid-encoded rfbO:54 gene cluster is required for biosynthesis of the O:54 antigen in Salmonella enterica serovar Borreze. Mol. Microbiol. 11:437-448. [DOI] [PubMed] [Google Scholar]
- 27.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. J., L. K. Romana, and P. R. Reeves. 1992. Sequence and structural analysis of the rfb (O antigen) gene cluster from a group C1 Salmonella enterica strain. J. Gen. Microbiol. 138:1843-1855. [DOI] [PubMed] [Google Scholar]
- 29.Le Minor, L. 1968. Lysogenie et classification des Salmonella. Int. J. Syst. Bacteriol. 18:197-201. [Google Scholar]
- 30.Le Minor, L., M. Chippaux, F. Pichinoty, C. Coynault, and M. Piechaud. 1970. Simple methods for the detection of tetrathionate-reductase in liquid cultures and in isolated colonies. Ann. Inst. Pasteur (Paris) 119:733-737. (In French.) [PubMed] [Google Scholar]
- 31.Li, Q., and P. R. Reeves. 2000. Genetic variation of dTDP-l-rhamnose pathway genes in Salmonella enterica. Microbiology 146:2291-2307. [DOI] [PubMed] [Google Scholar]
- 32.Liu, D., N. K. Verma, L. K. Romana, and P. R. Reeves. 1991. Relationships among the rfb regions of Salmonella serovars A, B, and D. J. Bacteriol. 173:4814-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lüderitz, O., A. M. Staub, and O. Westphal. 1966. Immunochemistry of O and R antigens of Salmonella and related Enterobacteriaceae. Bacteriol. Rev. 30:192-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luk, J. M., U. Kongmuang, P. R. Reeves, and A. A. Lindberg. 1993. Selective amplification of abequose and paratose synthase genes (rfb) by polymerase chain reaction for identification of Salmonella major serogroups (A, B, C2, and D). J. Clin. Microbiol. 31:2118-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malorny, B., C. Bunge, B. Guerra, S. Prietz, and R. Helmuth. 2007. Molecular characterisation of Salmonella strains by an oligonucleotide multiprobe microarray. Mol. Cell. Probes 21:56-65. [DOI] [PubMed] [Google Scholar]
- 36.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliott, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Layman, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268-1274. [DOI] [PubMed] [Google Scholar]
- 37.McQuiston, J. R., R. Parrenas, M. Ortiz-Rivera, L. Gheesling, F. Brenner, and P. I. Fields. 2004. Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. J. Clin. Microbiol. 42:1923-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oscar, T. P. 1998. Identification and characterization of Salmonella isolates by automated ribotyping. J. Food Prot. 61:519-524. [DOI] [PubMed] [Google Scholar]
- 39.Poirot, O., K. Suhre, C. Abergel, E. O'Toole, and C. Notredame. 2004. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucleic Acids Res. 32:W37-W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popoff, M. Y., J. Bockemuhl, F. W. Brenner, and L. L. Gheesling. 2001. Supplement 2000 (no. 44) to the Kauffmann-White scheme. Res. Microbiol. 152:907-909. [DOI] [PubMed] [Google Scholar]
- 41.Popoff, M. Y., and L. Le Minor. 1985. Expression of antigenic factor O:54 is associated with the presence of a plasmid in Salmonella. Ann. Inst. Pasteur Microbiol. 136B:169-179. [DOI] [PubMed] [Google Scholar]
- 42.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 43.Poppe, C., and K. A. McFadden. 1992. Elimination of a 45 MDal plasmid converts S. borreze to S. agona, p. 37-43. In Reports and communications of the Symposium on Salmonella and Salmonellosis. World Veterinary Poultry Association, Ploufragan/Saint-Brieuc, France.
- 44.Porwollik, S., E. F. Boyd, C. Choy, P. Cheng, L. Florea, E. Proctor, and M. McClelland. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 186:5883-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray, T., A. Mills, and P. Dyson. 1995. Tris-dependent oxidative DNA strand scission during electrophoresis. Electrophoresis 16:888-894. [DOI] [PubMed] [Google Scholar]
- 46.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 47.Reeves, P. R., and L. Wang. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109-135. [PubMed] [Google Scholar]
- 48.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 49.Samuel, G., and P. R. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503-2519. [DOI] [PubMed] [Google Scholar]
- 50.Samuel, G., J. P. Hogbin, L. Wang, and P. R. Reeves. 2004. Relationships of the Escherichia coli O157, O111, and O55 O-antigen gene clusters with those of Salmonella enterica and Citrobacter freundii, which express identical O antigens. J. Bacteriol. 186:6536-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shangkuan, Y. H., and H. C. Lin. 1998. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella typhi and other Salmonella species. J. Appl. Microbiol. 85:693-702. [DOI] [PubMed] [Google Scholar]
- 52.Soumet, C., G. Ermel, V. Rose, N. Rose, P. Drouin, G. Salvat, and P. Colin. 1999. Identification by a multiplex PCR-based assay of Salmonella typhimurium and Salmonella enteritidis serotypes from environmental swabs of poultry houses. Lett. Appl. Microbiol. 29:1-6. [DOI] [PubMed] [Google Scholar]
- 53.Szabo, E. A., and B. M. Mackey. 1999. Detection of Salmonella enteritidis by reverse transcription-polymerase chain reaction (PCR). Int. J. Food Microbiol. 51:113-122. [DOI] [PubMed] [Google Scholar]
- 54.Wang, L., L. K. Romana, and P. R. Reeves. 1992. Molecular analysis of a Salmonella enterica group rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics 130:429-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, L., and P. R. Reeves. 2000. The Escherichia coli O111 and Salmonella enterica O35 gene clusters: gene clusters encoding the same colitose-containing O antigen are highly conserved. J. Bacteriol. 182:5256-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, L., C. E. Briggs, D. Rothemund, P. Fratamico, J. B. Luchansky, and P. R. Reeves. 2001. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene 270:231-236. [DOI] [PubMed] [Google Scholar]
- 58.Xiang, S. H., A. M. Haase, and P. R. Reeves. 1993. Variation of the rfb gene clusters in Salmonella enterica. J. Bacteriol. 175:4877-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.