Abstract
In this study, nested PCR using novel primers targeting the pan-dermatophyte-specific sequence of the chitin synthase 1 gene (CHS1) was compared with KOH microscopy, culture isolation, and single-round PCR for diagnosis of 152 patients with clinically suspected onychomycosis. Results indicate that nested PCR may be considered the gold standard for the diagnosis of cases of onychomycosis for which the etiological agents are dermatophytes.
The incidence of onychomycosis has been rising sharply over the last few years. This is due to an increase in the number of immunocompromised patients, the extensive use of immunosuppressive chemotherapy, and an increase in the geriatric population. The prevalence of this disease ranges between 2 and 18% globally (10). Onychomycosis is most commonly caused by dermatophytes, although Candida spp. and nondermophytic molds have also been implicated (6). The treatment of onychomycosis would be most appropriate when the selection of antimicrobial agent is based on the identity of the causative agent. Griseofulvin is effective only for dermatophytic infections, with no activity against Candida spp. and nondermophytic molds. Terbinafine shows fungicidal activity against dermatophytes with a cure rate of 80 to 95% but shows only fungistatic activity against Candida albicans. For nondermatophytic mold infections, the role of terbinafine is not well defined and avulsion of nail combined with topical amorolfine lacquer may be effective for select patients (5).
Onychomycosis is diagnosed routinely by KOH microscopy; however, the disadvantages of this method are the high number of false positives and an inability to differentiate between dermatophytic and nondermatophytic infections. Conventionally, a definitive diagnosis depends on culture isolation, but this method is often associated with poor sensitivity and delayed results. Nail histopathology, immunochemistry, PCR, and flow cytometry have recently emerged as valuable tools, but these are practiced only at reference laboratories (2). PCR may prove to be a rapid, sensitive, and specific test, but the available techniques like PCR-restriction fragment length polymorphism analysis and single-round PCR are either too complex or not sensitive enough to detect the target sequence in small amounts of clinical samples (1, 3). In their case study, Nagao et al. detected Trichophyton rubrum by nested PCR targeting internal transcribed spacer gene 1 (ITS1) in a patient with trichophytia profunda acuta, which was negative by both KOH microscopy and culture (9). Single-round PCR using dermatophyte-specific primers targeting the chitin synthase 1 gene (CHS1) has been reported to be of use in detecting DNA from pure isolates of dermatophytes and skin scrapings of infected animals but not from other pathogenic fungi, bacteria, or healthy nails and hair (8).
The present study was performed to evaluate nested PCR targeting the CHS1 gene shared by three genera, i.e., Trichophyton, Epidermophyton, and Microsporum, in patients with clinically suspected cases of onychomycosis. This study was conducted at University Hospital, Banaras Hindu University, Varanasi, India. A total of 152 patients clinically suspected of having onychomycosis were included in the study irrespective of their age or gender. The study participants included some who had not clinically responded to 2 to 3 months of empirical treatment. The most common clinical presentation was distal lateral subungual onychomycosis (n = 98), followed by total dystrophic onychomycosis (n = 25), proximal subungual onychomycosis (n = 22), and white superficial onychomycosis (n = 7). Nail scrapings were collected after the affected area was cleaned with 70% alcohol. Samples from patients with distal lateral subungual onychomycosis and total dystrophic onychomycosis included the full thickness of the dystrophic nail plate along with the subungual debris. In patients with proximal subungual onychomycosis, the scrapings were collected from the infected proximal nail bed as close to the lunula as possible, while in patients with white superficial onychomycosis, superficial scrapings from the infected nail were collected.
The specimens were divided into three portions. The first portion of the specimen was examined microscopically using 20% KOH with 40% dimethyl sulfoxide. The second portion was cultured on Sabouraud's dextrose agar containing chloramphenicol (0.05%) with and without cycloheximide (0.5%) and incubated at 25°C for 4 to 6 weeks. Clinical isolates were identified on the basis of phenotypic characteristics of the colonies, microscopic examination of lactophenol cotton blue wet mounts, and physiological tests such as urease production, in vitro hair perforation, and nutritional requirement tests.
DNA extraction was performed on the third portion of nail scrapings by crushing them in liquid nitrogen. The crushed scrapings were suspended in 200 μl of Tris-EDTA buffer and subjected to repeated freezing and thawing. Then, 300 μl of 0.1% Triton X-100 (pH 8) and 2 μl of proteinase K solution (20 mg/ml) were added and incubated for 2 h at 65°C. The extracted DNA was purified by the phenol-chloroform- isoamyl alcohol (25:24:1) method and resuspended in 30 μl of Tris-EDTA buffer. First-round PCR was performed using primer pairs CHS1 1S (5′-CAT CGA GTA CAT GTG CTC GC-3′; nucleotides [nt] 70 to 89) and CHS1 1R (5′-CTC GAG GTC AAA AGC ACG CC-3′; nt 485 to 504). These primers amplify a 435-bp DNA fragment of the dermatophyte-specific CHS1 gene sequence of Arthroderma benhaemiae, a teleomorph of Trichophyton mentagrophytes (DDBJ accession no. AB003558) (9). Nested PCR was performed by designing a novel set of primers, JF2 (5′-GCA AAG AAG CCT GGA AGA AG-3′; nt 111 to 130) and JR2 (5′-GGA GAC CAT CTG TGA GAG TTG-3′; nt 378 to 398), amplifying a DNA fragment of 288 bp from the internal sequence of the amplicon obtained from first-round PCR.
The PCR mixture (25 μl) for first-round PCR contained 2.5 μl of 10× buffer (100 mM Tris-HCl, 500 mM KCl, and 0.8% [vol/vol] Nonidet P40; MBI Fermentas, Hanover, MD), 1.1 μl of (1.5 mM) MgCl2 (MBI Fermentas), 25 pmol each of primers CHS1 1S and CHS1 1R (Operon, Cologne, Germany), 1 μl of deoxynucleoside triphosphate mix (MBI Fermentas), 1 U of Taq DNA polymerase (MBI Fermentas), and 15 μl of DNA template. Deionized water was added subsequently to achieve the reaction volume. The reaction mixture was initially denatured at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 60 s, annealing at 60°C for 75 s, and extension at 72°C for 120 s. This was followed by a final extension step for 7 min at 72°C in a thermal cycler (Biometra, Goettingen, Germany). The PCR mixture for nested PCR consisted of 25 pmol of primers JF2 and JR2 along with a 1:6 diluted product of the primary cycle as the DNA template; the rest of the constituents were the same as those described above. The running conditions of nested PCR were similar to the first-round PCR except that an annealing temperature of 63°C and 40 cycles were used. Triple-distilled water and DNA of Trichophyton mentagrophytes were used as the negative and positive controls, respectively.
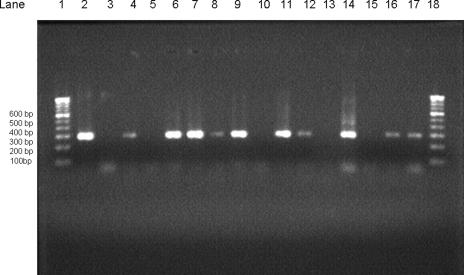
To document the amplified products, 5 μl of product from first-round PCR and nested PCR was electrophoresed on a 1.5% agarose gel (containing 1.5 μg/100 ml ethidium bromide) in Tris-borate-EDTA buffer, along with the tracking dye bromophenol blue, initially at 100 V for 5 min and then at 80 V for 60 min. Thereafter, bands were visualized under UV light (Fig. 1).
FIG. 1.
Results of nested PCR of clinical specimens from patients with onychomycosis. Lanes 1 and 18, 100-bp DNA ladder (molecular mass markers); lane 2, positive control (288 bp); lane 3, negative control; lanes 5, 10, 13, and 15, nested-PCR-negative specimens; lanes 4, 6, 7, 8, 9, 11, 12, 14, 16, and 17, nested-PCR-positive specimens.
For statistical analysis, the likelihood ratios for a positive test result (LR+) and a negative test result (LR−) were calculated (7). The significance between the two proportions was determined by using the z test (4).
Of the 152 patients with clinically suspected cases of onychomycosis, 63.4% (96/152) were positive for fungal elements by KOH microscopy. Dermatophytes were detected in 79.6% (121/152) of the cases by nested PCR and isolated in 25% (38/152) of the cases by culture. Nondermatophytic molds and Candida albicans were isolated in 11.8% (18/152) and 3.3% (5/152) of the cases, respectively, by culture. Among the dermatophytes isolated, Trichophyton rubrum was the most common isolate (55.26%, 21/38), followed by T. mentagrophytes (34.2%, 13/38), Trichophyton tonsurans (5.2%, 2/38), and Trichophyton violaceum (5.2%, 2/38). The overall positivity by first-round PCR was 51.3% (78/152). Of 114 specimens negative for dermatophyte isolation, 54 (47.3%) were positive by first-round PCR and 83 (72.8%) by nested PCR. Of the 83 specimens positive by nested PCR, nondermatophyte molds were cultured from 8 specimens, thus detecting cases with hidden mixed infections. Nested PCR was positive for 71.4% (40/56) of the KOH microscopy-negative specimens. In addition, all 83 patients on antifungal therapy were positive by nested PCR (Table 1). By statistical analysis, nested-PCR positivity was found to be significantly higher than that of KOH microscopy (P < 0.01), culture isolation (P < 0.001), or first-round PCR (P < 0.001).
TABLE 1.
Status of KOH microscopy, fungal culture, first-round PCR, and nested-PCR results for patients with suspected cases of onychomycosis
| No. of patients | Patients on treatment? | Test result for fungal detection
|
|||
|---|---|---|---|---|---|
| KOH microscopy | Culture showing growth of dermatophyte | First-round PCR | Nested PCR | ||
| 9 | Yes | − | − | − | + |
| 12 | No | + | + | − | + |
| 24 | No | + | + | + | + |
| 17 | Yes | + | − | + | + |
| 20 | Yes | + | − | − | + |
| 29 | Yes | − | − | + | + |
| 10a | No | + | − | − | − |
| 8a | Yes | + | − | + | + |
| 5b | No | + | − | − | − |
| 2 | No | − | + | − | + |
| 16 | No | − | − | − | − |
| No. (%) positivec | 83 (54.6) | 96 (63.4) | 38 (25) | 78 (51.3) | 121 (79.6) |
Culture positive for nondermatophytes.
Culture positive for Candida albicans.
Out of a total of 152 patients.
In the present study, nested PCR was observed to be more sensitive for the detection of dermatophytes than culture isolation, KOH microscopy, and single-round PCR. The lower sensitivity of single-round PCR compared to KOH microscopy was circumvented in the present study by the use of nested PCR. For an appropriate diagnostic test, desirable values for LR+ and LR− should be ≥10 and ≤0.1, respectively (7). By considering nested PCR as the gold standard, the LR+ values of KOH microscopy and culture were 1.36 and infinity, respectively, while the LR− values were 0.64 and 0.68, respectively.
It may therefore be concluded that nested PCR targeting the CHS1 gene may be considered the gold standard for detection of dermatophytes in patients with onychomycosis and can aid the clinician in initiating prompt and appropriate antifungal therapy. This technique may also play an important role in large-scale studies and in the management of problematic cases of onychopathies.
Acknowledgments
We acknowledge financial help extended through a laboratory grant by the Head, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, India, in completion of the study.
We thank N. Srinivas Acharya for critically reading the manuscript.
Footnotes
Published ahead of print on 15 August 2007.
REFERENCES
- 1.Arca, E., M. A. Saracli, A. Akar, S. T. Yildiran, Z. Kurumlu, and A. R. Gur. 2004. Polymerase chain reaction in the diagnosis of onychomycosis. Eur. J. Dermatol. 14:52-55. [PubMed] [Google Scholar]
- 2.Arrese, J. E., C. Pierard-Franchimont, and G. E. Pierard. 1999. Facing up to the diagnostic uncertainty and management of onychomycosis. Int. J. Dermatol. 38(Suppl. 2):7-12. [DOI] [PubMed] [Google Scholar]
- 3.Baek, S. C., H. J. Chae, D. Houh, D. G. Byun, and B. K. Cho. 1998. Detection and differentiation of causative fungi of onychomycosis using PCR amplification and restriction enzyme analysis. Int. J. Dermatol. 37:682-686. [DOI] [PubMed] [Google Scholar]
- 4.Dawson, B., and R. G. Trapp. 2001. Basic and clinical biostatistics, 3rd ed., p. 146-147. McGraw-Hill, Singapore, Malaysia.
- 5.Denning, D. W., E. G. V. Evans, C. C. Kibbler, M. D. Richardson, M. M. Roberts, T. R. Rogers, D. W. Warnock, and R. E. Warren. 1995. Fungal nail disease: a guide to good practice (report of a Working Group of the British Society for Medical Mycology). Br. Med. J. 311:1277-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elewski, B. E. 1998. Onychomycosis: Pathogenesis, Diagnosis, and Management. Clin. Microbiol. Rev. 11:415-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg, R. S., R. S. Daniels, W. D. Flanders, J. W. Eley, and J. R. Boring (ed.). 1996. Diagnostic testing, p. 77-89. In Medical epidemiology, 3rd ed. McGraw-Hill, New York, NY.
- 8.Kano, R., A. Hirai, M. Muramatsu, T. Watari, and A. Hasegawa. 2003. Direct detection of dermatophytes in skin samples based on sequences of the chitin synthase 1 (CHS1) gene J. Vet. Med. Sci. 65:267-270. [DOI] [PubMed] [Google Scholar]
- 9.Nagao, K., S. Takashi, O. Takashi, and N. Takeji. 2005. Identification of Trichophyton rubrum by nested PCR analysis from paraffin embedded specimen in trichophyton profunda acuta of the glabrous skin. Jpn. J. Med. Mycol. 46:129-132. [DOI] [PubMed] [Google Scholar]
- 10.Scher, R. K. 1999. Onychomycosis; therapeutic update. J. Am. Acad. Dermatol. 40:14-20. [DOI] [PubMed] [Google Scholar]