Abstract
Four (GACA)4 PCR fingerprinting sequences, used as markers to identify serotypes A and D and AD hybrids, were retrieved in four Cryptococcus neoformans genome databases. Their locations, both in serotype A and D genomes, were confirmed by chromosomal hybridization with specific probes. Two sequences were recognized to code for hypothetical functional proteins.
Analysis of Cryptococcus neoformans isolates by PCR fingerprinting with the (GACA)4 primer in comparison with mating type and DNA content showed that the strains might be clustered in four different groups. One includes haploid serotype A isolates (VN6), one includes haploid serotype D isolates (VN1), and the other two (VN3 and VN4) include diploid or aneuploid AD hybrid strains, which display fingerprint bands of both VN1 and VN6 genotypes and are heterozygous or homozygous at the mating type locus (1-4, 8). Four fingerprinting major bands (800, 540, 475, and 420 bp) were selected as markers to easily identify the four genotypes: VN1 presented the 540- and 420-bp bands; VN6 the 800-, 540-, and 475-bp bands; VN3 the 800-, 540-, and 420-bp bands; and VN4 all four bands. None of the above bands was present in Cryptococcus gattii (GACA)4 fingerprints (8). The nucleotide sequences of the four major bands were also determined, and comparison between the sequences of a number of C. neoformans isolates was performed (2). Finally, a multiplex PCR was set up using primers specific for each of the bands (1).
Recently, (GACA)4 primer PCR fingerprinting has been applied to genotype 311 isolates collected during a European prospective epidemiological study on cyptococcosis. Results showed, for the first time, a prevalence of AD hybrid strains in Europe that was largely underestimated in previous studies, as many of the strains were identified as serotype A or D by serotyping methods (5, 9).
At present, the characterization of the above-mentioned four major bands is possible due to the release of the full sequence of the C. neoformans genome (6). Therefore, the aim of the present study is (i) to identify where the four major band sequences are located in the C. neoformans genome, (ii) to verify if they are coding sequences or not, and (iii) to compare the results obtained by the analysis of both serotype A and D genomes.
The sequences of the four PCR fingerprinting major bands (GenBank accession numbers AF205275, AF205276, AF205260, and AF205261) were aligned by BLASTn algorithm (Washington University, St. Louis, MO) with the following C. neoformans genome databases: Duke Center for Genome Technology, Durham, NC (H99 serotype A strain); Broad Institute, Cambridge, MA (H99 serotype A strain); Stanford Genome Technology Center, Palo Alto, CA (B3501A serotype D strain); and The Institute for Genomic Research (TIGR), Rockville, MD (JEC21 serotype D strain).
The matched contigs were further aligned to the sequences of the primers used either in PCR fingerprinting or in the multiplex PCR typing method. The BLASTx analysis was performed by aligning the four band sequences to the cDNA database of TIGR for the serotype D genome and to that of the Broad Institute for the serotype A genome.
Chromosomal DNA extraction and pulsed-field gel electrophoresis were performed as reported elsewhere (4, 10). Chromosomal blotting was performed by the denaturing downward transfer method as previously described (7) on a positively charged nylon membrane (Hybond-N+; Amersham Biosciences, Bucks, United Kingdom). The four band sequences were detected by four specific probes synthesized by the amplification of H99 genomic DNA using the primers reported in Table 1. Purified amplicons were labeled by random priming using the Rediprime kit (Amersham Biosciences). Hybridization of the blotted membranes was performed in PerfectHyb Plus hybridization buffer (Sigma-Aldrich, Milano, Italy) at 65°C overnight. After washings, the radioactive signals were detected by autoradiography.
TABLE 1.
Characteristics of the primers used to synthesize the probes
| Probe | PCR fingerprinting band detected (bp) | Forward primer | Reverse primer | Taa (°C) | Product size (bp) |
|---|---|---|---|---|---|
| P420 | 420 | GGTTGCCCAAACCCTCA | CCTTGGCATCAGCAACACAT | 58 | 870 |
| P475 | 475 | TTGATGTGAATCTTTTATACACTTGC | AGCGTGGAGCGTGGAG | 58 | 535 |
| P540 | 540 | GAACCCACCGCCCTCTTC | ACCAGACGGCAACCTTCTTA | 58 | 838 |
| P800 | 800 | ATCGCGTAGCATTTGAGCTT | ACAAATGTGGCGATGTGAGT | 58 | 900 |
Ta, annealing temperature.
The present results showed that three of the four sequences were present in both serotype A and D genomes but that the alleles differed in some nucleotide sites. The fourth band sequence (800 bp) was identified only in the serotype A genome and was absent in serotype D, confirming that this DNA region is a good marker to identify serotype A strains (Table 2).
TABLE 2.
BLASTn alignment of the nucleotide sequences of the four PCR fingerprinting major bands with serotype A and serotype D genome databases
| Sequence (bp) | Duke Universitya (H99 strain)
|
Broad Instituteb (H99 strain)
|
Stanford Universityc (B3501A strain)
|
TIGRd (JEC21 strain)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identity (%) | SC-Ce | Code | Identity (%) | SC-C | Code | Identity (%) | SC-C | Code | Identity (%) | SC-C | Code | |
| 420 | 92 | 4-7 | chr4-piece7 | 92 | 9-84 | C1.84 | 99 | 8-119 | cneo011005.C119 | 98 | 4 | chr4 |
| 475 | 99 | 1-4 | chr1-piece4 | 99 | 1-5 | C1.5 | 89 | 1-1135 | cneo011005.C1135 | 85 | 1 | chr1 |
| 540 | 93 | 3-22 | chr3-piece22 | 93 | 3-32 | C1.35 | 99 | 2-592 | cneo011005.C592 | 99 | 3 | chr3 |
| 800 | 97 | 3-23 | chr3-piece23 | 97 | 3-32 | C1.32 | NMf | NM | ||||
Duke Center for Genome Technology (http://cgt.duke.edu/) and the Genome Sequence Centre, British Columbia Cancer Research Centre, Vancouver, BC, Canada (http://www.bcgsc.bc.ca/).
Stanford Genome Technology Center (http://www-sequence.stanford.edu).
SC-C, supercontig or chromosome-contig.
NM, no match.
Polymorphism of the two serotypes was found also at primer annealing sites of the (GACA)4 sequence. The different affinities of this primer for serotype A and D genomes explain why the 800-, 540-, and 475-bp bands are amplified in serotype A and the 540- and 420-bp bands are amplified in serotype D (Tables 3 and 4). Sequence analysis of primer annealing sites complementary to the oligonucleotides used in the multiplex PCR typing method showed that each pair of primers has a 100% or nearly 100% affinity to a single serotype (800, 475, and 420 bp) or both serotypes (540 bp) (Table 3), confirming that the primers were designed correctly and that the method is suitable for distinguishing serotype A from serotype D strains and their hybrid.
TABLE 3.
Alignment of primer sequences with contig matched in serotype A and serotype D genome databases
| Genome strain and band (bp) | Contig | Nucleotide identitya (%) for:
|
|||
|---|---|---|---|---|---|
| (GACA)4 primer
|
Multiplex PCR primers
|
||||
| Forward | Reverse | Forward | Reverse | ||
| H99 | |||||
| 420 | 1.84 | 10/16 (63) | 12/16 (75) | 15/17 (88) | 16/17 (94) |
| 475 | 1.5 | 12/16 (75) | 11/16 (69) | 18/18 (100) | 16/16 (100) |
| 540 | 1.35 | 11/16 (69) | 12/16 (75) | 16/17 (94) | 17/17 (100) |
| 800 | 1.32 | 12/16 (75) | 8/16 (50) | 17/17 (100) | 19/19 (100) |
| JEC21 | |||||
| 420 | C119 | 11/16 (69) | 13/16 (81) | 17/17 (100) | 17/17 (100) |
| 475 | C1135 | 12/16 (75) | 7/16 (44) | 11/18 (61) | 15/16 (94) |
| 540 | C592 | 12/16 (75) | 12/16 (75) | 17/17 (100) | 17/17 (100) |
| 800 | NMb | ||||
Number of identical sequences/total number of sequences.
NM, no match.
TABLE 4.
Nucleotide sequences at 5′ and 3′ ends of the four PCR fingerprinting major bands
| Strain (band [bp]) or primer | 5′ end
|
3′ end
|
Amplification resultc | ||
|---|---|---|---|---|---|
| Nucleotide sequencea | Nucleotide identity (%) | Nucleotide sequence | Nucleotide identity (%) | ||
| Primer | GACAGACAGACAGACA | TGTCTGTCTGTCTGTC | |||
| H99 (420) | ***AG*CA*A*AGACA | 63 | TGTCTG*C*GTC*GT* | 75 | − |
| JEC21 (420) | ***AG*CA*ACAGACA | 69 | TGTCTG*CTGTC*GT* | 81 | + |
| H99 (475) | *A**GACAGAC*GACA | 75 | TGTC*G*CTGTC**T* | 69 | + |
| JEC21 (475) | *A**GACAGAC*GACA | 75 | TGTC*G**T*****T* | 44 | − |
| H99 (540) | **C*G*CAGACAG*CA | 69 | TGTCTGT*TGTC*G** | 75 | + |
| JEC21 (540) | **CAG*CAGACAG*CA | 75 | TGTCTGT*TGTC***C | 75 | + |
| H99 (800) | *AC***CAGACAG*CA | 75 | TGT*TGTC****T*** | 50 | + |
| JEC21 (800) | NMb | NM | − | ||
*, nucleotide mismatch.
NM, no match.
−, no amplification; +, band amplification.
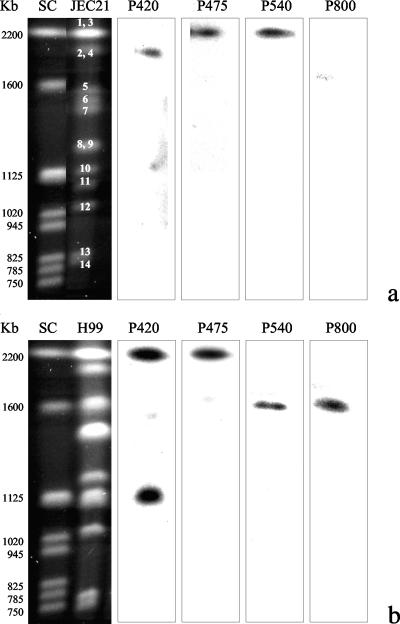
Hybridization of the JEC21 electrophoretic karyotype confirmed the data predicted by BLASTn analysis (Fig. 1a). The 420-bp sequence, expected to be located on chromosome 4, was actually detected on the second-highest-molecular-weight chromosomal band, which corresponds to chromosomes 2 and 4 overlapped. Similarly, the 475- and 540-bp sequences were detected on the highest-molecular-weight band, which contains both chromosomes 1 and 3. The 800-bp sequence, which did not show any match in the JEC21 database, was not detected by hybridization. As expected, the hybridization of the H99 genome located both the 540- and 800-bp sequences on the third chromosomal band and the 475-bp sequence on the first band. In contrast, the 420-bp sequence showed two sites of hybridization: one on the first band and the second on a band at 1,120 kb (Fig. 1b). These data suggest the presence of a DNA region in chromosome 3 of serotype A which is characteristic of the serotype A genome and can be detected by the amplification of the 800-bp sequence. The 475-bp sequence seems to have conserved its location both in serotype A and D genomes, while the other sequences show a divergent position, probably due to chromosomal rearrangements, which are known to occur frequently in C. neoformans. BLASTx analysis showed that only two of the four band sequences (420 and 540 bp) matched with hypothetical coding regions (Table 5). The 420-bp band sequence is included in a DNA region coding for a protein of unknown function, while the 540-bp sequence encodes a sequence that matches a portion of a protein used for DNA repair and recombination (PIF1). The remaining two band sequences (475 and 800 bp) did not match any of the cDNA present in the TIGR and Broad Institute databases.
FIG. 1.
Chromosomal locations of the four probes specific for the four PCR fingerprinting sequences on the electrophoretic karyotypes of JEC21 (a) and H99 (b) strains. Numbers on the chromosomal bands represent the chromosome numbers according to GenBank (www.ncbi.nlm.nih.gov). SC, Saccharomyces cerevisiae.
TABLE 5.
BLASTx alignment of the nucleotide sequences of the four PCR fingerprinting major bands with serotype A and serotype D cDNA databases
| Genome strain and sequence length (bp) | Protein type | Identity (%) | Supercontig or chromosomea | Code |
|---|---|---|---|---|
| H99b | ||||
| 420 | Conserved hypothetical protein | 100 | 9 | CNAG_04944.1 |
| 475 | NMd | NM | NM | NM |
| 540 | Conserved hypothetical protein | 76 | 3 | CNAG_01792.1 |
| 800 | NM | NM | NM | NM |
| JEC21c | ||||
| 420 | Expressed protein | 90 | 4 | 184.m04983 |
| 475 | NM | NM | NM | NM |
| 540 | DNA repair and recombination protein (PIF1) | 82 | 3 | 179.m00258 |
| 800 | NM | NM | NM | NM |
Supercontig for H99 genome and chromosome for JEC21 genome.
Broad Institute (http://www.broad.mit.edu/).
TIGR (http://www.tigr.org).
NM, no match.
In conclusion, 540- and 420-bp sequences seem to be DNA regions quite conserved, and therefore their amplification is expected to be reproducible. On the other hand, the 475- and 800-bp sequences are silent DNA regions, and thus it is likely that they are more variable than the other two sequences. This may explain the absence of the 800-bp sequence in the serotype D genome as well as the reason why in AD hybrid strains the 475-bp sequence is amplified in the VN4 genotype but not in the VN3 genotype.
In view of the above results the present study elucidates the identities and the locations of the four major bands amplified by PCR fingerprinting with the (GACA)4 primer and may help in the interpretation of future studies carried out using this typing method.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Cogliati, M., M. Allaria, A. M. Tortorano, and M. A. Viviani. 2000. Genotyping Cryptococcus neoformans var. neoformans with specific primers designed from PCR-fingerprinting bands sequenced using a modified PCR-based strategy. Med. Mycol. 38:97-103. [DOI] [PubMed] [Google Scholar]
- 2.Cogliati, M., M. Allaria, G. Liberi, A. M. Tortorano, and M. A. Viviani. 2000. Sequence analysis and ploidy determination of Cryptococcus neoformans var. neoformans. J. Mycol. Med. 10:171-176. [DOI] [PubMed] [Google Scholar]
- 3.Cogliati, M., M. C. Esposto, D. L. Clarke, B. L. Wickes, and M. A. Viviani. 2001. Origin of Cryptococcus neoformans var. neoformans diploid strains. J. Clin. Microbiol. 39:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogliati, M., M. C. Esposto, A. M. Tortorano, and M. A. Viviani. 2006. Cryptococcus neoformans population includes hybrid strains homozygous at mating-type locus. FEMS Yeast Res. 6:608-613. [DOI] [PubMed] [Google Scholar]
- 5.FIMUA Cryptococcosis Network. 2002. European Confederation of Medical Mycology (ECMM) prospective survey of cryptococcosis. Report from Italy. Med. Mycol. 40:507-517. [PubMed] [Google Scholar]
- 6.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marra, R. E., J. C. Hang, E. Fung, K. Nielsen, J. Heitman, R. Vilgalys, and T. M. Mitchell. 2004. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics 167:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viviani, M. A., H. Wen, A. Roverselli, R. Caldarelli-Stefano, M. Cogliati, P. Ferrante, and A. M. Tortorano. 1997. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. J. Med. Vet. Mycol. 35:355-360. [PubMed] [Google Scholar]
- 9.Viviani, M. A., M. Cogliati, M. C. Esposto, K. Lemmer, K. Tintelnot, M. F. Valiente, D. Swinne, A. Velegraki, R. Velho, and the ECMM Cryptococcosis Working Group. 2006. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res. 6:614-619. [DOI] [PubMed] [Google Scholar]
- 10.Wickes, B. L., T. D. E. Moore, and K. J. Kwon-Chung. 1994. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology 140:543-550. [DOI] [PubMed] [Google Scholar]