Abstract
We compared three novel methicillin-resistant Staphylococcus aureus (MRSA) genotyping methods with multilocus sequence typing (MLST) and spa typing to assess their utility for routine strain typing. The new methods were femA and nuc sequence typing and toxin gene profiling (TGP), using a multiplex-PCR-based reverse line blot assay to detect 13 pyrogenic superantigen and exfoliative toxin genes. Forty-two well-characterized MRSA strains, representing 15 MLSTs or 9 clonal clusters (CCs), were genotyped by all methods. Twenty-two spa, nine femA, and seven nuc sequence types were identified. The femA sequence types correlated exactly with CCs; nuc sequences types were less discriminatory but generally correlated well with femA types and CCs. Ten isolates contained none of 13 toxin genes; TGPs of the remainder comprised 1 to 5 toxin genes. The combination of spa typing and TGPs identified 26 genotypes among the 42 strains studied. A combination of two or three rapid, inexpensive genotyping methods could potentially provide rapid MRSA strain typing as well as useful information about clonal origin and virulence.
Methicillin-resistant Staphylococcus aureus (MRSA) genotyping is used to study its evolution and epidemiology and to assist in infection control (39). Different typing methods provide different information. Multilocus sequence typing (MLST) reveals slowly accumulating changes in conserved genes that reflect long-term evolutionary changes and can identify global spread of the relatively small number of successful clones (13). It has limited discriminatory power and is unsuitable for outbreak investigation, whereas pulsed-field gel electrophoresis is highly discriminatory and can identify recent changes. It is most widely used for outbreak investigation and infection control (9, 28). Both methods are relatively expensive and slow, and a number of rapid, inexpensive typing methods, based on sequence or length polymorphisms of variable genes or loci, have been described that are objective and relatively inexpensive. These include coa (41) and spa (38) sequence typing and the multilocus variable number tandem repeat assay (12, 29).
spa typing, which depends on differences in the number and sequence of tandem repeats in region X of the protein A gene (44), is discriminatory, rapid, inexpensive, and objective (25, 37, 41). The development of a shareable web-based database (www.spaServer.Ridom.de) (15) and the utility of spa typing for early-warning systems (31) have contributed to the rapid uptake of MRSA spa typing by diagnostic and public health laboratories.
In this study, we investigated the potential utility of two additional S. aureus gene polymorphisms for strain typing, namely, femA, one of several genes involved in the synthesis of the branched-peptide structure of S. aureus peptidoglycan (4), and nuc, which encodes an extracellular thermostable nuclease of S. aureus (5). Both are species-specific S. aureus genes; they have been widely used as PCR targets for identification (21), but their polymorphisms have not been widely investigated (14).
S. aureus produces numerous toxins, including enterotoxins or pyrogenic superantigens and exfoliative toxins, some of which are encoded by genes carried on staphylococcal pathogenicity islands and associated with certain clonal complexes (CCs), whereas genes encoding others, such as the Panton-Valentine leucocidin (PVL), are carried on bacteriophages and readily transferred between different lineages (26, 27). This suggests that a toxin gene profile (TGP) could help identify S. aureus CCs as well as providing information about virulence. Various molecular methods have been described for studying the distribution of staphylococcal toxins (2, 10).
We used 42 well-characterized MRSA strains to compare sequence polymorphisms of femA and nuc and TGPs, based on a multiplex PCR-based reverse line blot assay (mPCR/RLB) (22), with two established typing methods—namely, spa typing and MLST—to determine their potential utility for MRSA genotyping.
MATERIALS AND METHODS
S. aureus isolates.
We used 42 well-characterized reference and clinical S. aureus isolates in this study, as shown in Table 1, including 35 from various parts of Australia, provided by Philip Giffard, Cooperative Research Centre for Diagnostics, Queensland University of Technology, Brisbane, and Graeme Nimmo, Queensland Health Pathology Services, Princess Alexandra Hospital, Brisbane, Australia. Some have been used in several previous studies (40). Seven strains were provided by Herminia de Lencastre, Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras, Portugal, and also have been used in previous studies (33, 34) Two of these strains (MW2 and COL) have been fully sequenced (26). MLST and SCCmec typing results were provided by the donors of the strains (33, 34, 40).
TABLE 1.
Genotypes and spa types of 42 well-characterized methicillin-resistant S. aureus isolates used in this study
| Strain | GenBank accession no.a | Defined spa lengthb | spa typec | spa profilec | Clonal typed,e | Sources of Australian isolatesh |
|---|---|---|---|---|---|---|
| B827549 | EF094508 | 134 | t1784 | 07-34-33-13 | ST∼1-SCCmec-new | QHPS |
| HU25g | EF094528 | 182 | t138 | 08-16-02-25-17-24 | ST239-SCCmec-IIIA | |
| HDG2g | EF094527 | 182 | t421 | 15-12-16-02-25-17 | ST239-SCCmec-IIIB | |
| K704540f | EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST∼239-SCCmec-III | QHPS |
| K711532f | = EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST∼239-SCCmec-III | QHPS |
| AH13f | = EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST239-SCCmec-IIIA | AGAR |
| RDH81f | = EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST239-SCCmec-IIIA | AGAR |
| AH1f | = EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST128-SCCmec-IIIA | AGAR |
| RPAH 18f | = EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST239-SCCmec-III | AGAR |
| RPAH15f | = EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST239-SCCmec-III | AGAR |
| ANS46g | = EF094525 | 206 | t037 | 15-12-16-02-25-17-24 | ST239-SCCmec-III | |
| PC8f | EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST1-SCCmec-IV | AGAR |
| FH43f | = EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST∼1-SCCmec-IV | AGAR |
| SJOG 30f | = EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST1-SCCmec-IV | AGAR |
| RPH 85f | = EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST∼1-SCCmec-IV | AGAR |
| SN39f | = EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST∼1-SCCmec-new | AGAR |
| RHH58f | = EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST∼1-SCCmec-IV | AGAR |
| RHH10f | = EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST∼1-SCCmec-IV | AGAR |
| FH53f | = EF094507 | 206 | t127 | 07-23-21-16-34-33-13 | ST∼1-SCCmec-I | AGAR |
| RPH2f | EF094510 | 206 | t190 | 11-17-34-24-34-22-25 | ST8-SCCmec-new | AGAR |
| PAH 58f | EF094514 | 230 | t019 | 08-16-02-16-02-25-17-24 | ST30-SCCmec-IV | AGAR |
| PAH 1f | = EF094514 | 230 | t019 | 08-16-02-16-02-25-17-24 | ST30-SCCmec-IV | AGAR |
| MW2g | EF094526 | 230 | t128 | 07-23-23-21-16-34-33-13 | ST1-SCCmec-IV | |
| RBH98f | EF094522 | 230 | t202 | 11-17-23-17-17-16-16-25 | ST93-SCCmec-IV | AGAR |
| 13792-4492f | = EF094522 | 230 | t202 | 11-17-23-17-17-16-16-25 | ST∼93-SCCmec-IV | QHPS |
| IP01M1081f | EF094523 | 230 | t216 | 04-20-17-20-17-31-16-34 | ST59-SCCmec-IV | QHPS |
| 14176-5710f | EF094524 | 230 | t1959c | 15-21-12-16-02-25-17-16 | ST∼239-SCCmec-III | QHPS |
| B8-10f | EF094509 | 230 | t711 | 04-21-17-34-24-34-22-25 | ST∼8-SCCmec-IV | QHPS |
| J710566f | EF094516 | 254 | t065 | 09-02-16-34-13-17-34-16-34 | ST45-SCCmec-V | QHPS |
| RPH 74f | EF094517 | 254 | t123 | 09-02-16-34-13-16-34-16-34 | ST45-SCCmec-V | AGAR |
| IP01M2046f | EF094519 | 254 | t1958c | 08-21-17-13-13-new-34-33-34 | ST78-SCCmec-IV | QHPS |
| E804531f | EF094518 | 278 | t002 | 26-23-17-34-17-20-17-12-17-16 | ST5-SCCmec-IV | QHPS |
| CH97f | = EF094518 | 278 | t002 | 26-23-17-34-17-20-17-12-17-16 | ST73-SCCmec-IV | AGAR |
| BK2464g | = EF094518 | 278 | t002 | 26-23-17-34-17-20-17-12-17-16 | ST5-SCCmec-II | |
| IMVS 67f | EF094511 | 278 | t008 | 11-19-12-21-17-34-24-34-22-25 | ST8-SCCmec-V | AGAR |
| COLg | = EF094511 | 278 | t008 | 11-19-12-21-17-34-24-34-22-25 | ST250-SCCmec-I | |
| DEN2988g | = EF094511 | 278 | t008 | 11-19-12-21-17-34-24-34-22-25 | ST8-SCCmec-IVA | |
| F829549f | EF094521 | 278 | t186 | 07-12-21-17-13-13-34-34-33-34 | ST88-SCCmec-IV | QHPS |
| C801535f | EF094520 | 278 | t325 | 07-12-21-17-34-13-34-34-33-34 | ST88-SCCmec-new | QHPS |
| E822485f | EF094515 | 302 | t018 | 15-12-16-02-16-02-25-17-24-24-24 | ST36-SCCmec-II | QHPS |
| CH69f | EF094513 | 326 | t1963c | 26-23-13-17-31-29-17-25-17-25-16-28 | ST∼22-SCCmec-IV | AGAR |
| CH16f | EF094512 | 422 | t032 | 26-23-23-13-23-31-29-17-31-29-17-25-17-25-16-28 | ST22-SCCmec-IV | AGAR |
The relevant spa sequences have been submitted to GenBank; other strains with identical spa sequences are indicated by “=” and the accession no. for the corresponding submitted GenBank sequence.
Defined spa lengths are the distances from the start and end points, equal to 1156 and 1481 in GenBank sequence J01786, which correlates with the start and end point of the suggested 5′ and 3′ signature sequences (www.spaServer.Ridom.de). The full repetitive region sequence length can be calculated by adding together the lengths of sequences of individual repeats.
After comparison with spa database (www.spaServer.Ridom.de) and GenBank sequences, three new spa types sequences were identified and submitted to the spa database (www.spaServer.Ridom.de). Please refer to the spa database for spa type and profile nomenclature.
ST, MLST; SCCmec, staphylococcal cassette chromosome mec. Information provided by strain donors; ST∼, single nucleotide polymorphism type as described by Huygens et al. (18) using the computer program Minimum SNPs to compare with existing MLST data (17).
Clonal type refers to the combination of ST and SCCmec type.
Thirty-five Australian strains were provided by Philip Giffard, Cooperative Research Centre for Diagnostics, Queensland University of Technology, Brisbane, Australia, and Graeme Nimmo, Queensland Health Pathology Services, Princess Alexandra Hospital, Brisbane, and have been used in several previous studies (17, 18, 40).
Seven isolates were provided by Herminia de Lencastre, Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Oeiras, Portugal, and have been used in several previous studies (33, 34); spa types identified in this study were identical with those previously reported for these strains (de Lencastre, personal communication).
QHPS, Queensland Health Pathology Service (isolates from various diagnostic laboratories in Queensland); AGAR, Australian Group on Antibiotic Resistance (isolates from a study of community MRSA in Australia) (6).
DNA extraction.
DNA extraction was performed as described previously (23).
Toxin gene detection.
A well-established mPCR/RLB protocol developed in our laboratory (22) was used to detect 13 Staphylococcus aureus toxin genes. Target genes, primer and probe sequences, physical characteristics, and locations within selected GenBank sequences are shown in Table 2. All primers and probes had similar physical characteristics to allow simultaneous amplification and hybridization, respectively, in a multiplex reaction (22). Two gene-specific PCR primer pairs and two gene-specific probes were designed for each of 13 toxin genes. All primers were 5′ end biotinylated to allow detection of hybridization with a streptavidin peroxidase substrate. The probes were labeled with a 5′-end amine group to facilitate covalent linkage to the nylon membrane and to allow membranes to be stripped and reused repeatedly (22). Each multiplex reaction included nuc primers as the positive control for S. aureus and for quality control of DNA extraction and mPCR/RLB. All primers and probes were synthesized by Sigma-Aldrich (Sydney, Australia).
TABLE 2.
Primers and probes used in mPCR/RLB for detection of 13 toxin genes
| Primer/probea | Target | Tm °Cb | GenBank accession no. | Primer/probe sequence (5′-3′)c | Referencesd |
|---|---|---|---|---|---|
| nucSb | nuc | 65.68 | V01281 | 511GCG ATT GAT GGT GAT ACG GTT531 | 7 |
| nucAp | nuc | 61.36 | V01281 | 558CAT TGG TTG ACC TTT GTA CAT TAA 535 | This study |
| nucSp | nuc | 61.06 | V01281 | 745GAT GGA AAA ATG GTA AAC GAA G766 | This study |
| nucAb | nuc | 69.12 | V01281 | 789AGC CAA GCC TTG ACG AAC TAA AGC766 | 7 |
| seaSb | sea | 64.05 | M18970 | 487CCT TTG GAA ACG GTT AAA ACG507 | 3 |
| seaSp | sea | 68.83 | M18970 | 531GGA GTT GGA TCT TCA AGC AAG ACG 554 | 3 |
| seaAp | sea | 63.87 | M18970 | 613TCT GAA CCT TCC CAT CAA AAA C592 | 3 |
| seaAb | sea | 62.91 | M18970 | 691TTGA ATA CTG TCC TTG AGC ACC670 | 43 |
| sebSb | seb | 64.82 | M11118 | 634TCG CAT CAA ACT GAC AAA CG653 | 3 |
| sebSp | seb | 61.1 | M11118 | 662GTAT GTA TGG TGG TGT AAC TGA GC685 | 43 |
| sebAp | seb | 60.06 | M11118 | 831CA CCA AAT AGT GAC GAG TTA GG810 | 43 |
| sebAb | seb | 60.4 | M11118 | 924CAT GTC ATA CCA AAA GCT ATT CTC901 | 3 |
| secSb | sec | 62.5 | X05815 | 664G CTC AAG AAC TAG ACA TAA AAG CTA GG690 | 3 |
| secSp | sec | 63.13 | X05815 | 772AAC GG(/a)C AAT ACT TTT TGG TAT GAT795 | 3 |
| secAp | sec | 61.4 | X05815 | 885CTT CAC A(/t)CT TTT AGA ATC AAC CG863 | 43 |
| secAb | sec | 60.4 | X05815 | 935TCA AAA TCG GAT TAA CAT TAT CC913 | 3 |
| sedSb | sed | 60.2 | M28521 | 332CTA GTT TGG TAA TAT CTC CTT TAA ACG358 | 3 |
| sedSp | sed | 64.91 | M28521 | 360TAA AGC CAA TGA AAA CAT TGA TTC A384 | 3 |
| sedAp | sed | 60.85 | M28521 | 491CTT TTA TTT TCT CCT ATT ATT GG ATTTTT463 | 30 |
| sedAb | sed | 61.9 | M28521 | 653CAA TTA ATG CTA TAT CTT ATA GGG TAA ACA TC622 | 3 |
| seeSb | see | 63.41 | M21319 | 424C GAT TGA CCG AAG AAA AAA AAG445 | 30 |
| seeSp | see | 60.2 | M21319 | 479CTA CAG TAC CTA TAG ATA AAG TTA AAA CAA GC510 | 3 |
| seeAp | see | 66.87 | M21319 | 613TTT GCA CCT TAC CGC CAA AG594 | 3 |
| seeAb | see | 60.38 | M21319 | 659TAA CTT ACC GTG GAC CCT TC640 | 3 |
| segSb | seg | 66.14 | AF064773 | 229CAA CCC/T GAT CCT AAA TTA GAC GAA C253 | 2 |
| segSp | seg | 63.09 | AF064773 | 285GGG AAC TAT GGG T(/a)AA TGT AAT GAA TC310 | 2 |
| segAp | seg | 62.61 | AF064773 | 338CTT CCT TCA ACA GGT GGA GAC318 | 2 |
| segAb | seg | 62.91 | AF064773 | 485/401GGA ACG CCA AAA ATG TCT ACT T464/379 | 35 |
| sehSb | seh | 60.86 | U11702 | 407TTA GAA ATC AAG GTG ATA GTG GC 429 | 2 |
| sehSp | seh | 61.25 | U11702 | 454ACT GCT GAT TTA GCT CAG AAG TTT A 478 | 2 |
| sehAp | seh | 60.1 | U11702 | 575AGT GTT GTA CCT CCA TAT AGA C ATTC550 | 35 |
| sehAb | seh | 60.47 | U11702 | 641TTT TGA ATA CCA TCT ACC CAA AC619 | 2 |
| seiSb | sei | 63.01 | AY158703 | 396G GCC ACT TTA TCA GGA CAA TAC TT419 | 2 |
| seiSp | sei | 61.91 | AY158703 | 656A CA C(a)TG GTA AAG GC(t)A AAG AAT ATG679 | 2 |
| seiAp | sei | 62.26 | AY158703 | 726AAA ACT TAC AGG CAG TCC ATC TC704 | 2 |
| seiAb | sei | 58.23 | AY158703 | 818AAT TAT CAT TAG TTA CTA TCT ACA TAT GAT ATT TC784 | 35 |
| etaSb | eta | 61.39 | M17347 | 374CTA GTG CAT TTG TTA TTC AAG ACG397 | 3 |
| etaSp | eta | 69.51 | M17347 | 414CCA TGC AAA AGC AGA AGT TTC AGC 437 | 3 |
| etaAp | eta | 60.67 | M17347 | 492TGC A(/g)TT GAC ACC ATA GTA CTT ATT C468 | This study |
| etaAb | eta | 62.72 | M17347 | 794AAT GCT AAA TCA ACA CCT GC AC773 | 30 |
| etbSb | etb | 61.26 | M17348 | 190TAC CAC CTA ATA CCC TAA TAA TCC AA215 | 3 |
| etbSp | etb | 61.37 | M17348 | 286GAG ACA GTG CAT TAA ATG AAT AAC TTT312 | 3 |
| etbAp | etb | 62.41 | M17348 | 539GAT TTC TTC TGC GCT GTA TTC TT517 | This study |
| etbAb | etb | 61.16 | M17348 | 609C ATT ATC CGT AAT GTG TGT ATAAA GC584 | 43 |
| etdSb | etd | 61.75 | AB057421 | 5963GCT CGG ATA CCC TTA TAA CTT TTC5986 | This study |
| etdSp | etd | 62.2 | AB057421 | 6055CTG AGT CGG GAA ATT CTG G6073 | 43 |
| etdAp | etd | 61.47 | AB057421 | 6120CAA CAT GAA TAC CA0A CTA ACT CTC C6096 | This study |
| etdAb | etd | 61.88 | AB057421 | 6259CAT TAC TAA TGA GAC TGT AAT TCA GCT CT6231 | This study |
| tsstSb | tsst-1 | 65.22 | J02615 | 348AAG CCA ACA TAC TAG CGA AGG AAC371 | 3 |
| tsstSp | tsst-1 | 60.5 | J02615 | 394GGC GTT ACA AAT ACT GAA AAA TTA C418 | 30 |
| tsstAp | tsst-1 | 64.36 | J02615 | 495ATC GAA CTT TGG CCC(/a) ATA CTT T474 | 3 |
| tsstAb | tsst-1 | 61.03 | J02615 | 556GTA TTT GAG TTA GCT GAT GAC GAA533 | 43 |
| pvlSb | pvl | 65.29 | X72700 | 2651TTT TAG GCT CAA GAC AAA GCA AC2673 | This study |
| pvlAp | pvl | 65.3 | X72700 | 2731TAC CTC TGG ATA ACA CTG GCA TTT T2707 | 11 |
| pvlSp | pvl | 61.76 | X72700 | 2733CTT CAA TCC AGA ATT TAT TGG TGT 2756 | 11 |
| pvlAb | pvl | 65.8 | X72700 | 2783TTT GCA GCG TTT TGT TTT CG2764 | 11 |
S, sense; A, antisense; b, biotin labeled (all the primers were biotin labeled at the 5′ end); p, probe (all the probes were 5′ end C6 amine labeled).
Tm values were provided by the primer synthesizer (Sigma-Aldrich).
Boldface numbers represent the numbered base positions at which primer/probe sequences start and finish (starting at point “1” of the corresponding GenBank sequence). Underlined portions indicate modifications of published primer/probe sequences. The bases in parenthesis represent sequences with polymorphisms compared with GenBank sequences or our own sequencing results (for five probes with heterogeneous hybridization).
Primers and probes were used as previously published (some with modification) except, as indicated, those designed for this study.
The mPCR/RLB was performed as previously described (22) with the following modifications: each 25-μl reaction mixture contained 0.5 U Hotstar Taq polymerase (QIAGEN, Melbourne, Australia), and the mPCR annealing temperature was optimized to 55°C.
Sequencing, sequence analysis, and phylogenetic tree.
femA, nuc, and spa PCR primers were based on the published GenBank sequences using BioManager (http://biomanager.angis.org.au/). Sequencing was performed as described previously (24). For most targets, outer primers were used for amplification and inner primers for sequencing (Table 3).
TABLE 3.
Primers used for PCR sequencing of nuc, femA, and spa genes
| Primer | Target | Tm (°C) | GenBank accession no. | Primer sequence (5′-3′)c |
|---|---|---|---|---|
| nucS1a | nuc | 60.3 | V01281 | 226ATGACAGAATACTTATTAAGTGCTGG251 |
| nucS2b | nuc | 60.6 | V01281 | 232GAATACTTATTAAGTGCTGGCATATG257 |
| nucA1b | nuc | 63.9 | V01281 | 908TGACCTGAATCAGCGTTGTC889 |
| nucA2a | nuc | 63.7 | V01281 | 912TTATTGACCTGAATCAGCGTTG891 |
| femAS1a | femA | 64.1 | X17688 | 577ATGAAATTAATTAACGAGAGACAAATAGGAG607 |
| femAS2b | femA | 65.4 | X17688 | 591CGAGAGACAAATAGGAGTAATGATAATGAAG621 |
| femAA0b | femA | 67.3 | X17688 | 1868CTGTCTTTAACTTTTTTAAGTGCGGTATATGC1837 |
| femAAa | femA | 68.3 | X17688 | 1878CTAAAAAATTCTGTCTTTAACTTTTTTAAGTGCGG1844 |
| spaSa | spa | 71.7 | J01786 | 1077CTT CAT CCA AAG CCT TAA AGA CGA TCC TTC1106 |
| spaAa | spa | 71.4 | J01786 | 1543CAA TTT TGTCAG CAG TAG TGC CGT TTG1517 |
| spaSEQb | spa | 71.9 | J01786 | 1540TTT TGTCAG CAG TAG TGC CGT TTG CT1515 |
For most targets, outer primers were used for amplification and, less commonly, for sequencing.
Inner primers were mainly used for sequencing, since they gave better results.
Boldface numbers represent the numbered base positions at which primer sequences start and finish (starting at point “1” of the corresponding GenBank sequence).
The spa types were defined by reference to the shareable web-based database (www.spaServer.Ridom.de) (15). All spa repeat regions were submitted to the database, and spa types were assigned by the database by combining the sequences of all repeat regions.
Data obtained from different typing methods were recorded and stored in an Access file, which was imported into the BioNumerics software program (Applied Maths) with appropriate formatting. A phylogenetic tree was generated by using the categorical coefficient and clustered by the Ward algorithm.
Calculation of index of diversity.
Simpson's index of diversity was calculated for each individual genotyping method and for combinations of methods, as described by Hunter and Gaston (16).
Nucleotide sequence accession numbers.
The nearly full-length sequences (see below) of selected femA and nuc genes and partial spa sequences were deposited in GenBank with the following accession numbers: for femA, DQ103589 and DQ352456 to DQ352463; for nuc, DQ507377 to DQ507382; and for spa, EF094507 to EF094528. Eight S. aureus genome sequences were used for reference: AJ938182 (RF122), NC_002952 (MRSA252), AF144661 (Staphylococcus aureus subsp. anaerobius), NC_002745(N315), NC_002758 (Mu50), NC_003923 (MW2), NC_002951 (COL), and NC_002953 (MSSA476).
RESULTS AND DISCUSSION
Sequence polymorphisms of femA and nuc.
Nine femA sequence types with lengths of approximately 1,215 bp were identified, of which five were newly identified and four had been previously deposited in GenBank. A total of 39 polymorphism sites were found among those sequences. Seven nuc sequence types of approximately 700 bp were identified, with 28 polymorphisms sites. Four sequence types had not been previously identified.
spa types and TGPs.
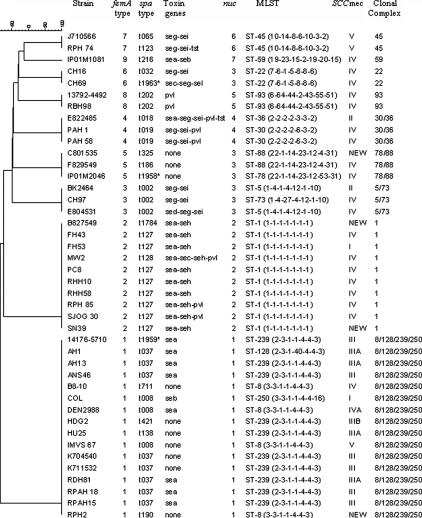
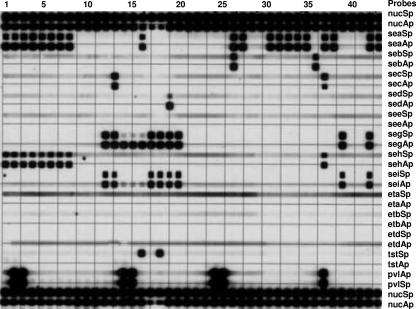
Twenty-two spa types were identified, and sequences of each new type were submitted to GenBank with accession numbers shown in Table 1. Nineteen of the spa types were already recorded in the spa database (www.spaServer.Ridom.de) (15), and three were new types, first identified in Australian strains (Table 1; Fig. 1). There were 14 different TGPs (Fig. 1 and 2).
FIG. 1.
Relatedness of 42 MRSA strains between different typing methods. *, strains CH 69, IPO1M2046, and 14176-5710 belonged to spa types t1963, t1958, and t1959, respectively, which have not been previously deposited in the database.
FIG. 2.
The 13 toxin gene profiles of the 41 strains. Lanes 1 to 42 show results for the following isolates, in order (see Table 1): FH43, SJOG 30, RPH85, B827549, SN39, RHH58, RHH10, FH53, B8-10, RPH2, IMVS67, CH16, CH69, PAH58, PAH1, E822485, J710566, RPH74, E804531, CH97, IP01M2046, C801535, F829549, RBH98, 13792-4492, IP01M1081, 14176-5710, K704540, K711532, AH13, RDH81, AH1, RPAH18, RPAH15, COL, MW2, DEN2988, BK2464, HDG2, HU25, a control strain, and ANS46.
Comparison of MLST, femA and nuc sequence types, spa types, and TGPs.
MLST are based on sequences of seven housekeeping genes (http://www.mlst.net/). Isolates with identical sequences for all seven genes are considered to be clonal and those with five or six matching genes to belong to the same CC (27). There were 15 MLST and nine CCs among the 42 strains studied (Table 1). The latter correlated exactly with femA sequence types, suggesting that femA sequencing may be a useful “shorthand” single-locus surrogate for MLST (Fig. 1). In future, informative single nucleotide polymorphisms in the femA sequence may be able to predict femA type and CC. One of the candidate methods is rolling-circle amplification, which has been used successfully in our laboratory (42).
There were seven nuc sequence types, which were therefore less informative. One sequence type was represented among three femA sequence types (and corresponding CCs).
The relationships between MLST and TGP varied (Fig. 1). No toxin genes were found in 10 isolates belonging to four sequence types (STs) (ST-8, -78, -88, and -239). One to five toxin genes were found in various combinations in the remaining isolates. Some STs included more than one TGP, e.g., ST-5, -22, -45, and -239 each included isolates with two different TGPs, which reflects the ability of mobile genetic elements on which toxin genes are carried to transfer laterally between clones.
However, some toxin genes are transferred vertically within specific CCs (32, 36). For example, all 10 ST-1 isolates (but none belonging to other STs) contained sea and seh; sea alone was found in another eight isolates belonging to CC 8/239. seg and sei, which are part of the enterotoxin gene complex (egc), were always present together, in 10 isolates spread among four CCs (ST-5/73, -22, -45, and -30/36). This is consistent with a previous report that egc is preferentially distributed among CCs 5 and 30/36 (19). In addition, we identified mutations in regions of seg and sei probes in isolates belonging to CC 30/36.
The Panton-Valentine leukocidin gene (pvl) was identified, with a variety of other toxin genes (depending on ST/CC), in eight isolates belonging to ST-1 (three isolates), ST-93 (two isolates), or CC 30/36 (three isolates); seven of eight PVL-containing isolates belonged to SCCmec type IV, which is generally associated with community-acquired MRSA. PVL is associated with necrotic skin and soft tissue lesions and, more recently, with life-threatening necrotizing pneumonia and sepsis due to community-acquired MRSA (8, 44).
There were 22 spa types among the 42 strains tested. When combined with TGP, some spa types were further subdivided, making a total of 26 genotypes. For example, isolates belonging to spa type t002 contained two TGPs (seg-sei and sed-seg-sei), and those belonging to t008 contained three (sea, seb, and none). The combination of these two methods thus provides a high level of discrimination, using relatively inexpensive, rapid methods.
Comparison of discriminatory powers of each genotyping method and various combinations (Table 4) showed that spa typing is the most discriminatory. The addition of TGPs alone or TGP plus SCCmec typing increases the discriminatory power, but there is little additional increase from additional femA or nuc sequence typing.
TABLE 4.
Comparison of discriminatory powers of each genotyping method and various combinations of methods for 42 MRSA strains using Simpson index of diversity
| Genotyping method(s) | No. of types | % of largest type | DIa |
|---|---|---|---|
| Individual | |||
| SCCmec | 9 | 45.2 | 0.764 |
| nuc sequence types | 7 | 38.1 | 0.77 |
| femA sequence types | 9 | 38.1 | 0.794 |
| TGPs | 14 | 23.8 | 0.88 |
| MLST | 15 | 23.8 | 0.882 |
| spa types | 22 | 19 | 0.926 |
| Combinations | |||
| femA-spa-TGP | 27 | 14.3 | 0.959 |
| TGP-spa | 27 | 14.3 | 0.959 |
| TGP-spa-nuc | 27 | 14.3 | 0.959 |
| TGP-spa-SCCmec | 30 | 9.5 | 0.98 |
| femA-spa-TGP-SCCmec | 31 | 9.5 | 0.981 |
| femA-spa-TGP-nuc-MLST-SCCmec | 34 | 9.5 | 0.987 |
DI, Simpson index, calculated according to the method of Hunter and Gaston (16).
Significance of sequence polymorphisms.
These results indicate that femA and to some extent nuc sequence types correlate closely with MLST in this set of MRSA isolates, suggesting that these genes evolve at a rate similar to that of housekeeping genes within CCs (20, 27). spa types were more discriminatory for strain typing but correlated less well with MLST or CCs.
Significant sequence variation in femA and nuc also has potential implications for their use as species-specific PCR targets for identification of S. aureus. The possibility of mutations needs to be considered in the design of probes and primers to avoid false-negative or inaccurate quantitative PCR results. Our results show that some mutations occurred in the region of primers used as species-specific primers (1).
There were significant sequence polymorphisms in femA and nuc genes, which have not been previously well studied, which has potential implications for their use as species-specific PCR targets for identification of S. aureus. Both correlated well with each other, and femA sequence types correlated with MLST/CCs. TGPs provide useful information about potential virulence and the evolutionary history of S. aureus strains and can increase the discriminatory power of femA and spa sequence typing. Prospective testing of unselected clinical isolates will be needed to adequately determine the optimal combination of methods for MRSA surveillance.
Acknowledgments
We sincerely thank the following colleagues for allowing us to study their isolates: Herminia de Lencastre, Philip Giffard, and Graeme Nimmo.
Fanrong Kong, Qinning Wang, and Yongwei Cai made similar contributions to this work and so would be seen as co-first authors.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Alarcon, B., B. Vicedo, and R. Aznar. 2006. PCR-based procedures for detection and quantification of Staphylococcus aureus and their application in food. J. Appl. Microbiol. 100:352-364. [DOI] [PubMed] [Google Scholar]
- 2.Becker, K., A. W. Friedrich, G. Lubritz, M. Weilert, G. Peters, and C. von Eiff. 2003. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 41:1434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, K., R. Roth, and G. Peters. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 36:2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger-Bachi, B., and S. Rohrer. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165-171. [DOI] [PubMed] [Google Scholar]
- 5.Brakstad, O. G., K. Aasbakk, and J. A. Maeland. 1992. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30:1654-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coombs, G. W., G. R. Nimmo, J. M. Bell, F. Huygens, F. G. O'Brien, M. J. Malkowski, J. C. Pearson, A. J. Stephens, and P. M. Giffard. 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa, A. M., I. Kay, and S. Palladino. 2005. Rapid detection of mecA and nuc genes in staphylococci by real-time multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 51:13-17. [DOI] [PubMed] [Google Scholar]
- 8.Denis, O., A. Deplano, H. De Beenhouwer, M. Hallin, G. Huysmans, M. G. Garrino, Y. Glupczynski, X. Malaviolle, A. Vergison, and M. J. Struelens. 2005. Polyclonal emergence and importation of community-acquired methicillin-resistant Staphylococcus aureus strains harbouring Panton-Valentine leucocidin genes in Belgium. J. Antimicrob. Chemother. 56:1103-1106. [DOI] [PubMed] [Google Scholar]
- 9.Deplano, A., R. De Mendonca, R. De Ryck, and M. J. Struelens. 2006. External quality assessment of molecular typing of Staphylococcus aureus isolates by a network of laboratories. J. Clin. Microbiol. 44:3236-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferry, T., D. Thomas, A. L. Genestier, M. Bes, G. Lina, F. Vandenesch, and J. Etienne. 2005. Comparative prevalence of superantigen genes in Staphylococcus aureus isolates causing sepsis with and without septic shock. Clin. Infect. Dis. 41:771-777. [DOI] [PubMed] [Google Scholar]
- 11.Francois, P., G. Renzi, D. Pittet, M. Bento, D. Lew, S. Harbarth, P. Vaudaux, and J. Schrenzel. 2004. A novel multiplex real-time PCR assay for rapid typing of major staphylococcal cassette chromosome mec elements. J. Clin. Microbiol. 42:3309-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert, F. B., A. Fromageau, L. Gelineau, and B. Poutrel. 2006. Differentiation of bovine Staphylococcus aureus isolates by use of polymorphic tandem repeat typing. Vet. Microbiol. 117:297-303. [DOI] [PubMed] [Google Scholar]
- 13.Gomes, A. R., S. Vinga, M. Zavolan, and H. de Lencastre. 2005. Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamels, S., J. L. Gala, S. Dufour, P. Vannuffel, N. Zammatteo, and J. Remacle. 2001. Consensus PCR and microarray for diagnosis of the genus Staphylococcus, species, and methicillin resistance. BioTechniques 31:1364-1366, 1368, 1370-1372. [DOI] [PubMed] [Google Scholar]
- 15.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huygens, F., J. Inman-Bamber, G. R. Nimmo, W. Munckhof, J. Schooneveldt, B. Harrison, J. A. McMahon, and P. M. Giffard. 2006. Staphylococcus aureus genotyping using novel real-time PCR formats. J. Clin. Microbiol. 44:3712-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huygens, F., A. J. Stephens, G. R. Nimmo, and P. M. Giffard. 2004. mecA locus diversity in methicillin-resistant Staphylococcus aureus isolates in Brisbane, Australia, and the development of a novel diagnostic procedure for the Western Samoan phage pattern clone. J. Clin. Microbiol. 42:1947-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarraud, S., G. Cozon, F. Vandenesch, M. Bes, J. Etienne, and G. Lina. 1999. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J. Clin. Microbiol. 37:2446-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kizaki, M., Y. Kobayashi, and Y. Ikeda. 1994. Rapid and sensitive detection of the femA gene in staphylococci by enzymatic detection of polymerase chain reaction (ED-PCR): comparison with standard PCR analysis. J. Hosp. Infect. 28:287-295. [DOI] [PubMed] [Google Scholar]
- 22.Kong, F., and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB)—a practical epidemiological and diagnostic tool. Nat. Protoc. 1:2668-2680. [DOI] [PubMed] [Google Scholar]
- 23.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsay, J. A., and M. T. Holden. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12:378-385. [DOI] [PubMed] [Google Scholar]
- 27.Lindsay, J. A., C. E. Moore, N. P. Day, S. J. Peacock, A. A. Witney, R. A. Stabler, S. E. Husain, P. D. Butcher, and J. Hinds. 2006. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macfarlane, L., J. Walker, R. Borrow, B. A. Oppenheim, and A. J. Fox. 1999. Improved recognition of MRSA case clusters by the application of molecular subtyping using pulsed-field gel electrophoresis. J. Hosp. Infect. 41:29-37. [DOI] [PubMed] [Google Scholar]
- 29.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyszton-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellmann, A., A. W. Friedrich, N. Rosenkotter, J. Rothganger, H. Karch, R. Reintjes, and D. Harmsen. 2006. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 3:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore, P. C., and J. A. Lindsay. 2001. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J. Clin. Microbiol. 39:2760-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira, D. C., C. Milheirico, S. Vinga, and H. de Lencastre. 2006. Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J. Antimicrob. Chemother. 58:23-30. [DOI] [PubMed] [Google Scholar]
- 35.Omoe, K., M. Ishikawa, Y. Shimoda, D. L. Hu, S. Ueda, and K. Shinagawa. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J. Clin. Microbiol. 40:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peacock, S. J., G. D. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruppitsch, W., A. Indra, A. Stoger, B. Mayer, S. Stadlbauer, G. Wewalka, and F. Allerberger. 2006. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2442-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla, S. K., M. E. Stemper, S. V. Ramaswamy, J. M. Conradt, R. Reich, E. A. Graviss, and K. D. Reed. 2004. Molecular characteristics of nosocomial and native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J. Clin. Microbiol. 42:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens, A. J., F. Huygens, J. Inman-Bamber, E. P. Price, G. R. Nimmo, J. Schooneveldt, W. Munckhof, and P. M. Giffard. 2006. Methicillin-resistant Staphylococcus aureus genotyping using a small set of polymorphisms. J. Med. Microbiol. 55:43-51. [DOI] [PubMed] [Google Scholar]
- 41.Tang, Y. W., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Kohner, L. M. Highsmith, H. Li, F. R. Cockerill III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 38:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong, Z., F. Kong, B. Wang, X. Zeng, and G. L. Gilbert. 2007. A practical method for subtyping of Streptococcus agalactiae serotype III, of human origin, using rolling circle amplification. J. Microbiol. Methods 70:39-44. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi, T., K. Nishifuji, M. Sasaki, Y. Fudaba, M. Aepfelbacher, T. Takata, M. Ohara, H. Komatsuzawa, M. Amagai, and M. Sugai. 2002. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 70:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamasaki, O., T. Yamaguchi, M. Sugai, C. Chapuis-Cellier, F. Arnaud, F. Vandenesch, J. Etienne, and G. Lina. 2005. Clinical manifestations of staphylococcal scalded-skin syndrome depend on serotypes of exfoliative toxins. J. Clin. Microbiol. 43:1890-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]