Abstract
Due to high cost, availability of human immunodeficiency virus type 1 (HIV-1) drug resistance testing in resource-poor settings is still limited. We therefore evaluated the usefulness of viral DNA extracted from either whole blood or dried blood spots (DBS). Samples were collected from 50 patients receiving therapy and 10 therapy-naïve patients. Amplification and sequencing of RNA and DNA was performed using an in-house assay. Protease (PR) and reverse transcriptase (RT) sequences of plasma viral RNA were obtained for 96.6% and 89.7%, respectively, of the 29 patients with a detectable viral load. For cellular viral DNA, useful PR and RT sequences were obtained for 96.6% and 93.1% of the whole-blood-cell samples and for 93.1% and 93.1% of the DBS samples, respectively. For the 31 patients with an undetectable viral load, PR and RT sequences were obtained for 67.7% and 61.3% of the whole-blood-cell DNA preparations and for 54.8% and 58.1% of the DBS DNA preparations, respectively. A good correlation between RNA and DNA sequences was found; most discordances were caused by the detection of mixed amino acids. Of the RT drug-resistant mutations, 13 (38.2%) were seen in RNA only, 6 (17.6%) in DNA only, and 15 (44.1%) in both. Repeated amplification and sequencing of DNA extracts revealed a lack of reproducibility for the detection of drug resistance mutations in a number of samples, indicating a possible founder effect. In conclusion, this study shows the feasibility of genotypic drug resistance testing on whole blood cells or DBS and its possible usefulness for HIV-1 subtyping or examining the overall distribution of drug resistance in a population. For individual patients, RNA sequencing was shown to be superior to DNA sequencing, especially for patients who experienced early treatment failure. The use of DNA extracted from whole blood or DBS for the detection of archived drug resistance mutations deserves further study.
Today, more than one million people in low- and middle-income countries have access to antiretroviral treatment (ART). Even though this is only 23% of the estimated 4.6 million human immunodeficiency virus type 1 (HIV-1)-infected individuals who are in need of highly active ART (HAART), it proves that the delivery of ART in resource-poor settings is feasible (41). Experiences from Europe and the United States, however, indicate that the emergence of HIV drug resistance remains an important obstacle to the long-term success of therapy. An adequate follow-up of patients on treatment, in order to identify cases in which therapy has failed, is essential to avoiding accumulation of drug resistance. Efforts to lower the prices for CD4 and viral load testing are being made, and alternative methods for measuring these parameters in resource-poor settings are being developed and evaluated (8, 12, 17). Unfortunately, genotypic resistance testing remains almost inaccessible due to its high cost, the need for specialized laboratories, and the logistical challenges, such as the requirements for a cold chain to transport samples from the field to reference laboratories. However, ART drug resistance testing in patients in whom treatment is failing remains very important, not only to guide a second line of treatment for individuals but also to allow the monitoring of the emergence and distribution of drug-resistant virus in the population.
In a previous study, we presented a sensitive method for combined detection of the presence of HIV-1 in plasma and subsequent sequencing of the protease (PR) and reverse transcriptase (RT) genes (36). Although this technique allows a significant reduction in the price of follow-up testing, the need for a proper infrastructure for collection and transportation of blood samples remains.
The aim of the current study was to evaluate the possibility of using cellular DNA instead of viral RNA for the detection of drug resistance mutations. As DNA is more stable than RNA, the use of DNA would eliminate the necessity for fast processing of the blood samples after collection and reduce the complexity of the manipulations. The fact that a reverse transcription step is no longer needed for the amplification of the genes of interest also reduces the overall cost of the procedure. Blood spots collected on filter paper have been used for DNA amplification purposes, but reports on the feasibility of performing genotypic analysis for HIV-1 drug resistance remain scarce (24, 33, 43). In resource-poor settings, dried blood spots (DBS) are the preferred method of sample collection in the field, as they do not require electricity or a cold chain. Moreover, once the samples are air dried on the paper, they can be handled as noninfectious and transported to the laboratory without the risk of infection transmission (19).
Whether the same resistance information can be obtained from DNA sequencing and RNA sequencing is still unclear. Results of published reports in this regard are very inconsistent (6, 10, 14, 21, 24, 34). The present study aims to investigate both the possibility of DNA sequencing from whole blood cells or from DBS and the usefulness of DNA sequencing for determination of drug resistance in patients from a resource-poor setting with a high level of HIV-1 subtype diversity.
MATERIALS AND METHODS
Patients and samples.
In May 2006, a total of 50 patients on HAART and 10 ART-naïve patients were randomly selected from the HIV Comprehensive Care Centre at Coast Province General Hospital in Mombasa, Kenya. The patients were seen during regular follow-up visits, and written informed consent for participation in the study was obtained. The study was approved by the ethics review committees of the University of Nairobi and the Ghent University. The mean age of the patients was 36.9 years (standard deviation, ±9.2 cells/mm3; range, 21 to 65 years), 38 (63%) of the patients were women, and 66% of the patients were in WHO clinical stage 3 or 4 (42). The mean CD4 count was 322 cells/mm3 (standard deviation, ±166 cells/mm3; range, 41 to 757 cells/mm3). Patients on HAART were treated with the current regimen for a median of 12 months (ranging from 1 week to 33 months). The current regimen of two nucleoside RT inhibitors (NRTIs) (stavudine [d4T]/zidovudine [AZT] and lamivudine [3TC]), and one nonnucleoside RT inhibitor (NNRTI) (nevirapine [NVP] or efavirenz [EFV]), was the first-line regimen for 48 patients. Two patients received a second-line protease inhibitor (PI; lopinavir/ritonavir [LPV/r])-based regimen.
Ten milliliters of EDTA blood was collected, and 50 μl of whole blood was immediately spotted four times on filter paper (Schleicher and Schuell 903). Filter papers were air dried overnight and stored at −20°C until processing. CD4 cell counts were performed (FACScount; Becton Dickinson Immunocytometry, Oxford, United Kingdom), and the remainder of the EDTA blood was centrifuged to collect the plasma and the buffy coat cells. Plasma and whole blood cells were stored at −80°C and −20°C, respectively. All samples were shipped to the AIDS Reference Laboratory at the University Hospital in Ghent, Belgium.
RNA viral load testing.
HIV RNA quantification was performed using the ultrasensitive Cobas Amplicor HIV-1 Monitor test, version 1.5 (Roche Molecular Systems, Branchburg, NJ), with a detection limit of 50 copies/ml.
DNA extraction.
DNA was extracted from the blood cells by use of the QIAamp DNA blood mini kit (QIAGEN, Venlo, The Netherlands), following the instructions of the manufacturer, and eluted in 50 μl elution buffer.
The procedure to extract DNA from the DBS was adopted from that of Fisher et al. (15). Briefly, one whole DBS was cut into small pieces and transferred to a 2-ml tube. One milliliter of phosphate-buffered saline solution (Cambrex Bio Science, Verviers, Belgium) with 0.1% Tween 20 (Sigma-Aldrich, Bornem, Belgium) was added. The mixture was subjected to a vortex briefly, and after 10 min of incubation at room temperature, the supernatant was removed. This wash step was repeated twice. After the final wash step, 200 μl of 5% Chelex-100 resin solution (Bio-Rad, Nazareth, Belgium) was added and the sample was incubated for 30 min at 56°C and another 30 min at 95°C. The supernatant was then transferred to a clean tube and stored at −20°C until processing.
Amplification of RNA and DNA extracts.
Amplification of HIV RNA was performed by nested RT-PCR as previously described (36). DNA extracts from the blood cells and the DBS were amplified by nested PCR using the same primer sets as the ones used for RNA amplification. The first round of amplification was carried out using 5 U/μl of Taq polymerase (Applied Biosystems Inc., Foster City, CA) in a 25-μl reaction volume containing buffer II (Applied Biosystems) with 25 mM MgCl2, 1.25 mM deoxynucleoside triphosphates (Amersham Biosciences, Buckinghamshire, United Kingdom), 0.1% bovine serum albumin (Roche Diagnostics, GmbH Mannheim, Germany), and 0.05 μM each of two forward primers (GAG2 [5′-GAGGAAGCTGCAGAATGGG-3′] and PR1 [5′-ATGATGCAGAGAGGCAATTT-3′]) and two reverse primers (RT137 [5′-TTCTGTATGTCATTGACAGTCCAGC-3′] and RT3303 [5′-TAAYTTYTGTATRTCATTGAC-3′]). Primers were selected to have a broad specificity for different HIV-1 subtypes.
Cycling conditions were 94°C for 5 min followed by 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min and a final extension for 7 min at 72°C. Two-microliter volumes of the outer RT-PCR products were used in two separate inner PCRs, one with nested PR primers (PR3 [5′-AGAGCCAACAGCCCCACCA-3′] and PR4 [5′-GGGCCATCCATTCCTGGCTT-3′]) and one with nested RT primers (RT1 [5′-CCAAAAGTTAAACAATGGCCATTGACAGA-3′] and RT4 [5′-AGTTCATAACCCATCCAAAG-3′]). The second-round PCR was carried out using Taq polymerase (Applied Biosystems Inc., Foster City, CA) in a 50-μl reaction volume containing Taq buffer II with 5 U/μl of Taq polymerase, 25 mM MgCl2, 1.25 mM deoxynucleoside triphosphates (Amersham Biosciences, Buckinghamshire, United Kingdom), and 0.05 μM each of the specific forward and reverse primers. The second-round PCR included an initial denaturation step at 94°C for 5 min followed by 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s and a final extension for 7 min at 72°C.
In each PCR run, a sample containing 10 copies of HIV-1 DNA (DNA equivalent of 10 8E5 cells) was included as a positive control. Ultrapure water was used as a negative control. Results were accepted only if the results of both controls were correct.
The PR primers amplify a 458-bp fragment spanning the whole PR gene. The RT primers amplify a 646-bp fragment spanning nucleotides corresponding to amino acids 27 to 227 of the RT gene. The amplification products were visualized after electrophoresis in a 1% agarose gel with ethidium bromide.
Genotyping of PR and RT genes.
Genotyping was performed using a homemade sequencing assay as described earlier (36). Direct sequencing of both sense and antisense strands was done with the dRhodamine Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems Inc., Foster City, CA). Sequencing reaction products were analyzed on an ABI310 or ABI3130XL genetic analyzer (Applied Biosystems Inc.). All validations and subsequent manipulations of the sequencing results, as well as the interpretations of the genotyping data and the subtyping, were performed using the Smartgene HIV software packages (Integrated Database Network System; Smartgene, Zug, Switzerland). The selection of drug resistance mutations was based on the Stanford algorithm (http://hivdb.stanford.edu).
Phylogenetic analyses.
Phylogenetic analyses were performed using version 3.6 of the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html) with a maximum-likelihood distance matrix and a transition-to-transversion ratio of 2.0. Tree diagrams were plotted with Treeview, version 1.4 (included in the PHYLIP package). Nucleotide differences were calculated using the Emboss infoalign software (http://embossgui.sourceforge.net/demo/infoalign.html).
Statistical analysis.
All statistical analyses were performed using SPSS 15.0 (SPSS, IL). Pearson's chi-square test was used to detect possible statistically significant differences between two groups of samples.
RESULTS
Viral load determination and RNA sequencing.
A detectable viral load (>50 copies/ml) was found in all 10 treatment-naive patients (mean, 35,555 copies/ml; range, 73 to >100,000 copies/ml) and in 19 of the 50 treated patients (mean, 20,377 copies/ml; range, 55 to >100,000 copies/ml).
Sequencing of the viral RNA was attempted on all 29 of these samples and was successful for PR in 28 (96.6%) and for RT in 26 (89.7%). The three patients for whom one or both failed had very low viral loads (73, 74, and 81 copies/ml, respectively). The viral load was undetectable in 31 patients, all treated.
Efficiency of DNA sequencing on whole blood cells and DBS.
Sequencing of the PR and RT genes from whole-blood-cell DNA was successful for 28 (96.6%) and 27 (93.1%), of the 29 blood-cell samples collected from patients with a detectable viral load, respectively, and for 21 (67.7%) and 19 (61.3%) of the 31 samples from patients with an undetectable viral load, respectively. For the DBS, the success rate was slightly lower, with 27 (93.1%) successful sequencing reactions for both PR and RT from the patients with a detectable viral load and 17 (54.8%) PR and 18 (58.1%) RT sequences from the patients with undetectable viral load. No statistically significant difference between the efficiency of genotyping on whole blood DNA and the efficiency of genotyping on DBS DNA was found (P values of 0.2743 for PR and 0.8311 for RT [Pearson's χ2 test]).
Subtype distribution.
For the 56 patients from whom PR and RT sequences were available, from DNA, RNA, or both, subtyping of the HIV-1 strains was performed by similarity search using the Smartgene subtyping tool. Results showed that the majority of patients was infected with a subtype A virus (n = 31); in order of prevalence, subtype A was followed by subtypes D (n = 7), CRF16_AD (n = 7), C (n = 6), and G (n = 1). Discordances between the subtype determined from the PR sequence and the subtype determined from the RT sequence were seen for four patients (PR/RT subtype combinations D/C, A/C, CRF_AE/C, and C/A). The results of the subtype determination were not influenced by the sample type.
Comparison of the nucleotide sequences from whole-blood DNA, DBS DNA, and plasma RNA.
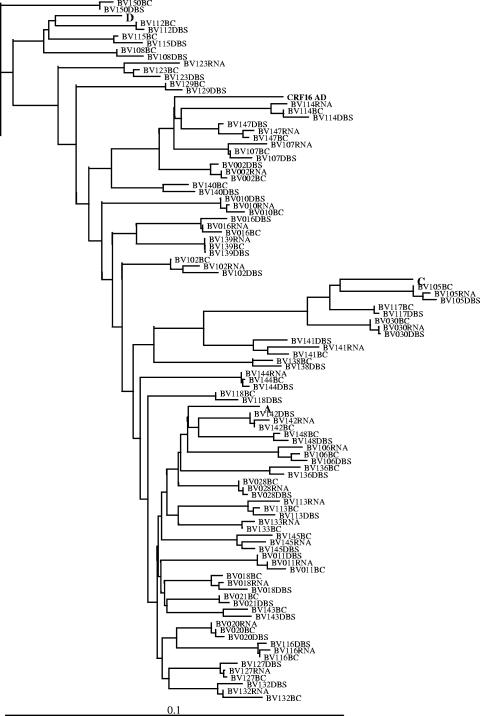
A phylogenetic tree was constructed using 100 PR and RT gene sequences from 38 patients (Fig. 1). The tree revealed a close clustering of the DNA and RNA sequences from the same patients in all cases. The mean nucleotide difference, expressed as the percentage of the total nucleotide length (857 bp), between paired DNA sequences of blood cells and DBS was 2.10% (range, 0.35% to 4.00%). The mean nucleotide difference between the sequences obtained from plasma RNA and blood-cell DNA and the mean nucleotide difference between the sequences obtained from plasma RNA and DBS DNA were 1.82% (range, 0.35% to 3.73%) and 1.90% (range, 0.58% to 3.50%), respectively. Mixtures of nucleotides (International Union of Pure and Applied Chemistry nomenclature) were considered differences. For comparison, the observed mean nucleotide differences between duplicate sequencing reactions on RNA and DNA were 1.45% (range, 0% to 3.37%) and 1.61% (range, 0% to 3.61%), respectively.
FIG. 1.
A phylogenetic tree was constructed based on 100 PR and RT gene sequences from 38 patients. Reference strains for subtypes A, C, D, and CRF16_AD were included in the phylogenetic analysis. The tree was rooted with a subtype B strain. BC, blood cells.
Drug resistance mutations in RNA and DNA.
Selection of drug resistance mutations was based on the recent update of the International AIDS Society-USA list (20). Only two patients showed primary mutations in the PR gene (corresponding to L33F and D30N).
The mutation resulting in L33F was detected consistently in all three samples from the same treatment-naïve patient (BV021). The mutation resulting in D30N was detected as part of a mixed population in one of the DNA sequences of patient BV011 (Table 1).
TABLE 1.
Overview of all patients with mutations at resistance-related positions in the PR gene
| Patient | ART (length of treatment) | VL (copies/ml)a | CD4 (cells/mm3) | Sampleb | Residue encoded in place of indicated amino acid
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10L | 13I | 16G | 20K | 30D | 33L | 36M | 46M | 63L | 71A | 73G | 77V | |||||
| BV021 | None | 73 | 474 | DBS | V | E | F | I/T | I | |||||||
| BC | V | E/G | F | T | I/V | |||||||||||
| RNA | V | E | F | T | I | |||||||||||
| BV016 | None | 5,330 | 233 | DBS | V | E | I | |||||||||
| BC | V | E | I | |||||||||||||
| RNA | V | E | I | |||||||||||||
| BV010 | None | 9,790 | 702 | DBS | V/L | I | ||||||||||
| BC | V | I | ||||||||||||||
| RNA | V | I | ||||||||||||||
| BV011 | None | 16,900 | 390 | DBS | V | V | I | T | ||||||||
| BC | V | V | N/D | I | ||||||||||||
| RNA | V | V | I | |||||||||||||
| BV028 | None | 21,600 | 473 | DBS | I/V | I | ||||||||||
| BC | V | I | ||||||||||||||
| RNA | V | I | ||||||||||||||
| BV018 | None | 35,400 | 246 | DBS | I | V | I | I/M | S | |||||||
| BC | I | V | I | |||||||||||||
| RNA | I | V | I | |||||||||||||
| BV030 | None | 66,300 | 109 | DBS | I | |||||||||||
| BC | I | |||||||||||||||
| RNA | I | |||||||||||||||
| BV002 | None | >100,000 | 136 | DBS | V | E | I | I | ||||||||
| BC | V | E | I | I | ||||||||||||
| RNA | V | E | I | I | ||||||||||||
| BV020 | None | >100,000 | 165 | DBS | I/V | V | I | P/L | ||||||||
| BC | I/V | V | I | L/V | ||||||||||||
| RNA | I/V | V | I | L/V | ||||||||||||
| BV107 | d4T + 3TC + EFV (6 mo) | 55 | 338 | DBS | V | E | R | I | T | |||||||
| BC | V | E | R | I | T | |||||||||||
| RNA | V | E | R | I | T | |||||||||||
| BV120 | AZT + 3TC + NVP (41 mo), d4T + 3TC + LPV/r (9 mo) | 81 | 226 | DBS | I | P | ||||||||||
| BC | I | P | ||||||||||||||
| RNA | I | P | ||||||||||||||
| BV142 | AZT + 3TC + NVP (2 mo) | 128 | 236 | DBS | V | R | I | P/L | ||||||||
| BC | V | R | I | |||||||||||||
| RNA | V | R | I | |||||||||||||
| BV145 | d4T + 3TC + EFV (33 mo) | 294 | 206 | DBS | V | I | ||||||||||
| BC | V | I | ||||||||||||||
| RNA | V | I | ||||||||||||||
| BV113 | d4T + 3TC + EFV (32 mo) | 1,030 | 467 | DBS | V | V | E | I | ||||||||
| BC | I | V | E | I | ||||||||||||
| RNA | V | V | E | I | ||||||||||||
| BV102 | d4T + 3TC + NVP (1 wk) | 1,250 | 111 | DBS | V | V | I | |||||||||
| BC | V/L | V | I | |||||||||||||
| RNA | V | V | I | |||||||||||||
| BV127 | d4T + 3TC + EFV (3 mo) | 1,690 | 314 | DBS | V | I | ||||||||||
| BC | V | I | ||||||||||||||
| RNA | V | I | ||||||||||||||
| BV132 | d4T + 3TC + EFV (2 wk) | 2,260 | 41 | DBS | R | I | ||||||||||
| BC | R | I | ||||||||||||||
| RNA | I/V | R | I | |||||||||||||
| BV144 | d4T + 3TC + NVP (2 mo) | 2,520 | 182 | DBS | I | |||||||||||
| BC | I | |||||||||||||||
| RNA | I | |||||||||||||||
| BV123 | d4T + 3TC + NVP (8 mo) | 3,060 | 321 | DBS | V | I | P | |||||||||
| BC | V | R | I | |||||||||||||
| RNA | V | R | I | |||||||||||||
| BV106 | d4T + 3TC + NVP (23 mo) | 3,520 | 300 | DBS | V | I | P | |||||||||
| BC | V | I | P | |||||||||||||
| RNA | V | I | P | |||||||||||||
| BV141 | d4T + 3TC + NVP (13 mo) | 10,400 | 388 | DBS | I | |||||||||||
| BC | I | |||||||||||||||
| RNA | I | |||||||||||||||
| BV114 | d4T + 3TC + EFV (34 mo), AZT + 3TC + LPV/r (1 mo) | 15,300 | 217 | DBS | V | E | R | I | ||||||||
| BC | V | E | R | I | ||||||||||||
| RNA | V | E | R | I | ||||||||||||
| BV105 | AZT + 3TC + EFV (9 mo) | 21,000 | 276 | DBS | I | P | ||||||||||
| BC | I/M | P | ||||||||||||||
| RNA | I | P | ||||||||||||||
| BV139 | d4T + 3TC + EFV (1 wk) | 26,300 | 556 | DBS | V | I | ||||||||||
| BC | V | I | ||||||||||||||
| RNA | V | I | ||||||||||||||
| BV147 | d4T + 3TC+EFV (13 mo) | 98,200 | 218 | DBS | V | E | I | |||||||||
| BC | I | V | E | I | ||||||||||||
| RNA | I/L | V | E | I | ||||||||||||
| BV116 | d4T + 3TC + NVP (22 mo) | >100,000 | 353 | DBS | V | V | R | I | ||||||||
| BC | V | V | R | I | ||||||||||||
| RNA | V | V | R | T | ||||||||||||
| BV133 | d4T + 3TC+NVP (8 mo) | >100,000 | 165 | DBS | NDc | ND | ND | ND | ||||||||
| BC | I/V | V | E | I | ||||||||||||
| RNA | V | V | E | I | ||||||||||||
VL, viral load.
RNA, plasma RNA; BC, blood-cell DNA; DBS, dried blood spot DNA.
NA, not applicable.
Secondary PR mutations were observed in samples from all patients. Most of these mutations (74 of the 83) were consistently detected in the RNA and in both of the DNA sequences, eight mutations were detected in DNA only, and one mutation was detected in RNA only.
No LPV-related PI resistance-associated mutations were observed in the two patients on a PI-based regimen (BV114 and BV120) (Table 1).
No NRTI or NNRTI resistance-associated mutations were observed in samples from the 10 treatment-naïve patients. For the 43 of 50 treated patients for whom at least one RT sequence (RNA and/or DNA) was available, NRTI or NNRTI resistance-associated mutations were detected in 13 samples (Table 2). A total of 34 nucleotide substitutions were seen; 10 of these were detected consistently in the RNA and in both DNA sequences, 5 were detected in the RNA and in one of the DNA sequences, 3 were detected only in the DNA from blood cells, and 2 were detected only in the DBS DNA (Table 2).
TABLE 2.
Overview of all patients with mutations at resistance-related positions in the RT gene
| Patient | ART (length of treatment) | VL (copies/ml)a | CD4 (cells/mm3) | Sampleb | Residue encoded in place of indicated amino acid
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 65K | 67D | 69T | 70K | 75V | 103K | 108V | 151Q | 181Y | 184M | 188Y | 190G | 215T | 219K | 225P | |||||
| BV145 | d4T + 3TC + EFV (33 mo) | 294 | 206 | DBS | N | ||||||||||||||
| BC | N | ||||||||||||||||||
| RNA | N | I | V | ||||||||||||||||
| BV113 | d4T + 3TC + EFV (32 mo) | 1,030 | 467 | DBS | |||||||||||||||
| BC | |||||||||||||||||||
| RNA | N | V | |||||||||||||||||
| BV132 | d4T + 3TC + EFV (2 wk) | 2,260 | 41 | DBS | N | ||||||||||||||
| BC | R | C | |||||||||||||||||
| RNA | N/T | K/N | C/Y | ||||||||||||||||
| BV123 | d4T + 3TC + NVP (8 mo) | 3,060 | 321 | DBS | |||||||||||||||
| BC | |||||||||||||||||||
| RNA | N | V | |||||||||||||||||
| BV106 | d4T + 3TC + NVP (23 mo) | 3,520 | 300 | DBS | |||||||||||||||
| BC | |||||||||||||||||||
| RNA | R/K | V | |||||||||||||||||
| BV141 | d4T + 3TC + NVP (13 mo) | 10,400 | 388 | DBS | |||||||||||||||
| BC | |||||||||||||||||||
| RNA | N | V | |||||||||||||||||
| BV114 | d4T + 3TC + EFV (34 mo), AZT + 3TC + LPV/r (1 mo) | 15,300 | 217 | DBS | |||||||||||||||
| BC | K/N | ||||||||||||||||||
| RNA | N | H | |||||||||||||||||
| BV105 | AZT + 3TC + EFV (9 mo) | 21,000 | 276 | DBS | N | V | |||||||||||||
| BC | K/N | M/V | |||||||||||||||||
| RNA | N | V | |||||||||||||||||
| BV147 | d4T + 3TC + EFV (13 mo) | 98,200 | 218 | DBS | I/V | K/N | I/V | K/E | |||||||||||
| BC | I/V | N | M/V | H/P | |||||||||||||||
| RNA | N | V | F | ||||||||||||||||
| BV116 | d4T + 3TC + NVP (22 mo) | >100,000 | 353 | DBS | R | N | V | ||||||||||||
| BC | N | V | |||||||||||||||||
| RNA | N | V | |||||||||||||||||
| BV133 | d4T + 3TC + NVP (8 mo) | >100,000 | 165 | DBS | N | K/M/Q/L | V | L | A | ||||||||||
| BC | N/T | M/V | L | A | |||||||||||||||
| RNA | N/T | M | V | L | A | ||||||||||||||
| BV112 | AZT + 3TC + NVP (1 mo) | <50 | 242 | DBS | |||||||||||||||
| BC | K/N | ||||||||||||||||||
| BV117 | d4T + 3TC + NVP (21 mo) | <50 | 328 | DBS | K/N | ||||||||||||||
| BC | K/N | ||||||||||||||||||
VL, viral load.
RNA, plasma RNA; BC, blood-cell DNA; DBS, dried blood spot DNA.
Thirteen mutations were found exclusively in the RNA. Six mutations were detected in one or both of the DNA sequences but not in the RNA. A mutation resulting in a replacement with residue 103N was seen in the DNA sequence of two patients on an NNRTI-based regimen but with an undetectable viral load.
The number of resistance mutations detected in the DNA samples of five treated patients with a viral load below 5,000 copies/ml (n = 4) was clearly lower than the number of resistance mutations detected in the DNA samples of six treated patients with a viral load of more than 5,000 copies/ml (n = 17), but the difference did not reach statistical significance (Pearson's chi-square P = 0.11). The numbers of resistance mutations in the RNA in both groups, on the other hand, were comparable, with 12 and 16 mutations found in the RNA from patients with a viral load below 5,000 copies/ml and from patients with a viral load above 5,000 copies/ml (Pearson's chi-square P = 0.83), respectively.
Reproducibility of the DNA sequencing.
Replicate amplification and sequencing reactions were performed on the DNA extracts of all individuals listed in Table 2, with the exception of one ART-naïve patient. The results showed the inconsistent detection of some mutations (Table 3). Five resistance mutations that originally were not detected in the DNA but were present in the RNA sample were picked up in at least one of the replicate DNA-sequencing reactions. However, despite replicate analysis, several important resistance mutations remained undetectable even after triplicate sequencing of the DNA: mutations resulting in the presence of 103N and 184V in patient BV141, mutations resulting in the presence of 103N and 184V in patient BV123, mutations resulting in the presence of 67N and 184V in patient BV113, and a mutation resulting in the presence of 65R in patient BV106.
TABLE 3.
Results of replicate amplification and sequencing reactions performed on DNA extracts
| Patient | ART (length of treatment) | VL (copies/ml)a | CD4 (cells/mm3) | Sampleb | Residue encoded in place of indicated amino acid
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 65K | 67D | 69T | 70K | 75V | 101K | 103K | 108V | 151Q | 181Y | 184M | 188Y | 190G | 215T | 219K | 225P | |||||
| BV145 | d4T + 3TC + EFV (33 mo) | 294 | 206 | DBS1 | N | |||||||||||||||
| DBS2 | N/K | |||||||||||||||||||
| DBS3 | N/K | M/V | ||||||||||||||||||
| BC1 | N | |||||||||||||||||||
| BC2 | N | V | ||||||||||||||||||
| BC3 | N | |||||||||||||||||||
| RNA | N | I | V | |||||||||||||||||
| BV113 | d4T + 3TC + EFV (32 mo) | 1,030 | 467 | DBS1 | ||||||||||||||||
| DBS2 | ||||||||||||||||||||
| DBS3 | ||||||||||||||||||||
| BC1 | ||||||||||||||||||||
| BC2 | ||||||||||||||||||||
| BC3 | ||||||||||||||||||||
| RNA | N | V | ||||||||||||||||||
| BV132 | d4T + 3TC + EFV (2 wk) | 2,260 | 41 | DBS1 | N | |||||||||||||||
| DBS2 | N | C | ||||||||||||||||||
| DBS3 | R | C | ||||||||||||||||||
| BC1 | R | C | ||||||||||||||||||
| BC2 | R | C | ||||||||||||||||||
| BC3 | N | C | ||||||||||||||||||
| RNA | N/T | K/N | C/Y | |||||||||||||||||
| BV123 | d4T + 3TC + NVP | 3,060 | 321 | DBS1 | ||||||||||||||||
| (8 mo) | DBS2 | |||||||||||||||||||
| DBS3 | ||||||||||||||||||||
| BC1 | ||||||||||||||||||||
| BC2 | K/R | |||||||||||||||||||
| BC3 | ||||||||||||||||||||
| RNA | N | V | ||||||||||||||||||
| BV106 | d4T + 3TC + NVP | 3,520 | 300 | DBS1 | ||||||||||||||||
| (23 mo) | DBS2 | V | ||||||||||||||||||
| DBS3 | H/D | |||||||||||||||||||
| BC1 | ||||||||||||||||||||
| BC2 | ||||||||||||||||||||
| BC3 | ||||||||||||||||||||
| RNA | R/K | V | ||||||||||||||||||
| BV141 | d4T + 3TC + NVP | 10,400 | 388 | DBS1 | ||||||||||||||||
| (13 mo) | DBS2 | |||||||||||||||||||
| DBS3 | ||||||||||||||||||||
| BC1 | ||||||||||||||||||||
| BC2 | ||||||||||||||||||||
| BC3 | ||||||||||||||||||||
| RNA | N | V | ||||||||||||||||||
| BV114 | d4T + 3TC + EFV | 15,300 | 217 | DBS1 | ||||||||||||||||
| (34 mo), AZT + | DBS2 | N | I/V | |||||||||||||||||
| 3TC + LPV/r | DBS3 | N | A/G | |||||||||||||||||
| (1 mo) | BC1 | K/N | ||||||||||||||||||
| BC2 | K/N | T/A | ||||||||||||||||||
| BC3 | N | A | H | |||||||||||||||||
| RNA | N | H | ||||||||||||||||||
| BV105 | AZT + 3TC + EFV | 21,000 | 276 | DBS1 | N | V | ||||||||||||||
| (9 mo) | DBS2 | N | I | V | ||||||||||||||||
| DBS3 | ||||||||||||||||||||
| BC1 | N/K | V/M | ||||||||||||||||||
| BC2 | N | I/V | V | |||||||||||||||||
| BC3 | N | I/V | V/M | |||||||||||||||||
| RNA | N | V | ||||||||||||||||||
| BV147 | d4T + 3TC + EFV | 98,200 | 218 | DBS1 | I/V | K/N | I/V | K/E | ||||||||||||
| (13 mo) | DBS2 | N | ||||||||||||||||||
| DBS3 | ||||||||||||||||||||
| BC1 | I/V | N | M/V | H/P | ||||||||||||||||
| BC2 | N | V | F | H | ||||||||||||||||
| BC3 | D | N | H | |||||||||||||||||
| RNA | N | V | F | |||||||||||||||||
| BV116 | d4T + 3TC + NVP | >100,000 | 353 | DBS1 | R | N | V | |||||||||||||
| (22 mo) | DBS2 | I/V | N | I/V | V | |||||||||||||||
| DBS3 | N | V | ||||||||||||||||||
| BC1 | N | V | ||||||||||||||||||
| BC2 | N | V | ||||||||||||||||||
| BC3 | N | V | ||||||||||||||||||
| RNA | N | V | ||||||||||||||||||
| BV133 | d4T + 3TC + NVP | >100,000 | 165 | DBS1 | N | M/Q/K/L | V | L | A | |||||||||||
| (8 mo) | DBS2 | K/E | V | L | A | |||||||||||||||
| DBS3 | ||||||||||||||||||||
| BC1 | N/T | M/V | L | A | ||||||||||||||||
| BC2 | V | L/F | A | |||||||||||||||||
| BC3 | N | M | V | L | A | |||||||||||||||
| RNA | N/T | M | V | L | A | |||||||||||||||
| 65K | 67D | 69T | 70K | 75V | 101K | 103K | 108V | 151Q | 181Y | 184M | 188Y | 190G | 215T | 219K | 225P | |||||
| BV112 | AZT + 3TC + NVP | <50 | 242 | DBS1 | K | |||||||||||||||
| (1 mo) | DBS2 | K/N | ||||||||||||||||||
| DBS3 | K | |||||||||||||||||||
| BC1 | K/N | |||||||||||||||||||
| BC2 | K | |||||||||||||||||||
| BC3 | K/N | |||||||||||||||||||
| RNA | ||||||||||||||||||||
| BV117 | d4T + 3TC + NVP | <50 | 328 | DBS1 | K/N | |||||||||||||||
| (21 mo) | DBS2 | N | ||||||||||||||||||
| DBS3 | N | |||||||||||||||||||
| BC1 | K/N | |||||||||||||||||||
| BC2 | N | |||||||||||||||||||
| BC3 | N | |||||||||||||||||||
| RNA | ||||||||||||||||||||
VL, viral load.
DBS1 to DBS3, DBS samples 1 to 3, respectively; BC1 to BC3, blood-cell DNA samples 1 to 3, respectively; RNA, plasma RNA.
Replicate sequencing confirmed the presence of the mutation resulting in the presence of 103N in the two patients with an undetectable viral load while on an NNRTI-based HAART regimen. The mutation resulting in the presence of 103N was consistently detected in all replicates of patient BV117 and in half of the replicates from patient BV112 (Table 3).
Reproducibility of the RNA-sequencing reactions.
To evaluate the influence of sampling on RNA sequencing, replicate reactions were performed on four RNA samples (from patients BV113, BV123, BV132, and BV141). Results were much more consistent than those for DNA. In samples from patients BV123 and BV141, the same resistance mutations were detected in all reactions. For patient BV113, the mutation resulting in a D67N substitution was detected in only two of the four replicates. For patient BV132, the mutation resulting in a K103N substitution was detected in only two of the five replicates (results not shown).
DISCUSSION
In the last 25 years, DBS have been used for various purposes, including detection of HIV antibodies (2, 5, 18, 23, 31), detection of HIV antigen (22, 32), early diagnosis of perinatally infected infants (4, 15, 28, 30, 35), quantification of CD4+ T cells (26), and quantification of viral plasma RNA (1, 3, 7, 9, 16, 25, 29). Few studies examined the possibilities of performing drug resistance testing on dried blood, plasma, or serum on filter paper (24, 33, 43). Because of the ease to collect, store, and transport DBS, use of DBS is the ideal method for blood sampling in resource-poor rural settings. This, combined with the possibility of directly amplifying the genes of interest, without the need for a reverse transcription step, therebyreducing both complexity and cost of genotyping procedures (price reduction of 12 U.S. dollars/test), is an important advantage in view of efforts to increase the accessibility of resistance testing in resource-poor regions.
The results of the study presented confirmed the feasibility of extracting and subsequently sequencing HIV-1 DNA from DBS as shown by others (24, 43). The sensitivity of our in-house genotyping assay for sequencing DNA from DBS was only slightly lower than the sensitivity for sequencing DNA extracted from whole blood cells. Besides allowing the successful sequencing of the PR and RT gene in 93.1% of the patients with a detectable viral load, the DBS also allowed PR and RT gene sequencing of samples from more than 50% of the patients with an undetectable viral load. The reduced sensitivity compared to that of the patients with a detectable viral load suggests a correlation between the viral DNA contributions in DBS and the plasma viral load, as was reported by McNulty et al. (24). Although one might expect that in patients with a low CD4 count, the total number of HIV-infected cells per volume of blood is limited, no relation was seen between low CD4 counts and failure to amplify viral DNA from DBS or failure to detect resistant strains in the DNA (results not shown). The possibility of obtaining sequence information from patients with an undetectable viral load can be valuable for epidemiological studies and subtype determination and might allow the detection of mutations selected during previous suboptimal treatment regimens, as is discussed below.
If all nucleotides are considered, irrespective of the importance of their position for drug resistance, good correlations between the nucleotide sequences of DBS DNA, whole-blood-cell DNA, and RNA were obtained, with differences that only slightly exceeded the differences observed between replicate sequencing reactions starting from the same DNA or RNA extract. For the detection of mutations at positions associated with drug resistance, however, superiority of RNA sequencing over DNA sequencing was observed. Relying on only the DNA sequence would lead to a misinterpretation of the assumed phenotypic resistance in 6 of the 11 patients with significant resistance in the virus in the plasma, irrespective of whether blood cells or DBS sequences were considered.
The feasibility of using viral DNA for the establishment of drug resistance is still controversial. Some studies reported more mutations in the plasma RNA (6, 10, 21, 24), and others reported more mutations in the cellular DNA (14, 34). Most of these discordances may be attributed to differences in the selection of the samples used for the evaluation. While plasma RNA represents the population of short-lived actively replicating virus, viral DNA from infected cells is composed of a heterogeneous mix of DNA from acutely infected, actively virus-producing cells as well as quiescent cells that form the viral reservoir (13, 27, 40). Due to the limited capacities of current population-based sequencing genotyping methods, minor variants can remain undetectable (38). The virus replicating in plasma in the early stage of resistance development most probably derives from a small number of cells in the reservoir, and therefore, detection of resistant virus in plasma will precede detection in the cells. Since all patients in this study initiated HAART very recently and about half of the 11 patients in whom drug resistance mutations were detected still had a viral load of less than 5,000 copies/ml at the time of sampling, they can be considered early treatment failures. Although the number of patients is limited, a correlation between low viral load and a reduced capacity for the detection of resistance mutations in DNA compared to that of resistance mutations in RNA was observed, further strengthening the hypothesis of a relationship between duration of treatment failure and the ability to detect drug resistance in DNA, as was suggested by Bi et al. (6).
Despite a lack of sensitivity in the early phase of treatment failure, sequencing of viral DNA can be valuable, especially for patients with previous episodes of treatment failure that were not documented by resistance analysis. Interruption of a failing regimen or a failing drug will result in an overgrowth of the drug-resistant variant by the more fit wild type (13, 27, 40). In the cellular reservoir on the other hand, the resistant strains will remain detectable for longer time. Venturi et al. (39) demonstrated the kinetics of resistant and wild-type virus in the RNA and DNA compartment very clearly. They found more mutations in the RNA samples than in the DNA samples in the patients on a failing treatment, while those undergoing treatment interruption showed more mutations in the DNA samples. The same higher level of sensitivity of DNA for the detection of drug resistance mutations in a group of patients undergoing treatment interruption was reported by Devereux et al. (14). Previous exposure to EFV explains the mutation resulting in the presence of 190A that was found in the DNA but not in the RNA of patient BV114 one month after a switch from EFV to LPV. All the other patients that were selected for this study were assumed to be on their first line of HAART. Nevertheless, programs aimed at evaluating the influence of a short course of NVP or AZT on mother-to-child HIV-1 transmission have run in this region. Previous exposure to NVP therefore seems the most plausible explanation for the observed 103N mutation in the DNA of patient BV112. This, however, cannot explain the resistance pattern for BV117, a male patient. Surprisingly, both patients BV112 and BV117 showed undetectable viral loads while on NVP-based HAART. The possibility of obtaining an undetectable viral load with an NNRTI-based regimen despite the presence of the mutation resulting in K103N has been observed before (11).
The mutation resulting in the presence of 65R seen in samples from patients BV106 and BV132 is typically known as a tenofovir-associated mutation, but its selection by d4T, especially in patients infected with non-B subtypes, has been reported (37). For patient BV132, the 65R variant was seen in one of the DNA samples only. From the sequencing results for the plasma RNA isolated from this patient, it appears that a mixture of wild-type and mutant strains is present, and this may be the cause for the mutation resulting in the presence of 65R to be missed. The other NRTI or NNRTI resistance-associated mutations that were detected in the DNA sequences only (those resulting in the presence of 70R, 75I, 108I, 219E, and 225H) are considered to have only a minor impact on the drug resistance (http://hivdb.stanford.edu).
The finding of PI-associated mutations in the DNA of two patients who were not on a PI regimen was unexpected. Although the occurrence of 33F in the absence of other PI resistance mutations is rare, we assume that its presence in an isolate from patient BV021 is due to a natural polymorphism. The mutations resulting in the presence of 30N and 46I are seen as part of a mixed population in one of the DNA sequences of patient BV011. We cannot exclude the possibility that this was due to sequencing errors.
The discordances seen between the replicate DNA amplification and sequencing reactions indicate that DNA genotyping, especially genotyping performed on DNA preparations containing small amounts of viral DNA, is prone to a founder effect.
In conclusion, the results of this study demonstrate that whole-blood DNA or DBS DNA sequencing can be a very useful tool for epidemiological studies aimed at analyzing the HIV subtype distribution or the overall distribution of drug resistance mutations in a population. For the follow-up of individual patients, on the other hand, replacement of RNA sequencing by DNA sequencing cannot be recommended, due to the high number of missed mutations, especially in the early phase of treatment failure. The use of DNA and DBS sequencing for the detection of drug resistance mutations selected during previous drug exposure deserves further study.
Acknowledgments
We thank Nancy De Cabooter and Jacqueline Reynaerts for their technical assistance in the laboratory.
Kim Steegen is supported by the Flemish Interuniversity Council.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Alvarez-Muñoz, M. T., S. Zaragoza-Rodriguez, O. Rojas-Montes, G. Palacios-Saucedo, G. Vazquez-Rosales, A. Gomez-Delgado, J. Torres, and O. Munoz. 2005. High correlation of human immunodeficiency virus type-1 viral load measured in dried-blood spot samples and in plasma under different storage conditions. Arch. Med. Res. 36:382-386. [DOI] [PubMed] [Google Scholar]
- 2.Arya, S. C. 1993. Stability of human immunodeficiency virus type 1 antibodies in whole blood-impregnated filter papers under various tropical conditions. J. Clin. Microbiol. 31:765-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayele, W., R. Schuurman, T. Messele, W. Dorigo-Zetsma, Y. Mengistu, J. Goudsmit, W. A. Paxton, M. P. de Baar, and G. Pollakis. 2007. Use of dried spots of whole blood, plasma, and mother's milk collected on filter paper for measurement of human immunodeficiency virus type 1 burden. J. Clin. Microbiol. 45:891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, I. A., K. D. Drennan, A. J. Melvin, K. M. Mohan, A. M. Herz, J. Alarcon, J. Piscoya, C. Velazquez, and L. M. Frenkel. 2001. Simple, sensitive, and specific detection of human immunodeficiency virus type 1 subtype B DNA in dried blood samples for diagnosis in infants in the field. J. Clin. Microbiol. 39:29-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beebe, J. L., and L. C. Briggs. 1990. Evaluation of enzyme-linked immunoassay systems for detection of human immunodeficiency virus type 1 antibody from filter paper disks impregnated with whole blood. J. Clin. Microbiol. 28:808-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, X., H. Gatanaga, S. Ida, K. Tsuchiya, S. Matsuoka-Aizawa, S. Kimura, and S. Oka. 2003. Emergence of protease inhibitor resistance-associated mutations in plasma HIV-1 precedes that in proviruses of peripheral blood mononuclear cells by more than a year. J. Acquir. Immune Defic. Syndr. 34:1-6. [DOI] [PubMed] [Google Scholar]
- 7.Brambilla, D., C. Jennings, G. Aldrovandi, J. Bremer, A. M. Comeau, S. A. Cassol, R. Dickover, J. B. Jackson, J. Pitt, J. L. Sullivan, A. Butcher, L. Grosso, P. Reichelderfer, and S. A. Fiscus. 2003. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: measurement, precision, and RNA stability. J. Clin. Microbiol. 41:1888-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bürgisser, P., P. Vernazza, M. Flepp, J. Boni, Z. Tomasik, U. Hummel, G. Pantaleo, J. Schupbach, et al. 2000. Performance of five different assays for the quantification of viral load in persons infected with various subtypes of HIV-1. J. Acquir. Immune Defic. Syndr. 23:138-144. [DOI] [PubMed] [Google Scholar]
- 9.Cassol, S., M. J. Gill, R. Pilon, M. Cormier, R. F. Voigt, B. Willoughby, and J. Forbes. 1997. Quantification of human immunodeficiency virus type 1 RNA from dried plasma spots collected on filter paper. J. Clin. Microbiol. 35:2795-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew, C. B., S. J. Potter, B. Wang, Y. M. Wang, C. O. Shaw, D. E. Dwyer, and N. K. Saksena. 2005. Assessment of drug resistance mutations in plasma and peripheral blood mononuclear cells at different plasma viral loads in patients receiving HAART. J. Clin. Virol. 33:206-216. [DOI] [PubMed] [Google Scholar]
- 11.Coffie, P., D. Ekouevi, M. L. Chaix, B. Tonwe-Gold, S. Toure, I. Viho, C. Amani-Bosse, V. Leroy, C. Rouzioux, F. Dabis, and MTCT-Plus Initiative of ICAP. 2007. Short-course zidovudine and lamivudine or single-dose nevirapine-containing PMTCT compromises 12-month response to HAART in African women, Abidjan, Côte d'Ivoire. 14th Conf. Retrovir. Opportunistic Infect., abstr. 93LB.
- 12.Crowe, S., S. Turnbull, R. Oelrichs, and A. Dunne. 2003. Monitoring of human immunodeficiency virus infection in resource-constrained countries. Clin. Infect. Dis. 37:S25-S35. [DOI] [PubMed] [Google Scholar]
- 13.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 14.Devereux, H. L., C. Loveday, M. Youle, C. A. Sabin, A. Burke, and M. Johnson. 2000. Substantial correlation between HIV type 1 drug-associated resistance mutations in plasma and peripheral blood mononuclear cells in treatment-experienced patients. AIDS Res. Hum. Retrovir. 16:1025-1030. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, A., C. Lejczak, C. Lambert, J. Servais, N. Makombe, J. Rusine, T. Staub, R. Hemmer, F. Schneider, J. C. Schmit, and V. Arendt. 2004. Simple DNA extraction method for dried blood spots and comparison of two PCR assays for diagnosis of vertical human immunodeficiency virus type 1 transmission in Rwanda. J. Clin. Microbiol. 42:16-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiscus, S. A., D. Brambilla, L. Grosso, J. Schock, and M. Cronin. 1998. Quantitation of human immunodeficiency virus type 1 RNA in plasma by using blood dried on filter paper. J. Clin. Microbiol. 36:258-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiscus, S. A., B. Cheng, S. M. Crowe, L. Demeter, C. Jennings, V. Miller, R. Respess, and W. Stevens. 2006. HIV-1 viral load assays for resource-limited settings. PLoS Med. 3:e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortes, P., J. Menitove, A. Ross, R. Steece, K. Cabrian, C. Ferrera, P. A. Perkins, J. Sturge, R. Lealos, and M. S. Krieger. 1989. Evaluation of blood collected on filter paper for detection of antibodies to human immunodeficiency virus type 1. J. Clin. Microbiol. 27:1380-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.IATA. 2007. IATA dangerous goods regulations, 48th ed. IATA, Montreal, Canada.
- 20.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 21.Koch, N., N. Yahi, F. Ariasi, J. Fantini, and C. Tamalet. 1999. Comparison of human immunodeficiency virus type 1 (HIV-1) protease mutations in HIV-1 genomes detected in plasma and in peripheral blood mononuclear cells from patients receiving combination drug therapy. J. Clin. Virol. 37:1595-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, C. C., K. D. Seidel, R. W. Coombs, and L. M. Frenkel. 2005. Detection and quantification of human immunodeficiency virus type 1 p24 antigen in dried whole blood and plasma on filter paper stored under various conditions. J. Clin. Microbiol. 43:3901-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindhardt, B. O., I. C. Bygbjerg, K. Ulrich, H. D. Petersen, I. Lausen, and B. Frederiksen. 1987. Detection of antibodies to human immunodeficiency virus (HIV) in eluates from whole blood impregnated filter paper discs. J. Virol. Methods 18:73-77. [DOI] [PubMed] [Google Scholar]
- 24.McNulty, A., C. Jennings, D. Bennett, J. Fitzgibbon, J. W. Bremer, M. Ussery, M. L. Kalish, W. Heneine, and J. G. Garcia-Lerma. 2007. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J. Clin. Microbiol. 45:517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mwaba, P., S. Cassol, A. Nunn, R. Pilon, C. Chintu, M. Janes, and A. Zumla. 2003. Whole blood versus plasma spots for measurement of HIV-1 viral load in HIV-infected African patients. Lancet 362:2067-2068. [DOI] [PubMed] [Google Scholar]
- 26.Mwaba, P., S. Cassol, R. Pilon, C. Chintu, M. Janes, A. Nunn, and A. Zumla. 2003. Use of dried whole blood spots to measure CD4+ lymphocyte counts in HIV-1-infected patients. Lancet 362:1459-1460. [DOI] [PubMed] [Google Scholar]
- 27.Noë, A., J. Plum, and C. Verhofstede. 2005. The latent HIV-1 reservoir in patients undergoing HAART: an archive of pre-HAART drug resistance. J. Antimicrob. Chemother. 55:410-412. [DOI] [PubMed] [Google Scholar]
- 28.Nyambi, P. N., K. Fransen, H. De Beenhouwer, E. N. Chomba, M. Temmerman, J. O. Ndinya-Achola, P. Piot, and G. van der Groen. 1994. Detection of human immunodeficiency virus type 1 (HIV-1) in heel prick blood on filter paper from children born to HIV-1-seropositive mothers. J. Clin. Microbiol. 32:2858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Shea, S., J. Mullen, K. Corbett, I. Chrystie, M. L. Newell, and J. E. Banatvala. 1999. Use of dried whole blood spots for quantification of HIV-1 RNA. AIDS 13:630-631. [DOI] [PubMed] [Google Scholar]
- 30.Panteleeff, D. D., G. John, R. Nduati, D. Mbori-Ngacha, B. Richardson, J. Kreiss, and J. Overbaugh. 1999. Rapid method for screening dried blood samples on filter paper for human immunodeficiency virus type 1 DNA. J. Clin. Microbiol. 37:350-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pappaioanou, M., M. Kashamuka, F. Behets, S. Mbala, K. Biyela, F. Davachi, J. R. George, T. A. Green, T. J. Dondero, W. L. Heyward, et al. 1993. Accurate detection of maternal antibodies to HIV in newborn whole blood dried on filter paper. AIDS 7:483-488. [DOI] [PubMed] [Google Scholar]
- 32.Patton, J. C., G. G. Sherman, A. H. Coovadia, W. S. Stevens, and T. M. Meyers. 2006. Ultrasensitive human immunodeficiency virus type 1 p24 antigen assay modified for use on dried whole-blood spots as a reliable, affordable test for infant diagnosis. Clin. Vaccine Immunol. 13:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plantier, J. C., R. Dachraoui, V. Lemee, M. Gueudin, F. Borsa-Lebas, F. Caron, and F. Simon. 2005. HIV-1 resistance genotyping on dried serum spots. AIDS 19:391-397. [DOI] [PubMed] [Google Scholar]
- 34.Sarmati, L., E. Nicastri, I. Uccella, G. D'Ettorre, S. G. Parisi, L. Palmisano, C. Galluzzo, E. Concia, V. Vullo, S. Vella, and M. Andreoni. 2003. Drug-associated resistance mutations in plasma and peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected patients for whom highly active antiretroviral therapy is failing. J. Clin. Microbiol. 41:1760-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherman, G. G., G. Stevens, S. A. Jones, P. Horsfield, and W. S. Stevens. 2005. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J. Acquir. Immune Defic. Syndr. 38:615-617. [DOI] [PubMed] [Google Scholar]
- 36.Steegen, K., E. Demecheleer, N. De Cabooter, D. Nges, M. Temmerman, P. Ndumbe, K. Mandaliya, J. Plum, and C. Verhofstede. 2006. A sensitive in-house RT-PCR genotyping system for combined detection of plasma HIV-1 and assessment of drug resistance. J. Virol. Methods 133:137-145. [DOI] [PubMed] [Google Scholar]
- 37.Sungkanuparph, S., W. Manosuthi, S. Kiertiburanakul, B. Piyavong, and W. Chantratitra. 2007. Tenofovir resistance among HIV-infected patients failing a fixed-dose combination of stavudine + lamivudine + nevirapine in a resource-limited setting. 14th Conf. Retrovir. Opportunistic Infect., abstr. 663. [DOI] [PubMed]
- 38.Van Laethem, K., K. Van Vaerenbergh, J. C. Schmit, S. Sprecher, P. Hermans, V. De Vroey, R. Schuurman, T. Harrer, M. Witvrouw, E. Van Wijngaerden, L. Stuyver, M. Van Ranst, J. Desmyter, E. De Clercq, and A. M. Vandamme. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for detection of resistance in mixed HIV-1 genotypic populations. J. Acquir. Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]
- 39.Venturi, G., L. Romano, T. Carli, P. Corsi, L. Pippi, P. E. Valensin, and M. Zazzi. 2002. Divergent distribution of HIV-1 drug-resistant variants on and off antiretroviral therapy. Antivir. Ther. 7:245-250. [PubMed] [Google Scholar]
- 40.Verhofstede, C., A. Noe, E. Demecheleer, N. De Cabooter, F. Van Wanzeele, B. Van Der Gucht, D. Vogelaers, and J. Plum. 2004. Drug-resistant variants that evolve during nonsuppressive therapy persist in HIV-1-infected peripheral blood mononuclear cells after long-term highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 35:473-483. [DOI] [PubMed] [Google Scholar]
- 41.WHO. December 2006, posting date. AIDS epidemic update. World Health Organization, Geneva, Switzerland. http://data.unaids.org/pub/EpiReport/2006/2006_EpiUpdate_en.pdf.
- 42.WHO. 2006. WHO case definitions of HIV for surveillance and revised clinical staging and immunological classification of HIV related disease in adults and children. World Health Organization, Geneva, Switzerland. http://www.who.int/hiv/pub/guidelines/HIVstaging150307.pdf.
- 43.Ziemniak, C., A. George-Agwu, W. J. Moss, S. C. Ray, and D. Persaud. 2006. A sensitive genotyping assay for detection of drug resistance mutations in reverse transcriptase of HIV-1 subtypes B and C in samples stored as dried blood spots or frozen RNA extracts. J. Virol. Methods 136:238-247. [DOI] [PubMed] [Google Scholar]