Abstract
The control of outbreaks of calicivirus infection in high-density, high-throughput populations is a challenge to both human and veterinary medicine. In such populations, the prevalence of infection is, in part, dependent on the levels of biosecurity and how this affects virus transmission. Here we show how longitudinal analysis of feline calicivirus (FCV) infection in an animal rescue shelter can be used as a model to examine the dynamics of calicivirus transmission and evolution in such environments. FCV was isolated from 33 of 116 cats sampled over a 15-month period (overall prevalence, 28%). Sequence analysis of the immunodominant variable regions of the viral capsid gene identified 16 strains circulating in the shelter, with no single strain appearing to predominate. The majority of these strains were introduced into the shelter from the community and did not appear to be transmitted within the population. However, for three of these strains, putative transmission events within the shelter were identified. The rates of evolution within hypervariable regions of the FCV capsid gene in individual cats ranged from 0.05 to 1.4% per week, with the highest rates generally being found in animals that either acquired the virus while in the shelter or were undergoing acute infection. These data suggest that despite the high prevalence and presence of multiple strains of FCV within the shelter, the spread of such pathogens may be restricted by various control measures, including good hygiene and biosecurity.
The family Caliciviridae is an important family of human and animal viral pathogens, causing acute outbreaks of gastroenteritis in humans (the genera Norovirus and Sapovirus) and vesicular and other diseases in animals (the genus Vesivirus, e.g., feline calicivirus [FCV], and the genus Lagovirus, e.g., rabbit hemorrhagic disease virus). In humans, noroviruses frequently cause acute outbreaks of disease, particularly when large groups of people come together, such as in health care institutions, schools, hotels, and cruise ships (9, 10, 18, 35, 38). Similarly, in animals, outbreaks of calicivirus disease are also common when groups of animals come together in close proximity, for example, outbreaks of oral and respiratory diseases due to FCV infection in cat shelters (1, 11, 23).
Animal shelters are not dissimilar to human health care institutions in that they generally aim to implement strict biosecurity and hygiene measures. Such measures include quarantine and vaccination of the animals on arrival, the placement of diseased animals in isolation wards, and the use of hygiene and design features aimed at minimizing pathogen transmission. Like hospitals, animal shelters often have high turnovers, with large numbers of hosts leaving on a regular basis and being replaced by new animals from a wide variety of sources. This provides an ideal opportunity for multiple virus introductions from the community, which may in time lead to transmission events within the shelter environment.
For many caliciviruses, there are limited opportunities by which transmission events in such high-density environments can be identified and monitored. For human caliciviruses, investigations of transmission events are generally limited to outbreaks of disease that occur over relatively short periods of time (3, 9). In contrast, for FCV, such investigations can be carried out over extended periods of time, since a large proportion of cats continually shed virus, regardless of their disease status (5, 6). Such cats are easily detected, and the variable nature of the virus enables transmission events between individuals to be identified and the evolution of the virus to be effectively monitored by sequence analysis (6, 7, 25).
Here we report on how the molecular analysis of FCV infection in an animal shelter can be used as a model for examining calicivirus transmission and evolution in a high-density, high-turnover environment.
MATERIALS AND METHODS
Study population and sample collection.
Samples were collected from an animal shelter which housed an average of 120 cats at any one time and which was made up of three separate areas: an admissions block, a main accommodation unit, and an isolation block. Each area was designed to minimize the risk of disease transmission by the use of solid partitions between individual pens and the use of hygiene measures within each area to reduce the risk of pathogen transmission via fomites and personnel.
Due to the unknown health and immune status of cats entering a rescue shelter, it is common practice to quarantine incoming animals away from the others. In this study, incoming cats were quarantined in the admissions unit for an average of 1 week before being transferred to the main accommodation units for rehoming. During their period in the admissions unit, all cats were examined by a veterinary surgeon, and the majority were vaccinated with a commercially available live attenuated vaccine, which incorporated strain F9 as the FCV component. It should be noted, however, that vaccination is not immediately effective, and some cats may leave the admissions unit without the prior development of immunity. In addition, although vaccination generally reduces the incidence of disease, it does not prevent infection (12). Cats that appeared unhealthy either at admission or at any other time during their stay in the shelter were transferred to the isolation unit.
The sampling strategy used in this study was a longitudinal analysis of a proportion of the cats passing through the shelter. Sampling was carried out by cattery workers and took place over a 15-month period. Staff at the rescue shelter aimed to recruit the first three cats that arrived in the shelter on a weekly basis until approximately 100 cats had been recruited. Five oropharyngeal swab specimens were taken from each recruited cat: as soon as possible after arrival (generally within 24 h), 1 week after the first swab, before transfer to the main accommodation from the admissions block, 1 week after being moved to the main accommodation, and before rehoming. Fewer swabs were taken from some cats, which were rehomed quickly, although these were still included in the study.
Virus isolation.
Oropharyngeal swabs were collected, placed into 2 ml of virus transport medium, and sent by first class post to the Department of Veterinary Pathology University of Liverpool. Viruses were isolated on feline embryo A cells or Crandall-Reese feline kidney cells, as described previously (16, 37).
Molecular analysis.
In order to investigate the diversity and relationships of the FCV strains in the shelter, reverse transcription-PCR and sequence analyses were attempted with 48 representative FCV samples. These were selected from cats that were positive on only one occasion (18 samples) and from cats that were positive on more than one occasion, in which the first and the last isolates were included (15 cats, two samples per cat).
RNA extraction, reverse transcription-PCR, and consensus sequencing.
Viral RNA was extracted from positive samples (on the second passage or less; QIAmp viral RNA mini kit; QIAGEN) and transcribed into cDNA (Superscript III; Invitrogen) by using 500 ng of primer P2 (5′-CCTCACCTARACCAGTGTAACC-3′) (6), according to the manufacturers’ instructions. Negative controls consisting of mock-infected cell cultures were processed simultaneously. A 529-nucleotide region of the FCV capsid gene, equivalent to residues 6406 to 6934 of FCV strain F9 (4) and including variable immunodominant regions of the FCV capsid gene (19, 32, 33), was amplified by using 100 ng each of forward primer P1 (5′-CCGTTTGTGTTTCAAGCAAACCG-3′) and reverse primer P2, as described previously (6).
Amplicons were purified (QIAquick PCR purification kit; QIAGEN) and sequenced bidirectionally by PCR with primers P1 and P2 (6), according to standard protocols (ABI Prism BigDye Terminator, version 3.0, cycle sequencing kit; Applied Biosystems). For each amplicon, a consensus sequence was produced by using the ChromasPro program, version 1.32 (Technelysium Ptl. Ltd.). Ambiguities consistently identified in both strands were included in the consensus sequence. All primer sites were removed prior to analysis, resulting in a final useable sequence of 420 nucleotides, corresponding to codons 383 to 522 of the FCV capsid (33). We have previously shown this approach to be 99.3 to 100% reproducible (6).
Nucleotide sequence and phylogenetic analyses.
Sequence alignments and phylogenetic analyses were performed by using the GROWTREE program from version 8 of the Wisconsin package of the Genetics Computer Group (8) (Jukes-Cantor distance analysis and neighbor joining), and were viewed by using the TREEVIEW program (21). Support for individual nodes was sought by bootstrap analysis with 1,000 repetitions (MEGA software, version 3.0) (17). For comparison, the published sequence of the virus strain present in the live attenuated vaccines used in the animal center, strain F9 (GenBank accession number M86379), was added to the subsequent analysis.
RESULTS
Characteristics of study sample.
A total of 116 shelter cats were recruited over the 15-month study period, representing 24% of the cats passing through the shelter during this time.
Viral prevalence.
From the 116 cats included in this study, a total of 428 swabs were obtained (average number per cat, 3.7; range, 1 to 6), of which 59 (13.8%) were positive. Of the 116 cats recruited, FCV was isolated from 33 (28%) at some time during their stay in the shelter. Of these 33 cats, the first swab obtained following their arrival was positive for 19 cats (58%), and 12 cats (36%) became positive at some point after their arrival. The remaining two cats (6%) were positive on their first sampling, although it was not clear from the records exactly when these swabs were taken in relation to the time of the cats’ arrival.
Phylogenetic analysis.
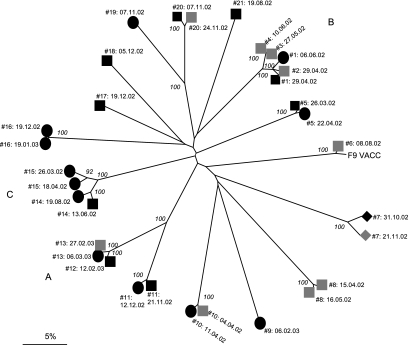
Of the 48 representative FCV isolates, PCR products and consensus sequences were successfully obtained from 32 (67%), representing 19 cats (Fig. 1) (GenBank accession numbers EF616546 to EF616577). Several other primer pairs flanking the region of interest were unsuccessful in increasing the PCR success rate in this study (data not presented). The variable nature of FCV and, indeed, caliciviruses in general often precludes the design of cross-reactive primers that amplify variable domains, such that PCR success rates are rarely ever 100% (5, 13, 30). Phylogenetic analysis based on these 32 consensus sequences identified 16 FCV strains within the shelter during the course of the study, with no single strain appearing to predominate (Fig. 1). It is possible that other strains were present but were not detected by the primers used. Fourteen of these strains were separated by nucleotide distance values of 24% to 47%, with such values being comparable to those reported previously for distinct strains of FCV (6). The remaining two sets of strains were separated by nucleotide distance values of 16.7 to 17.3% (viruses isolated from cats 19 and 20 [17.1 to 17.3% distant] and viruses isolated from cats 11, 12, and 13 [16.7 to 17.3% distant]) (Fig. 1). Such distances are not generally found in the general cat population but tend to be identified particularly within colonies in which the infection is endemic, where viruses are continuously evolving and in some cases are evolving into new strains (6, 27).
FIG. 1.
Unrooted neighbor-joining tree of 33 FCV consensus sequences (including the published sequence FCV F9 [GenBank accession number M86379]) for a 420-nucleotide region of the FCV capsid gene. Black squares, virus isolated from swab obtained at arrival in admissions; gray squares, virus isolated after arrival in admissions; black circles, virus isolated after arrival in main accommodation; black diamonds, virus isolated on arrival swab in isolation; gray diamonds, virus isolated after arrival in isolation. Each virus sequence is identified by the number of the cat from which it was obtained and the date (day.month.year) on which virus sample was taken. A, B, and C, the groups of cats involved in the transmission events within the shelter. Evolutionary distances were calculated by using the Jukes-Cantor model, with the scale bar indicating percentage divergence. Numbers at major nodes are the bootstrap values ≥80% of 1,000 replicates (MEGA program, version 3.0).
A number of isolates clustered together as members of the same strains with distance values of <6% (Fig. 1) (6, 28). These clusters included virus samples obtained from 11 of the 15 cats for which paired samples were available, suggesting that each of these 11 cats remained infected with their own strain of virus for the duration of its stay in the shelter. Swabs from seven of these cats (cats 1, 11, 12, 14, 17, 18, and 20) were positive on their arrival, suggesting that they brought the virus into the shelter, whereas 3 cats (cats 13, 15, and 10) appeared to acquire the infection within the shelter. The first swab from one cat (cat 16) was positive, although in this case it was not clear from the record when the swab was taken in relation to the cat's arrival. In addition, clustering of viruses obtained from different cats also occurred, suggesting that transmission events were occurring in the shelter (events A to C) (Fig. 1).
Transmission events.
The first transmission event (event A) involved two cats; cat 12 was positive on arrival and appeared to transmit virus to cat 13 within the admissions unit. Cat 13 continued to shed virus for at least a further 7 days (Fig. 1; Table 1). The second transmission event (event B) involved four cats (cats 1 to 4) that were shedding the same viral strain over a 6-week period while in the admissions unit: the first swab following arrival from cat 1 was positive, whereas the other three cats appeared to acquire the infection while they were in the unit (Fig. 1; Table 1). It was not clear, however, which cat within the admissions unit was the “source” cat for this transmission event. A third “putative” transmission event (event C) involved two cats that were shedding the same strain of FCV but that were present in the cattery at different times: cat 15 appeared to acquire the infection after admission to the shelter, while cat 14 appeared to be shedding the same virus in its first swab following its arrival, some 2 months later (Fig. 1; Table 1).
TABLE 1.
Shedding patterns of the cats involved in the three putative transmission events during their stay in the shelter
| Cat identifier (groupa) | Pattern in month no.b:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 12 (A) | +c− | ||||||||||||||
| 13 (A) | −−+c | +c | − | ||||||||||||
| 1 (B) | +c | −++ | +c | ||||||||||||
| 2 (B) | −−+c | −− | |||||||||||||
| 3 (B) | −−− | +c | − | ||||||||||||
| 4 (B) | −− | +c− | |||||||||||||
| 15 (C) | −−+c | +c | |||||||||||||
| 14 (C) | +c− | −+c | |||||||||||||
Groups of cats involved in the putative transmission events and whose viruses group together on the basis of phylogenetic analysis (Fig. 1).
Month 1 is equal to the start of the study (March 2002), and month 15 is equal to the end of the study (May 2003). +, FCV isolated; −, no virus isolated.
Virus isolates that were amplified and sequenced.
One cat (cat 6) became positive 2 months after its arrival in the shelter with a virus closely related to the F9 vaccine strain (distance, 1.6%) (Fig. 1). This cat was vaccinated with a live attenuated vaccine within approximately 2 weeks of arrival in the cattery. An insert of 9 nucleotides previously identified in strain F9 (4) was also found within the isolate obtained from this cat (data not presented).
Evolution rates.
Estimates for the rate of evolution were carried out for the paired virus samples from the 11 cats that appeared to be infected with their own strain of virus during their stay in the shelter, regardless of whether their first swab following their arrival was FCV positive or whether they putatively acquired the FCV infection during their stay (Table 2). Evolution rates ranged from 0.05% to 1.4% per week, with the greatest rate of change observed in two of three cats which acquired infection within the cattery (cats 10 and 15; 0.8% and 0.8%, respectively) and in cat 7 (1.4%), which was placed in isolation on its arrival and which was known to be undergoing acute upper respiratory tract disease at the time of sampling, again suggesting recent infection.
TABLE 2.
Nucleotide distance and estimated evolution rates of FCV per week for 22 paired virus isolates obtained from 11 cats that appeared to be persistently infected with their own strain of virus during their stay in the shelter
| Cat identifier | Time of FCV shedding in the shelter | Time between samples (days) | Nucleotide distancea (%) | Estimated evolution rate per wk (%) |
|---|---|---|---|---|
| 16 | Unknown | 31 | 0.2 | 0.05 |
| 13 | Acquired | 8 | 0.2 | 0.2 |
| 14 | Arrival | 67 | 1.9 | 0.2 |
| 8 | Arrival | 31 | 0.9 | 0.2 |
| 5 | Arrival | 27 | 0.9 | 0.2 |
| 11 | Arrival | 21 | 1.0 | 0.3 |
| 1 | Arrival | 38 | 2.1 | 0.4 |
| 20 | Arrival | 17 | 1.3 | 0.5 |
| 15 | Acquired | 23 | 2.6 | 0.8 |
| 10 | Acquired | 7 | 0.8 | 0.8 |
| 7 | Arrival (acutely infected) | 21 | 4.2 | 1.4 |
Determined by Jukes-Cantor distance analysis.
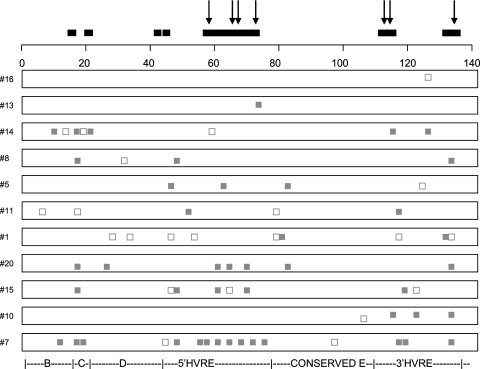
The patterns of observed nucleotide substitutions for the paired viral strains isolated from these 11 cats demonstrated that the majority of observed substitutions were associated with changes in the predicted amino acid sequence (Fig. 2). These nonsynonymous changes occurred in regions that have previously been predicted to be evolving under positive selection (6) and that colocalize with previously mapped linear and conformational B-cell epitopes (32, 34) (Fig. 2). Interestingly, the highest number of nonsynonymous substitutions was observed in cat 7 while it was undergoing an acute episode of upper respiratory tract disease.
FIG. 2.
Schematic diagram indicating the patterns of nucleotide substitutions in the FCV capsid gene for virus strains isolated from the 11 cats that appeared to be persistently infected with their own strain of virus. Gray squares, nonsynonymous substitutions; open squares, synonymous substitutions; downward-pointing arrows, positions of codon mutations known to disrupt neutralizing B-cell epitopes (32, 34); black bars, regions of FCV that have previously been identified as predicted to be evolving under positive selection (6). The regions of the capsid gene are indicated below the alignment (33). The first codon is equivalent to codon 383 of the FCV capsid protein (33).
DISCUSSION
The control of outbreaks of calicivirus infections in environments where large numbers of infectious and susceptible hosts are brought together in close proximity is a challenge to both human and veterinary medicine. In these high-density environments the prevalence of infection is dependent, in part, upon the levels of biosecurity and viral transmission. In this study we have shown how the molecular analysis of FCV infection in an animal shelter can be used as a model system to examine calicivirus transmission and evolution in high-density, high-throughput environments.
We identified 16 strains circulating in the shelter during the study period, with no single strain appearing to predominate. The majority of these strains appeared to be present as a result of multiple virus introductions into the shelter from the community over time. Such multiple introductions are a reflection of the relatively high proportion (up to ∼50%) of the cat population that sheds the virus asymptomatically for prolonged periods (2, 5, 6, 23, 30) and highlights the challenge that this virus poses to disease control in such environments. A previous cross-sectional study has also identified multiple FCV strains present in an animal shelter, but the long-term dynamics of transmission were not examined (30). In human calicivirus infections, molecular analyses of viruses obtained from similar environments have identified only one or two strains that tend to predominate (9, 15, 38).
In situations in which cats are housed in groups together but no measures are taken to reduce transmission, cross infection is common (6). In contrast, in the present study, longitudinal analysis demonstrated that although transmission events appeared to be occurring within the shelter, they were relatively rare. This was most likely due to the biosecurity measures being implemented, although clearly they were not completely effective.
In the first transmission event (event A), the putative “source” cat was identified, but in two cases (events B and C) the precise transmission chain could not be determined, although it appeared that the cats were being infected with viral strains that were circulating within the shelter. However, as only a proportion of the cats present in the shelter were sampled, not all intervening transmission events would necessarily have been identified. In addition, not all viruses that were isolated were successfully amplified. It is also possible that some cats involved in transmission events were shedding virus at levels that fell below the sensitivity of the cell culture test (36). Alternatively, the same strain may have been introduced into the shelter on several different occasions from the community. Indeed, retrospective analysis of data for the two cats involved in event C showed that they had both originated from the same geographical location, suggesting that rather than representing a transmission event, they may have both independently introduced this strain into the shelter.
One cat (cat 6) appeared to be infected with a variant of FCV vaccine strain F9 6 weeks after being vaccinated with a live F9-based vaccine. Retrospective analysis of the shedding patterns of this cat showed that FCV was isolated from this cat only postvaccination; two swabs taken before vaccination were negative for FCV. It seems likely, therefore, that this cat became infected via vaccination. Previous studies have reported the shedding of live vaccine virus following the incorrect administration of the vaccine or licking of the injection site (22, 24). However, we cannot rule out the possibility that this cat became infected with a circulating strain of the vaccine virus, since not all cats present in the shelter were included in the study. Viruses closely related to vaccine strain F9 have also been reported in other groups of cats (5, 14, 29). Although the circulation of vaccine virus is generally considered to be a rare event, its role in the continued high prevalence of FCV remains unclear and may warrant further investigation.
In this study there was little evidence for the presence of clinical disease, suggesting that the vaccine was providing protection against disease in the majority of cats in the shelter, despite the many different strains identified. Current vaccines do aim to prevent clinical disease, although they do not prevent infection (12). The present study highlights the current debate in FCV vaccinology over concern associated with the use of live vaccines and the need to use broadly cross-reactive vaccines to prevent infection as well as disease (26).
In the present study, relatively high rates of evolution of FCV were found during an early or newly acquired infection. Such a finding has not previously been reported in cats undergoing a natural infection. However, high rates of evolution have previously been observed in cats during an early experimental infection, followed by lower rates of evolution as the infection progressed (31). In contrast, the lower rates of evolution of FCV in cats that were already positive on arrival were not dissimilar to those previously reported in persistently infected cats within households where the infection is endemic (6). More detailed studies with larger numbers of samples would be required to assess more accurately how patterns of evolution might change in individuals over the course of infection. In addition, evolution rates will also depend on the region of the genome selected, and therefore, full capsid sequencing may give a more meaningful analysis.
It has been shown previously that such evolution in the capsid region of the FCV genome appears to be driven by immune selection (6, 31), and this finding is supported by the results of the present work. It is therefore possible that such selection pressure is enhanced during early infection. In other caliciviruses, immune selection has also been suggested as a mechanism that drives evolution (20). However, it is likely that, in addition to the stage of infection, the evolution of caliciviruses is influenced by a number of other host and viral factors.
Acknowledgments
This study was funded by The Blue Cross and the Petplan Charitable Trust.
We thank Chris McCracken and Ruth Ryvar for excellent technical assistance and the staff at the rescue shelter for swabbing the cats.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Bannasch, M. J., and J. E. Foley. 2005. Epidemiologic evaluation of multiple respiratory pathogens in cats in animal shelters. J. Feline Med. Surg. 7:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binns, S. H., S. Dawson, A. J. Speakman, L. E. Cuevas, C. A. Hart, C. J. Gaskell, K. L. Morgan, and R. M. Gaskell. 2000. A study of feline upper respiratory tract disease with reference to prevalence and risk factors for infection with feline calicivirus and feline herpesvirus. J. Feline Med. Surg. 2:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brachet, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 43:4659-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, M. J., I. D. Milton, J. Meanger, M. Bennett, R. M. Gaskell, and P. C. Turner. 1992. The complete nucleotide sequence of a feline calicivirus. Virology 190:443-448. [DOI] [PubMed] [Google Scholar]
- 5.Coyne, K. P., S. Dawson, A. D. Radford, P. J. Cripps, C. J. Porter, C. M. McCracken, and R. M. Gaskell. 2006. Long term analysis of feline calicivirus prevalence and viral shedding patterns in naturally infected colonies of domestic cats. Vet. Microbiol. 118:12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coyne, K. P., R. M. Gaskell, S. Dawson, C. J. Porter, and A. D. Radford. 2007. Evolutionary mechanisms of persistence and diversification of a calicivirus within endemically infected natural host populations. J. Virol. 81:1961-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyne, K. P., F. C. Reed, C. J. Porter, S. Dawson, R. M. Gaskell, and A. D. Radford. 2006. Recombination of feline calicivirus within an endemically-infected cat colony. J. Gen. Virol. 87:921-926. [DOI] [PubMed] [Google Scholar]
- 8.Deveraux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle, K. E. 2004. Mutation in a Lordsdale norovirus epidemic strain as a potential indicator of transmission routes. J. Clin. Microbiol. 42:3950-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 11.Gaskell, R. M., A. D. Radford, and S. Dawson. 2006. Feline infectious respiratory disease, p. 145-154. In E. A. Chandler, C. J. Gaskell, and R. M. Gaskell (ed.), Feline medicine and therapeutics, 3rd ed. Blackwell Publishing, Oxford, United Kingdom.
- 12.Gaskell, R. M., A. D. Radford, and S. Dawson. 2004. Vaccination, p. 13-18. In E. A. Chandler, C. J. Gaskell, and R. M. Gaskell (ed.), Feline medicine and therapeutics, 3rd ed. Blackwell Publishing, Oxford, United Kingdom.
- 13.Green, J., C. I. Gallimore, J. P. Norcott, D. Lewis, and D. W. Brown. 1995. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J. Med. Virol. 47:392-398. [DOI] [PubMed] [Google Scholar]
- 14.Horimoto, T., Y. Takeda, K. Iwatsuki-Horimoto, S. Sugii, and T. Tajima. 2001. Capsid protein gene variation among feline calicivirus isolates. Virus Genes 23:171-174. [DOI] [PubMed] [Google Scholar]
- 15.Ike, A. C., S. O. Brockmann, K. Hartelt, R. E. Marschang, M. Contzen, and R. M. Oehme. 2006. Molecular epidemiology of norovirus in outbreaks of gastroenteritis in southwest Germany from 2001 to 2004. J. Clin. Microbiol. 44:1262-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles, J. O., S. Dawson, R. M. Gaskell, C. J. Gaskell, and C. E. Harvey. 1990. Neutralisation patterns among recent British and North American feline calicivirus isolates from different clinical origins. Vet. Rec. 127:125-127. [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 18.McCarthy, M., M. K. Estes, and K. C. Hyams. 2000. Norwalk-like virus infection in military forces: epidemic potential, sporadic disease, and the future direction of prevention and control efforts. J. Infect. Dis. 181(Suppl. 2):S387-S391. [DOI] [PubMed] [Google Scholar]
- 19.Neill, J. D. 1992. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among calicivirus capsid proteins. Virus Res. 24:211-222. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson, M., K. O. Hedlund, M. Thorhagen, G. Larson, K. Johansen, A. Ekspong, and L. Svensson. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77:13117-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, N. C., and K. F. Hawkins. 1995. Mechanisms for persistence of acute and chronic feline calicivirus infections in the face of vaccination. Vet. Microbiol. 47:141-156. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen, N. C., R. Sato, J. E. Foley, and A. M. Poland. 2004. Common virus infections in cats, before and after being placed in shelters, with emphasis on feline enteric coronavirus. J. Feline Med. Surg. 6:83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Povey, R. C. 1977. Feline respiratory disease—which vaccine? Feline Practice 7:12-16. [Google Scholar]
- 25.Radford, A. D., M. Bennett, F. McArdle, S. Dawson, P. C. Turner, M. A. Glenn, and R. M. Gaskell. 1997. The use of sequence analysis of a feline calicivirus (FCV) hypervariable region in the epidemiological investigation of FCV related disease and vaccine failures. Vaccine 15:1451-1458. [DOI] [PubMed] [Google Scholar]
- 26.Radford, A. D., S. Dawson, K. P. Coyne, C. J. Porter, and R. M. Gaskell. 2006. The challenge for the next generation of feline calicivirus vaccines. Vet. Microbiol. 117:14-18. [DOI] [PubMed] [Google Scholar]
- 27.Radford, A. D., S. Dawson, R. Ryvar, K. Coyne, D. R. Johnson, M. B. Cox, E. F. Acke, D. D. Addie, and R. M. Gaskell. 2003. High genetic diversity of the immunodominant region of the feline calicivirus capsid gene in endemically infected cat colonies. Virus Genes 27:145-155. [DOI] [PubMed] [Google Scholar]
- 28.Radford, A. D., S. Dawson, C. Wharmby, R. Ryvar, and R. M. Gaskell. 2000. Comparison of serological and sequence-based methods for typing feline calicivirus isolates from vaccine failures. Vet. Rec. 146:117-123. [DOI] [PubMed] [Google Scholar]
- 29.Radford, A. D., L. Sommerville, R. Ryvar, M. B. Cox, D. R. Johnson, S. Dawson, and R. M. Gaskell. 2001. Endemic infection of a cat colony with a feline calicivirus closely related to an isolate used in live attenuated vaccines. Vaccine 19:4358-4362. [DOI] [PubMed] [Google Scholar]
- 30.Radford, A. D., L. M. Sommerville, S. Dawson, A. M. Kerins, R. Ryvar, and R. M. Gaskell. 2001. Molecular analysis of isolates of feline calicivirus from a population of cats in a rescue shelter. Vet. Rec. 149:477-481. [DOI] [PubMed] [Google Scholar]
- 31.Radford, A. D., P. C. Turner, M. Bennett, F. McArdle, S. Dawson, M. A. Glenn, R. A. Williams, and R. M. Gaskell. 1998. Quasispecies evolution of a hypervariable region of the feline calicivirus capsid gene in cell culture and in persistently infected cats. J. Gen. Virol. 79(Pt. 1):1-10. [DOI] [PubMed] [Google Scholar]
- 32.Radford, A. D., K. Willoughby, S. Dawson, C. McCracken, and R. M. Gaskell. 1999. The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. J. Virol. 73:8496-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seal, B. S., J. F. Ridpath, and W. L. Mengeling. 1993. Analysis of feline calicivirus capsid protein genes: identification of variable antigenic determinant regions of the protein. J. Gen. Virol. 74:2519-2524. [DOI] [PubMed] [Google Scholar]
- 34.Tohya, Y., N. Yokoyama, K. Maeda, Y. Kawaguchi, and T. Mikami. 1997. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J. Gen. Virol. 78(Pt. 2):303-305. [DOI] [PubMed] [Google Scholar]
- 35.van Duynhoven, Y. T., C. M. de Jager, L. M. Kortbeek, H. Vennema, M. P. Koopmans, F. van Leusden, W. H. van der Poel, and M. J. van den Broek. 2005. A one-year intensified study of outbreaks of gastroenteritis in The Netherlands. Epidemiol. Infect. 133:9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wardley, R. C. 1976. Feline calicivirus carrier state. A study of the host/virus relationship. Arch. Virol. 52:243-249. [DOI] [PubMed] [Google Scholar]
- 37.Wardley, R. C., and R. C. Povey. 1977. The pathology and sites of persistence associated with three different strains of feline calicivirus. Res. Vet. Sci. 23:15-19. [PubMed] [Google Scholar]
- 38.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 190:27-36. [DOI] [PubMed] [Google Scholar]