Abstract
We report on a case of fungemia due to fluconazole-resistant Candida nivariensis (MIC, ≥128 μg/ml). Internal transcribed spacer PCR followed by microchip gel electrophoresis with a blood culture that tested positive revealed a unique pattern different from those of other pathogenic yeasts.
CASE REPORT
A 70-year-old woman with rheumatoid arthritis was hospitalized for the treatment of a nutritional disorder in September 2006. The patient had received total parenteral nutrition for 2 months before admission. On day 20, the patient had a temperature of 37.8°C, which continued for 5 days. Cultures of urine and stool samples yielded Candida albicans, while cultures of the patient's catheter tip and blood were negative. Although no source of infection was clinically apparent, she was treated with intravenous fluconazole (400 mg daily) for 27 days. On day 77, however, the patient developed a high fever (temperature, 40°C). Blood was drawn for culture, and the central venous catheter was removed. Hematological investigations revealed a hemoglobin level of 11.6 g/dl, a platelet count of 139 × 109/liter, and a white cell count of 6.6 × 109/liter. The C-reactive protein concentration was 6 mg/liter, and the (1→3)-β-d-glucan level was 153.7 μg/liter. The patient was positive for serum Candida antigen by the Unimedi Candida monotest (Unitica Ltd., Tokyo, Japan). The Bact/Alert blood culture system (bioMerieux, Inc., Durham, NC) signaled microbial growth on the day after blood collection. As Gram staining of the blood culture showed yeast, intravenous fluconazole (400 mg daily) was commenced as empirical therapy. The culture of the catheter tip also grew yeast in pure culture. Despite fluconazole administration, the patient remained febrile, and a culture of blood performed during fluconazole treatment was again positive for yeast. Therefore, the treatment was changed to intravenous voriconazole (400 mg daily) and micafungin (150 mg daily) for 7 days, based on the results of broth microdilution susceptibility testing performed with a commercial kit (6). The MICs of the isolate were as follows: 0.5 μg/ml for amphotericin B, 2.0 μg/ml for flucytosine, 2 μg/ml for miconazole, ≥16 μg/ml for itraconazole, ≥128 μg/ml for fluconazole, 4 μg/ml for voriconazole, and 0.06 μg/ml for micafungin. The fever resolved 4 days later, and the patient was given micafungin for a further 10 days. There was no evidence of relapse during the next month, when she was transferred to another hospital.
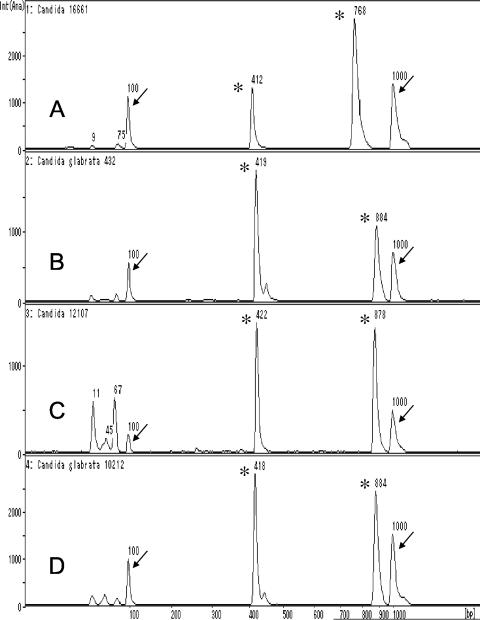
Nuclear DNA extraction from the yeast-positive blood culture bottles and PCR amplification of internal transcribed spacers (ITSs; ITS-PCR), followed by microchip gel electrophoresis, are routinely performed in our laboratory, as described previously (3, 4), with the following changes: (i) the use of a MagExtract DNA extraction kit was omitted, and (ii) PCR cycling was set at 25 cycles. It took approximately 50 min to extract DNA from the positive blood culture bottles. The PCR procedure required 35 min, and electrophoresis took 5 min. Therefore, the overall turnaround time of the ITS-PCR was approximately 1.5 h. ITS-PCR of the three isolates (two from blood cultures and one from a catheter tip culture) with universal primers ITS1 and ITS4 yielded fragments 765 to 768 bp in length, while universal primers ITS3 and ITS4 yielded fragments 411 to 414 bp in length. The ITS-PCR pattern of the isolate was not found in our in-house ITS-PCR database (3). The three isolates were identified as Candida glabrata with the ID32C yeast identification system (bioMerieux, Marcy l'Etoile, France). The ID32C code obtained was 0001-0002-01, which showed an excellent match with the profile for C. glabrata in the APILAB database (identity, 96.9%; typicity index, 0.94). However, the ITS-PCR pattern differed from those of C. glabrata K432, C. glabrata K12107, and C. glabrata K10212: the sizes (mean ± standard deviation) of the PCR products from the blood isolate (Candida nivariensis K16661) were 413 ± 1.2 and 770 ± 2.0 bp, and the sizes of the products from three C. glabrata strains were 420 ± 2.0 and 875 ± 7.2 bp (Fig. 1). The mean amplicon sizes were obtained from the results of five experiments with each strain. Furthermore, the colonies of the isolates on CHROMagar Candida medium were white, whereas those of C. glabrata are pink. The ITS2 region was sequenced to confirm the identity of the isolate by using an ABI Prism 377 automated DNA sequencer (Applied Biosytems, Foster City, CA) with a BigDye Terminator cycle sequencing kit (Applied Biosystems). The primers used to sequence the ITS regions were ITS1, ITS2, ITS3, and ITS4 (7). The ITS2 sequences were compared by using the nucleotide-nucleotide BLAST program (BLASTN) with default settings, and the isolate showed a high level of similarity (99% nucleotide identity; 218 of 219 nucleotides) with the sequence of a C. nivariensis strain (GenBank accession no. AY620957); the ITS2 region of the patient blood isolate differed from that of the C. nivariensis type strain (CBS 9983) by only 1 bp (a conversion of T to A at position 80). The lengths of the ITS1 and ITS2 regions of the blood isolate were 283 bp and 232 bp, respectively. In contrast, the length of ITS1 of C. glabrata ranged from 399 to 402 bp, and that of ITS2 ranged from 233 to 238 bp (GenBank accession nos. AB032177, AF167993, AM492797, and AM429798). The sequences of three C. glabrata control strains revealed 99 to 100% nucleotide identities with those of C. glabrata reference starins (GenBank accession nos. AF218995, AY939793, and AF218966).
FIG. 1.
ITS-PCR patterns of C. nivariensis K16661 (A), C. glabrata K432 (B), C. glabrata K12107 (C), and C glabrata K10212 (D). Arrows indicate DNA size markers of 100 bp (left) and 1,000 bp (right), and asterisks indicate PCR products (A, 412 and 768 bp; B, 419 and 884 bp; C, 422 and 878 bp; and D, 418 and 884 bp). Int (Ana), intensity (analysis).
Candida nivariensis is a recently described pathogenic yeast closely related to C. glabrata, and only four cases of C. nivariensis infection have been reported: three from Spain (1) and one from Indonesia (GenBank accession no. EF056322). Candida nivariensis isolates have been misidentified as C. glabrata by commercial identification kits (1). The same misidentification was made for the strains isolated from the patient in the present study. CHROMagar Candida medium seems to be useful for the differentiation of C. glabrata and C. nivariensis. However, a PCR-based technique is needed for the rapid and specific identification of C. nivariensis (1, 2). It is well known that C. glabrata and C. krusei are resistant or intermediately susceptible to fluconazole (5), but no susceptibility testing results for C. nivariensis have been reported. Although the blood isolate was resistant to fluconazole, further studies are needed to determine whether C. nivariensis strains are resistant to azole antifungal agents.
In conclusion, if a patient has previously been treated with an azole, the possibility of C. nivariensis infection should be considered, as shown in the present study. ITS-PCR pattern analysis not only can reliably identify C. nivariensis but also is straightforward and can be completed within 1.5 h from the time that a positive test result is obtained with blood culture bottles.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Alcoba-Florez, J., S. Méndez-Álvarez, J. Cano, J. Guarro, E. Pérez-Roth, and M. del Pilar Arévalo. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcoba-Florez, J., M. del Pilar Arévalo, F. J. González-Paredes, J. Cano, J. Guarro, E. Pérez-Roth, and S. Méndez-Álvarez. 2005. PCR protocol for specific identification of Candida nivariensis, a recently described pathogenic yeast. J. Clin. Microbiol. 43:6194-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita, S., Y. Senda, S. Nakaguchi, and T. Hashimoto. 2001. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J. Clin. Microbiol. 39:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita, S., Y. Senda, T. Iwagami, and T. Hashimoto. 2005. Rapid identification of staphylococcal strains from positive-testing blood culture bottles by internal transcribed spacer PCR followed by microchip gel electrophoresis. J. Clin. Microbiol. 43:1149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappas, P. G., J. H. Rex, J. D. Sobel, S. G. Filler, W. E. Dismukes, T. J. Walsh, and J. E. Edwards. 2004. Guidelines for treatment of candidiasis. Clin. Infect. Dis. 38:161-189. [DOI] [PubMed] [Google Scholar]
- 6.Pfaller, M. A., S. Arikan, M. Lozano-Chiu, Y.-S. Chen, S. Coffman, S. A. Messer, R. Rennie, C. sand, T. Heffner, J. H. Rex, J. Wang, and N. Yamane. 1998. Clinical evaluation of the ASTY colorimetric microdilution panel for antifungal susceptibility testing. J. Clin. Microbiol. 36:2609-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White, T. J., T. Burns, S. Lee, and J. Taylor. 1990. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Snisky, and T. J. White (ed.), PCR protocols. A guide to methods and applications. Academic Press, Inc., San Diego, CA.