Abstract
The hemagglutinin (H) protein of measles virus (MV) mediates attachment to cellular receptors. The ectodomain of the H spike is thought to consist of a membrane-proximal stalk and terminal globular head, in which resides the receptor-binding activity. Like other paramyxovirus attachment proteins, MV H also plays a role in fusion promotion, which is mediated through an interaction with the viral fusion (F) protein. The stalk of the hemagglutinin-neuraminidase (HN) protein of several paramyxoviruses determines specificity for the homologous F protein. In addition, mutations in a conserved domain in the Newcastle disease virus (NDV) HN stalk result in a sharp decrease in fusion and an impaired ability to interact with NDV F in a cell surface coimmunoprecipitation (co-IP) assay. The region of MV H that determines specificity for the F protein has not been identified. Here, we have adapted the co-IP assay to detect the MV H-F complex at the surface of transfected HeLa cells. We have also identified mutations in a domain in the MV H stalk, similar to the one in the NDV HN stalk, that also drastically reduce fusion yet do not block complex formation with MV F. These results indicate that this domain in the MV H stalk is required for fusion but suggest either that mutation of it indirectly affects the H-dependent activation of F or that the MV H-F interaction is mediated by more than one domain in H. This points to an apparent difference in the way the MV and NDV glycoproteins interact to regulate fusion.
Measles virus (MV) is the only human pathogen of the Morbillivirus genus in the family Paramyxoviridae (17). Paramyxoviruses, like other enveloped viruses, enter cells by fusing their lipid membranes with those of target host cells (23). The membrane fusion process is brought about by interactions between cellular receptors and viral glycoproteins (22). The surfaces of measles virions and infected cells contain two types of viral transmembrane glycoproteins, the hemagglutinin (H) and fusion (F) proteins (17). MV H binds to cellular protein receptors, either CD46 or CD150 (11, 14, 34), and is an important determinant of viral tropism (49). MV F, with a contribution from the H protein, mediates virion-to-cell and cell-to-cell membrane fusion (48).
The MV H protein is a type II glycoprotein believed to exist as a tetramer composed of a pair of dimers, in which the monomers are disulfide linked. MV H has a short N-terminal cytoplasmic tail followed by a transmembrane domain and a large C-terminal ectodomain (1). It is believed that the ectodomain consists of a stalk region that supports a globular domain containing the receptor recognition and antigenic regions of the protein (24).
MV glycoprotein-promoted membrane fusion is thought to involve a series of conformational changes in the F protein from a metastable prefusion state to a highly stable postfusion state (38). These changes occur at neutral pH at the surface of the target membrane and are believed to be triggered by receptor recognition by the H protein. In addition to bringing the two membranes into close proximity prior to F activation, there is evidence that MV H is involved in triggering F activation through a specific interaction between the two proteins. First, expression of both MV H and F is required for membrane fusion (48). Second, although the H glycoproteins of MV and canine distemper virus (CDV) are, to a certain extent, interchangeable, fusion promotion is most efficient when the H and F proteins are derived from the same virus and strain (3, 5, 40, 44, 47). Third, monoclonal antibodies (MAbs) to MV H that block infectivity and inhibit fusion without interfering with receptor binding have been characterized previously (15, 19).
The exact mechanism by which MV H receptor recognition triggers the conformational changes in F remains unknown. Based on the requirement for expression of MV or CDV glycoproteins from the same virus and strain for maximum fusion efficiency, it has been suggested that fusion promotion requires a physical interaction between specific domains of the proteins (5, 44). Indeed, a heterooligomeric interaction between MV H and F in the endoplasmic reticulum has been detected and is proposed to play an integral role in regulating fusion (36).
While multiple regions of both H and F have been implicated in fusion promotion, it has not been established which domains on the two proteins directly mediate the interaction between them. In addition to the residues that mediate receptor binding, there are two regions of MV H that have been suggested to have a role in fusion promotion. Region 244 to 250 has been identified as a linear epitope in the globular region recognized by an MAb that inhibits syncytium formation but not receptor recognition (15). Based on this, it was suggested that the region constitutes either a functional or a physical interface between MV H and F. However, this was not tested directly and it remains possible that the MAb blocks fusion by an indirect mechanism, such as prevention of conformational changes in MV H that are integral to the fusion process.
Additionally, a mutation at residue I98 in the MV H stalk was found to be responsible for the lack of syncytium formation in a persistently infected cell line (20). Examination of the sequence surrounding this residue reveals a motif, spanning the region I84 to L105, which, analogous to a corresponding domain in the Newcastle disease virus (NDV) hemagglutinin-neuraminidase protein (HN) stalk (41), possesses some properties of a heptad repeat (HR). In this region, every seventh residue is hydrophobic; however, unlike a true HR, the residues in the fourth (or d) positions are not hydrophobic. The domain in NDV HN has, nonetheless, been termed an HR (41). This region in MV H is of potential interest because mutations in the corresponding domain in the NDV HN protein interfere with both fusion and the interaction with F, as detected in a coimmunoprecipitation (co-IP) assay (27, 28). Mutations in NDV HN that reduced fusion activity to less than 20% of that of the wild-type (wt) protein could not be detected in a complex with NDV F at the cell surface. This raises the possibility that mutation of I98 in the MV H protein may also interfere with fusion by disrupting the H-F interaction.
To further explore the roles of regions 244 to 250 and 84 to 105 in MV H function, we have performed site-directed mutational analyses of these two regions and determined the effects on several aspects of the fusion process. On the one hand, alanine-scanning mutagenesis does not support the idea that the region 244 to 250 constitutes a functional interface between H and F. Mutation of residues in this region does not significantly alter either receptor binding or fusion promotion by MV H. On the other hand, several site-directed mutations in the stalk of MV H do modulate hemadsorption (HAd) activity. This suggests that changes in the HR-like domain can alter the structure and/or orientation of the globular head domain. More importantly, several mutations in the HR-like domain specifically decrease fusion with no discernible effect on attachment, consistent with the region playing a role in mediating the fusion helper activity of MV H. However, these mutations do not abolish the H-F interaction at the cell surface, as detected by a cell surface co-IP assay. Thus, either the domain is not directly involved in mediating the interaction with F or the interaction is mediated by more than one domain.
MATERIALS AND METHODS
Recombinant plasmids and site-directed mutagenesis.
The H and F genes of the Edmonston MV strain were gifts of Michael Oldstone (The Scripps Research Institute). MV H was released from the vector in which it was supplied by SacII digestion, and MV F was released by XhoI/SacII digestion. Both genes were ligated into the pBluescript SK(+) (pBSK) expression vector (Stratagene, La Jolla, CA). The H and F genes of the Ondersterpoort CDV strain were gifts of Shmuel Rozenblatt (Tel Aviv University). Both genes were released from the vectors in which they were provided by NcoI/StuI digestion and ligated into pBSK at the same sites, previously added by mutagenesis.
Site-directed mutagenesis was performed as described previously (6). Briefly, a single-stranded DNA template was rescued by R408 helper phage (Stratagene, La Jolla, CA) in CJ236 cells. Mutagenesis primers (Integrated DNA Technologies, Coralville, IA) were annealed to the template and extended with T4 DNA polymerase, and the ends were ligated with T4 DNA ligase (Roche). The mutagenesis reaction products were transformed into MV1190 cells (Bio-Rad, Hercules, CA) that were then selected for ampicillin resistance. Identification of colonies carrying mutated genes was facilitated by screening for the presence of a unique restriction site introduced by each mutagenic primer. Multiple clones were characterized for each mutant DNA, and the presence of the desired mutation was confirmed by DNA sequencing.
Antibodies.
Polyclonal antiserum (Fcyt) to a peptide mimicking a region in the cytoplasmic tail of MV F (4) was initially a gift of Roberto Cattaneo and was later prepared (Proteintech Group, Inc., Chicago, IL). A mixture of two MAbs to the MV H protein was purchased commercially (Chemicon, Temicula, CA). A hybridoma producing another anti-H MAb, called B2, was a gift of Paul Rota. The MAbs recognize both native and denatured H.
Transient expression.
HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) with high glucose, l-glutamine, and pyridoxine hydrochloride supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 0.1 mM MEM nonessential amino acid solution, 4 U/ml penicillin, and 4 μg/ml streptomycin. wt and mutated proteins were expressed using a vaccinia virus-T7 (vTF7-3) RNA polymerase expression system, which drives expression of genes under the control of the T7 promoter in pBSK (16). Transfections were performed by a modification of previously described methods (29). Briefly, cells were seeded in six-well plates at 3.5 × 105 per well 1 day prior to transfection. The monolayers were infected with vTF7-3 at a multiplicity of infection of 0.5 and incubated for 1 h at 37°C. The cells were transfected with 0.5 μg of each DNA complexed with dimethyldioctadecylammonium bromide (DDAB) in OptiMEM. After a 5-h incubation at 37°C, cell maintenance medium was added to each well and the cells were returned to 37°C for at least 20 h. For co-IP assays, cells were seeded at 3 × 105 per well, DMEM was added immediately following addition of the DNA-DDAB complexes, and the transfected cells were incubated for a maximum of 16 h at 37°C.
Quantitation of cell surface MV H.
Flow cytometric analysis was used to quantitate cell surface expression as previously described (29). Anti-MV H hybridoma supernatant was harvested from B2 cells. Transfected HeLa monolayers were washed twice with PBS-FCS (phosphate-buffered saline [PBS] containing 5% FCS) and then incubated at room temperature for 30 min with hybridoma supernatant. The cells were washed twice with PBS-FCS and then incubated with a 1:200 dilution of fluorescein isothiocyanate conjugated to either goat anti-mouse immunoglobulin G or goat anti-rabbit immunoglobulin G in PBS-FCS for 30 min. After two additional washes with PBS-FCS, the monolayers were detached with 0.0625 mM EDTA in PBS, pelleted by centrifugation, and washed once with PBS-FCS. The cells were fixed in PBS with 1% FCS plus 1% paraformaldehyde prior to analysis. Expression level is presented as mean fluorescent intensity minus background labeling of control cells transfected with vector alone.
HAd assay.
HAd activity of transfected HeLa cells was determined by measuring the ability of the expressed MV H protein to adsorb African green monkey (AGM) erythrocytes via CD46 (18) (Three Springs Scientific, Inc., Perkasie, PA) by use of a modification of a previously described protocol (8). These assays were performed at 37°C to reduce nonspecific binding of the erythrocytes to the monolayers. MV H-expressing monolayers were incubated for 30 min with a 2% suspension of erythrocytes in prewarmed PBS supplemented with 1% CaCl2 and 1% MgCl2 (PBS+). Monolayers were washed with warm PBS+ and then incubated for an additional 10 min at 37°C to allow release of nonspecifically bound erythrocytes. The cells were washed with warm PBS+, and then adsorbed erythrocytes were lysed in 50 mM NH4Cl. Lysates were cleared by centrifugation, and absorbance was quantified at 540 nm with a Spectra Max 250 microplate spectrophotometer (Molecular Devices, Sunnyvale, CA), with background obtained with cells expressing vector alone subtracted.
Content-mixing assay for fusion.
The abilities of H and F proteins to promote fusion were quantitated using a modification of the content-mixing assay described by Nussbaum et al. (31) and Bagai and Lamb (2). Monolayers of HeLa effector cells were infected with vTF7-3 and cotransfected with H and a trypsin-activated cleavage site mutant form of F (Fcsm). Fcsm was created by mutating the cleavage site from RRHKR to RNHNR, as described by Maisner et al. (25). Monolayers of HeLa target cells were infected with wt vaccinia at a multiplicity of infection of 10 and transfected with 1 μg per well of plasmid pGINT7β-gal, which carries the β-galactosidase gene under the control of a T7 promoter.
At 20 to 22 h posttransfection, cells were removed from the wells by treatment with 0.05% trypsin and 0.53 mM EDTA (Gibco) and washed with DMEM. The two cell populations were combined in replicate wells of a 96-well microtiter plate at an effector-to-target ratio of 2:1. Cells were incubated at 37°C for 5 h and then lysed for at least 30 min with 10 μl of 10% IGEPAL CA-630 (Sigma). Then, 50 μl of the lysate was mixed with 20 μl of the β-galactosidase substrate 16 mM chlorophenol red-β-d-galactopyranoside and incubated briefly at room temperature. The extent of fusion was quantitated by determination of the absorbance at 590 nm with a Spectra Max 250 microplate spectrophotometer, with background obtained with cells expressing MV Fcsm alone subtracted.
Lipid-mixing assay.
The abilities of MV H and F mutants to promote lipid mixing were assessed by the transfer of octadecyl rhodamine b chloride (R18; Invitrogen) from AGM erythrocytes to HeLa cells cotransfected with MV H and F genes, using a modification of a protocol described by Morris et al. (30). Freshly labeled erythrocytes were prepared just prior to each experiment. Briefly, to label the erythrocytes, 20 ml of a 2% suspension was incubated with 30 μl of 100 μM R18 in ethanol in cold PBS+ for 15 min in the dark at room temperature. Unbound R18 was removed by the addition of 30 ml DMEM with 5% FCS and an additional 15-min incubation. The erythrocytes were washed three times with DMEM with 5% FCS and twice with PBS+ followed by resuspension in 20 ml PBS+.
Lipid-mixing assays were performed using HeLa cells at 18 h posttransfection with the MV H and F genes. For each assay, R18-labeled erythrocytes were added to each monolayer of transfected HeLa cells. After incubation for 30 min on ice in the dark to allow erythrocyte binding by MV H, fusion between the cell monolayers and erythrocytes was initiated by transferring cells to 37°C. After the desired amount of time, the cell monolayers were washed with warm PBS+ to remove unbound erythrocytes. Images were acquired immediately with a 20× objective by using fluorescent microscopy and OPEN Lab software (Improvision, Inc., Cambridge, MA).
co-IP.
The abilities of wt and mutated MV H proteins to co-IP with MV Fcsm were assayed using a modification of protocols of Yao et al. (50) and Deng et al. (8). At 16 h posttransfection, cells were starved for 1 h at 37°C in DMEM lacking cysteine and methionine. The cells were then labeled for 5 h at 37°C with 100 μCi/ml of Expre35S35S-cysteine-methionine labeling mix (Dupont-New England Nuclear, Boston, MA). Then, the cells were washed three times and incubated for 30 min on ice with cold PBS-CM (PBS supplemented with 0.1 mM CaCl2 and 1 mM MgCl2). Cell surface proteins were biotinylated with sulfo-NHS-SS-biotin (Pierce, Rockford, IL) dissolved in cold PBS-CM for 30 min on ice with gentle agitation. Excess biotinylating reagent was removed with PBS-CM washes, and then the labeled cells were lysed with 0.4 ml DH lysis buffer (50 mM HEPES, pH 7.3 [United States Biochemical, Cleveland, OH], 10 mM lauryl maltoside [United States Biochemical], 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride) for 45 min on ice.
Lysates from three wells of a six-well plate were combined and split into two equal aliquots into tubes containing the appropriate antibodies (either Fcyt alone or Fcyt plus anti-H MAbs). The immunoprecipitation reaction mixtures were incubated for 90 min at 4°C and then cleared by centrifugation. Antigen-antibody complexes were collected from the supernatants with bovine serum albumin-blocked Immunopure immobilized protein G beads for 1 h at 4°C and then washed with DH lysis buffer. The protein-bead complexes were boiled for 5 min in 10 μl of 10% sodium dodecyl sulfate (SDS), and the released proteins were resuspended in DH lysis buffer. The protein G beads were removed by centrifugation, and then the supernatants were incubated with immobilized streptavidin beads (Pierce) overnight at 4°C. The beads were washed twice with DH lysis buffer and then resuspended in reducing buffer for analysis by SDS-polyacrylamide gel electrophoresis. The percentage of total cell surface H coimmunoprecipitated with F was quantified using a Bio-Rad Fluor-S multi-imager (Hercules, CA).
RESULTS
Alanine-scanning mutagenesis of residues 244 to 250 in MV H: cell surface expression and functional characteristics.
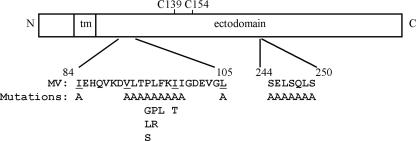
As shown in Fig. 1, residues 244 to 250 are located in the ectodomain of MV H. This region has been proposed to form the “topographical or functional interface” between MV H and F (15). With the goal of characterizing the roles of these residues in the fusion-related functions of MV H, including the receptor recognition and fusion helper activities, proteins carrying alanine substitutions for residues S244, E245, L246, S247, Q248, L249, S250, S244/L246, and S244/E245 were prepared and characterized (Fig. 1).
FIG. 1.
Diagram of MV H structure, including the amino acid sequences of the HR-like region (residues 84 to 105) and region 244 to 250. Individual mutations introduced into these regions are listed beneath the wt sequence. tm, transmembrane; C139 and C154, cysteines 139 and 154 involved in intermonomer disulfide bonds.
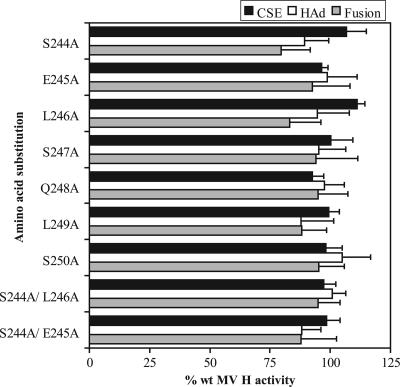
The cell surface expression of each mutated protein was quantitated by flow cytometry with an MAb that recognizes the ectodomain of MV H. As shown in Fig. 2, all of the mutated proteins were detected at the cell surface at levels similar to that of wt MV H, ranging from 92 to 111% of the wt expression level. The receptor-binding activities of the proteins were evaluated by assaying their abilities to adsorb AGM erythrocytes. HAd levels ranged from 88% for L249A to 107% for S250A. Additionally, the ability of mutated MV H proteins to promote membrane fusion with the homologous F protein was quantitated using a content-mixing assay. All of the mutated proteins promote cell-to-cell fusion at levels similar to that of wt MV H, ranging from 80 to 95% of the wt level.
FIG. 2.
Cell surface expression (CSE) and functional characteristics of the MV H proteins with alanine substitutions in region 244 to 250. CSE was quantitated by flow cytometry with MAbs specific for antigenic sites in the ectodomain of the protein. HAd activity was quantitated by the ability of the transfected HeLa cells to adsorb AGM erythrocytes. The abilities of the mutated H proteins to complement MV Fcsm in the promotion of membrane fusion were determined by a content-mixing assay. For each assay, the background detected in cells transfected with vector has been subtracted. All data points represent the means of at least three independent experiments and are expressed relative to the activities of the wt proteins.
Two additional proteins carrying double alanine substitutions, S244A/L246A and S244A/E245A, were prepared and characterized. The rationale for these choices was the fact that the alanine mutations at positions 244 and 246 exhibited the lowest levels of fusion, albeit only slightly, and that, in early experiments, the E245A mutation exhibited the third-lowest level of fusion. However, both sets of double mutations resulted in levels of fusion similar to that of the wt H protein (95% and 88% of wt activity, respectively). Similar results were obtained for cell surface expression and receptor binding. Thus, contrary to previous predictions (15), these results suggest that no residue in this region has a significant role in the receptor-binding activity or fusion promotion activity of MV H.
Mutation of an HR-like domain in the stalk of MV H: cell surface expression and functional characteristics of MV H proteins carrying mutations for the heptadic residues.
Examination of the sequence of the MV H stalk reveals an HR-like domain spanning the region I84 to L105 (Fig. 1). As shown in Fig. 3, alignment of the MV H stalk sequence with those of other paramyxoviruses reveals that, within the family, there is positional conservation of these domains. Although the HR-like domains are not conserved with respect to length or number of repeats, there is a striking conservation of 2 residues near the middle of each region: a proline that is completely conserved, as well as a leucine that is conserved in type. These residues are located between the second and third heptadic residues in the domain at positions 94 and 95, though the PL motif is not positionally conserved relative to the HRs between family members.
FIG. 3.
Comparison of the amino acid sequences in the HR-like domains in the stalks of various paramyxovirus attachment glycoproteins. The numbers (1 to 4) above the sequences indicate the heptadic residues, which are also in bold print. The highly conserved proline and leucine residues are indicated in large print. The sequences and relevant references are as follows: MV (1), CDV (21), NDV (26), hPIV3 (12), and mumps virus (MuV) (46).
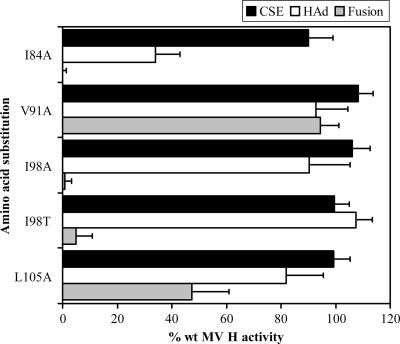
The roles of the heptadic residues (I84, V91, I98, and L105) in the fusion process were examined by the introduction of an alanine substitution at each of these positions in the MV H protein and by characterization of the mutated proteins. Based on the results of Hummel and Bellini (20), an additional protein with an I98T substitution was tested.
Initially, the abilities of MV H proteins with amino acid substitutions at the heptadic positions to complement MV F in fusion promotion were quantitated. H proteins carrying the I84A, I98A, I98T, and L105A mutations exhibit significantly reduced fusion promotion activities of 0%, 0.7%, 5%, and 47% of the activity of wt H, respectively (Fig. 4). The V91A mutation results in only a very slight reduction in fusion promotion (94% that of wt H). The reduction in fusion promotion by the mutated proteins cannot be attributed to reduced cell surface expression, which ranges from 90 to 108% of the wt level for these proteins (Fig. 4).
FIG. 4.
Cell surface expression (CSE) and functional characteristics of the MV H proteins with mutations in the heptadic residues of the HR-like domain. Assays were performed as described in the legend to Fig. 2. All data points represent the means of at least three independent experiments and are expressed relative to the activities of the wt proteins.
To determine if the basis for the reduced fusion promotion by the mutated proteins is a decreased ability to recognize receptors, HAd activity was quantitated. Only one of the four mutated proteins with deficiencies in membrane fusion promotion, the I84A-mutated protein, also has significantly reduced HAd activity (34% that of wt H). Thus, the reduced fusion promotion exhibited by the I84A-mutated protein seems to correlate with an alteration in the structure and/or orientation of the globular head domain. The other proteins, carrying the I98A, I98T, and L105A mutations, have HAd activities ranging from 82 to 108% of wt H activity, indicating that the fusion deficiency of these mutated proteins is not a result of decreased recognition of receptors. Importantly, the phenotype of the H protein carrying the I98T mutation is consistent with the results of Hummel and Bellini (20), and an I98A-mutated protein exhibits similar properties.
Cell surface expression and functional characteristics of MV H proteins carrying mutations at nonheptadic positions in the HR-like domain.
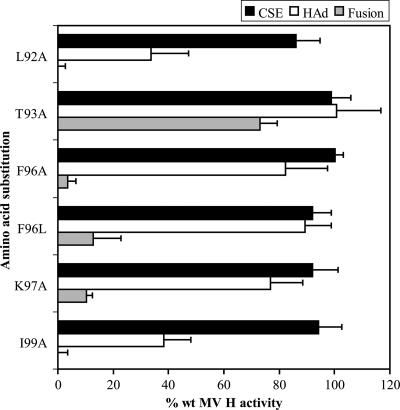
To further probe the role of the HR-like region in fusion promotion, MV H proteins with a P94A, -G, -L, or -S substitution, as well as those with an L95A, -P, or -R substitution, were prepared and characterized. The remaining residues between the second and third heptadic hydrophobic residues were also each individually replaced with alanine residues. Thus, individual mutated proteins, carrying an L92A, T93A, F96A, or K97A substitution, were constructed. Two additional mutations, F96L and I99A, were also evaluated.
As shown in Fig. 5, H proteins carrying individual P94A, -G, -L, and -S mutations exhibit fusion promotion activities of 3%, 0%, 2.8%, and 7.9%, respectively, of the level of wt H activity. While each of the mutated proteins has a cell surface expression level similar to that of wt H, ranging from 95% to 102%, the HAd activities of the proteins are significantly reduced to levels of 36%, 36%, 44%, and 48% of wt H levels, respectively. These results suggest that amino acid substitutions at P94 alter the structure and/or orientation of the globular head domain, in which the receptor-binding activity resides.
FIG. 5.
Cell surface expression (CSE) and functional characteristics of the MV H proteins with mutations at P94 and L95 in the HR-like domain. Assays were performed as described in the legend to Fig. 2. All data points represent the means of at least three independent experiments and are expressed relative to the activities of the wt proteins.
MV H proteins with amino acid substitutions at L95 were also tested for the ability to complement MV F in fusion promotion (Fig. 5). Each of the mutated proteins exhibits a significant deficiency in the ability to promote membrane fusion, with activities ranging from 0.5% to 32% of the wt H level. Each of the proteins carrying a substitution at L95 was expressed at the cell surface at a level similar to that of wt H (95% to 109% of the wt level). While L95A and -R mutations do not significantly decrease receptor recognition by MV H, the mutation of L95 to proline strongly reduces it to a level of 34% of wt H. These results suggest that the identity of the amino acid at position 95 may play a role in fusion promotion. However, it appears that the drastic proline mutation may alter the structure and/or orientation of the globular domain of MV H.
Alanine substitutions at the remaining positions between the second and third heptadic residues of the HR-like domain, including L92, T93, F96, and K97, were tested for their effect on the functions of MV H. Subsequently, F96 was also mutated to leucine. Another mutation, I99A, which is located between the third and fourth heptadic residues, was also characterized. Each of the mutated proteins is expressed at the cell surface at a level similar to that of wt H, ranging from 86% to 100% of the wt level (Fig. 6). All but one of these mutations, T93A, significantly decrease fusion promotion. Two of the amino acid substitutions, L92A and I99A, also decrease receptor recognition to 34% and 38% of the wt level, respectively. Substitutions at the two remaining residues, F96 and K97, result in significant deficiencies in fusion promotion not attributable to corresponding reductions in receptor recognition. Together with the data obtained for the conserved P94 and L95 residues, these results suggest that the nonheptadic residues in the HR domain in the stalk of MV H are important for both the structure and fusion promotion functions of H.
FIG. 6.
Cell surface expression (CSE) and functional characteristics of the MV H proteins with mutations within the IRs of the HR-like domain. Assays were performed as described in the legend to Fig. 2. All data points represent the means of at least three independent experiments and are expressed relative to the activities of the wt proteins.
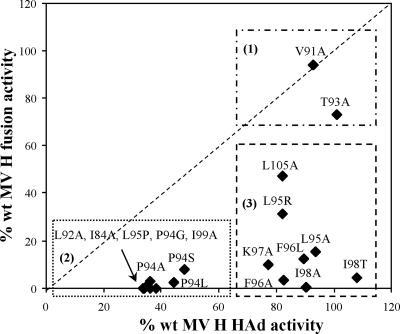
Comparison of HAd versus fusion promotion activity segregates amino acid substitutions in the MV H HR into three groups.
A plot of HAd versus fusion promotion activity of MV H proteins carrying amino acid substitutions in the HR-like domain reveals that the mutated proteins can be divided into three groups based on their functional activities (Fig. 7). The first group (V91A and T93A) includes amino acid substitutions that do not significantly alter either receptor recognition or fusion promotion. The second group (I84A, L92A, P94A, etc.) is characterized by strong deficiencies in both receptor binding and fusion promotion. The third group (L95A, F96A, I98A, etc.) exhibits significant deficiencies in fusion promotion activities not attributable to diminished receptor-binding activity. All of the MV H proteins carrying mutations in the HR-like domain have cell surface expression levels similar to that of wt H. Additionally, representative proteins from each group displayed sucrose sedimentation profiles similar to that of wt H, with the majority sedimenting as a tetramer but with a smaller amount of dimer also present (data not shown).
FIG. 7.
Correlation of ability to promote fusion with HAd activity of MV H proteins with mutations in the HR-like domain in the stalk. Based on this plot, the mutated proteins can be divided into three groups: (1) no significant deficiencies in receptor binding or fusion promotion, (2) significant deficiencies in both activities, and (3) significant deficiencies in fusion promotion but not receptor binding.
Detection of an interaction between MV H and F at the cell surface.
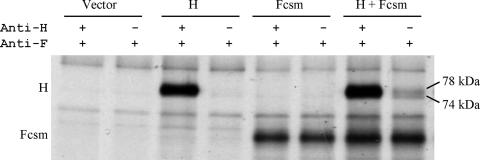
If MV H and F interact during the promotion of fusion, it should be possible to co-IP the two proteins from the surface of infected cells with an antibody to only one of them. As the MV H and F proteins are expressed and functional in HeLa cells, these cells were used to develop a co-IP assay to look for an interaction between the proteins at the cell surface. For this assay, Fcsm, which requires the addition of exogenous trypsin for activation, was used in order to prevent comparison of fusing and nonfusing monolayers. The method used for co-IP is analogous to that used in our laboratory for studies of NDV glycoprotein interactions (8, 27, 28). Figure 8 shows that the two glycosylation isoforms of MV H, with apparent molecular sizes of 74 and 78 kDa (32, 33), are coimmunoprecipitated with Fcsm. This demonstrates the detection of the formation of a complex between MV H and F at the cell surface. Approximately 21% (±9%) of the total amount of MV H can be coimmunoprecipitated with Fcsm by using Fcyt antibody.
FIG. 8.
Detection of formation of a complex of wt MV H and Fcsm at the surface of HeLa cells by using a co-IP assay. The surface proteins of transfected cells were biotinylated, and then the cells were lysed with lauryl maltoside. The lysates were split into two equal aliquots and subjected to the co-IP protocol by using a mixture of two MAbs against the H protein and a polyclonal antiserum against the cytoplasmic tail of F (the first lane in each pair) or the F antiserum alone (the second lane in each pair). Immunoprecipitates were displayed on reducing SDS-polyacrylamide gels.
Figure 8 also shows critical controls. The first lane for each pair shows the maximum amounts of the two proteins that can be immunoprecipitated from the cell surface for each sample. The first pair of lanes shows that neither protein is present in control cells transfected with empty vector. The second pair of lanes demonstrates that MV H does not immunoprecipitate with the F antiserum in the absence of the F protein, and the third pair shows that the protein that co-IPs with F is not present in cells that have not been transfected with the MV H gene. These controls establish the specificity of the co-IP of H by the anti-F antibody.
Amino acid substitutions in the HR domain of MV H do not prevent formation of the complex with F at the cell surface.
Mutations in the intervening region (IR) between the repeats in the stalk of the NDV HN protein have been shown to modulate fusion and interfere with formation of the HN-F complex at the cell surface (28). In order to investigate this as a possible explanation for the phenotypes of the MV H proteins carrying point mutations in this domain, several of the mutated proteins were tested for the ability to interact with MV F at the cell surface.
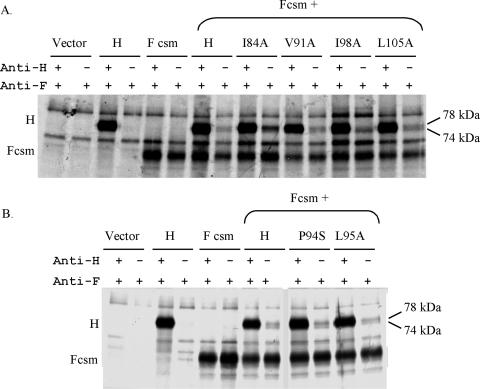
Figure 9A shows the results of a co-IP assay for each of the proteins carrying alanine substitutions for the heptadic hydrophobic residues: I84, V91, I98, and L105. Despite the significant deficiencies in receptor binding and/or fusion promotion associated with three of the four mutations, each of the mutated proteins efficiently co-IPs with MV F. Indeed, for these mutated proteins, there seems to be an inverse relationship between the extent of fusion and the extent of co-IP (Table 1). I84A-mutated H, which fails to promote detectable fusion, co-IPs three times more efficiently than wt H; I98A-mutated H, for which fusion is barely detectable (<1% of the wt level), co-IPs twice as efficiently as wt H. Moreover, L105A-mutated H fuses approximately half as efficiently yet co-IPs two-thirds more efficiently than the wt protein, whereas V91A-mutated H both fuses and co-IPs at levels similar to wt levels. Thus, complete or near-complete elimination of the fusion-promoting activity of MV H, resulting from mutations for heptadic residues in the HR-like domain in its stalk, does not eliminate the ability of the H protein to interact with the homologous F protein at the cell surface; on the contrary, it seems to stabilize the H-F interaction. It is also interesting to note that there is variability in the amounts of the 78-kDa protein that co-IP with F for each of these mutated proteins (Fig. 9). However, this variability does not appear to correlate with functional deficiencies in either receptor binding or membrane fusion promotion.
FIG. 9.
Detection of formation of complexes between MV H mutants with MV Fcsm at the cell surface. (A) Co-IP of MV H proteins carrying mutations in the heptadic residues with MV F. (B) Co-IP of MV H proteins carrying mutations at P94 and L95 with MV F.
TABLE 1.
Percent co-IP of mutated H proteins compared to their HAd and fusion-promoting activities
| Mutation | Groupa | % wt HAd | % wt fusion | % wt co-IPb |
|---|---|---|---|---|
| V91A | 1 | 92.8 | 94.4 | 119 |
| T93A | 1 | 100.7 | 73.1 | 140 |
| I84A | 2 | 34.1 | 0 | 299 |
| L92A | 2 | 33.5 | 0 | 180 |
| P94S | 2 | 48.0 | 7.9 | 115 |
| I99A | 2 | 38.2 | 0 | 141 |
| L95A | 3 | 93.6 | 15.5 | 93 |
| F96A | 3 | 82.2 | 3.5 | 65 |
| F96L | 3 | 89.4 | 12.8 | 125 |
| K97A | 3 | 76.9 | 10.3 | 109 |
| I98A | 3 | 90.4 | 0.7 | 200 |
| L105A | 3 | 81.9 | 47.3 | 168 |
From Fig. 7.
Percentage of total mutated H that co-IPs with antibody to F divided by percentage of wt H that co-IPs, multiplied by 100.
To determine if the deficiencies in receptor recognition and fusion promotion by mutated proteins carrying substitutions at the conserved P94 and L95 residues correlate with an inability to interact with MV F at the cell surface, the abilities of P94S- and L95A-mutated H, each of which promotes fusion at less than 20% of the wt level, to co-IP with MV F were determined. As shown in Fig. 9B, both of the mutated proteins retained the ability to co-IP with MV F, despite their drastic loss of fusion-promoting activity. P94S- and L95A-mutated H, which fuse at 8 and 16% of the wt level (Fig. 5), co-IP about as efficiently as wt H, i.e., 114.5% and 93.4%, respectively (Table 1). Thus, the apparent specific loss of fusion helper function resulting from the L95A mutation (no significant effect on HAd) (Fig. 5) is not due to an effect on the ability of H to interact with F. Interestingly, the inverse relationship between fusion and co-IP exhibited by the group of heptadic mutations is not manifested by these two mutations.
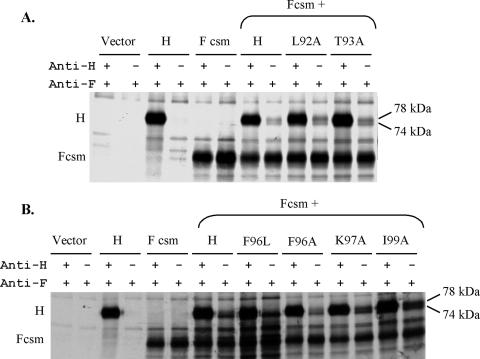
Mutated proteins carrying amino acid substitutions at the remaining residues between the second and third heptadic residues of the domain, as well as a protein with a substitution at the first amino acid between the third and fourth heptadic residues, were also tested for the ability to interact with MV F at the cell surface. Similarly to the above-mentioned results, each of the mutated proteins, including L92A and T93A (Fig. 10A), as well as F96L, F96A, K97A, and I99A (Fig. 10B), can be coimmunoprecipitated efficiently with MV F at the cell surface. Again, as for the mutated proteins with results shown in Fig. 9A, those with no detectable fusion, L92A- and I99A-mutated H, exhibited increased co-IP (180% and 141%, respectively) relative to that of the wt protein (Table 1). F96A-, F96L-, and K97A-mutated proteins, which exhibit comparatively minimal fusion-promoting activities, ranging from 3.5% to 12.8% that of the wt, co-IP at levels similar to or less than that of wt H (65%, 125%, and 109%, respectively). The T93A-mutated protein fuses at 73.1% of the wt level and co-IPs at 140% of the wt level.
FIG. 10.
Co-IP of MV H proteins carrying mutations in the IR with MV F. (A) Co-IP of L92A- and T93A-mutated proteins. (B) Co-IP of proteins carrying a mutation at position 96, 97, or 99. The experiments were performed as described in the legend to Fig. 8.
Taken together, the results of these co-IP assays indicate that individual amino acid substitutions in the HR-like domain, some of which drastically reduce or even eliminate fusion, do not prevent formation of the MV H-F complex at the cell surface. An interaction at the cell surface between H and F can be detected for mutated H proteins from all three groups identified in Fig. 7. On the contrary, the data indicate that a complete loss of fusion-promoting activity correlates with a stabilization of the H-F complex at the cell surface.
The HR domain is not the sole determinant of the specificity of the MV H-F interaction.
Alignment of the MV and CDV H sequences reveals that there are four residues that differ between the proteins in the HR domain (Fig. 3). These differences are at positions 84, 85, 89, and 103. In order to determine if the HR domain mediates the specificity of the interaction between H and F, these sites were mutated in MV H to the corresponding residues of CDV H. An MV H protein carrying all of the mutations I84V, E85H, K89I, and V103I was prepared and characterized. The mutated protein promotes membrane fusion at approximately 55% of the extent of wt H activity. The reduction in fusion activity does not correspond to a decrease in cell surface expression (107% that of wt H) or receptor binding (99% that of wt H). The mutated protein is unable to promote fusion when coexpressed with CDV F (0% wt MV H-F fusion activity). These data are consistent with the results of the co-IP assays, indicating that the MV H HR-like domain is not the sole mediator of the specific physical association between H and F at the cell surface that is required for fusion promotion.
Amino acid substitutions in the HR-like domain of MV H that abolish fusion also abolish lipid mixing.
It is thought that, during the membrane fusion process, activation of F and insertion of the fusion peptide into the target membrane initially lead to merger of the lipid bilayers, often called hemifusion, before fusion pore formation and content mixing. Mutation of the conserved proline, P111, between the HRs in the stalk of the human parainfluenza virus 3 (hPIV3) attachment protein does not alter receptor binding but has been found to decrease the rate of triggering of the F protein into its activated form (38). Insertion of the fusion peptide into the target membrane and lipid mixing were shown to occur, but more slowly than when F was triggered by wt HN-expressing cells. A lipid-mixing assay with R18-labeled erythrocytes was used to determine if this is also the case for the MV H proteins carrying mutations in the HR that abolished fusion without altering receptor binding.
Figure 11 shows the results of a lipid-mixing assay performed with wt MV F coexpressed with the H proteins carrying substitutions P94A, L95A, I98A, and I98T, each of which drastically decreases fusion. The controls for this experiment show that there is dye transfer from the labeled erythrocytes to cells expressing wt H and F, but not when H is expressed with Fcsm. As expected, based on its defect in receptor binding, the P94A-mutated H protein does not trigger lipid mixing. Despite detectable HAd of the labeled red blood cells, dye transfer is not detectable for the remaining mutants tested with this assay. These results suggest that membrane fusion promotion by these mutated proteins is blocked before MV H triggers F activation to initiate insertion of the fusion peptide into the target membrane.
FIG. 11.
Promotion of lipid mixing by MV H proteins with mutations in the HR-like domain. The extent of lipid mixing is shown by the spread of the dye from R18-labeled AGM erythrocytes into the cell membranes of transfected HeLa cells. Labeled erythrocytes were added to each monolayer, and then the cells were incubated on ice for 30 min. Fusion between the transfected cells and erythrocytes was initiated by transferring the cells to 37°C. After 30 min, the cells were washed, and then images were acquired immediately with a 20× objective by using fluorescent microscopy.
DISCUSSION
Identification of the domains of the paramyxovirus attachment and fusion proteins that mediate the interaction between them necessary for fusion is an important part of understanding the mechanism of membrane fusion. Two regions of MV H have been proposed to mediate its fusion helper function, one, including residue I98, in the stalk and another, including residues 244 to 250, in the head (15, 17, 20). Despite the potential importance of these regions of the H and F proteins in fusion promotion, their role in fusion has not been established.
The contention that region 244 to 250 in MV H constitutes the F-interactive domain is based on its recognition by an MAb that inhibits membrane fusion without altering receptor recognition activity (13, 15). However, the involvement of these residues in fusion promotion was not tested directly. The results of alanine-scanning mutagenesis of the residues in this region and characterization of the mutated proteins suggest that no single residue in this region is critical for either receptor binding or fusion promotion.
Region 244 to 250 is positioned on the model of MV H in a loop that protrudes into the putative center of symmetry of the tetramer (10). It is possible that antibodies directed against this region may inhibit fusion by interfering with the stability of the H tetramer. Antibody binding in this region could also inhibit membrane fusion indirectly by interfering with conformational changes in H that take place after receptor binding or by sterically blocking a nearby region.
Chimera and mutagenesis studies of the attachment proteins of other paramyxoviruses suggest that the stalk region both determines the specificity of the HN-F interaction and directly mediates it (7-9, 27, 28, 42, 43). Site-directed mutagenesis of individual residues within the stalk has identified specific amino acids involved in the interaction with F, in the process identifying a region that may contribute to the F-interactive domain (28, 39). These residues are located in domains known as HRs (38), though they do not completely fulfill the requirements of that motif. In most paramyxoviruses, the HR-like domain in the attachment protein stalk consists of two motifs separated by an IR containing a highly conserved proline-leucine doublet. Unlike those of most other paramyxoviruses, the morbillivirus attachment proteins have 6 residues in the analogous region, such that the entire domain constitutes a single, more extended HR-like domain, albeit with a helix-disrupting or kink-forming proline in the middle. A similar motif containing an LXP domain has been described previously for the MV F protein (35).
Through construction and testing of MV H chimeras for fusion promotion, an amino acid difference at position 98, which is located at a core position in the HR-like domain, was found to be responsible for the lack of fusion in a persistently infected cell line (20). While it was demonstrated that this residue is critical for fusion promotion, it was not determined if mutation of it affects receptor recognition activity. Although it is located in the stalk, the mutation could conceivably alter the conformation of the regions of the globular domain that are responsible for receptor binding. Indeed, substitutions for some of the individual heptadic hydrophobic residues in NDV HN, as well as residues in the IR between them, diminish fusion promotion but also alter receptor recognition and neuraminidase activity (28, 41, 45).
Here, we have confirmed that alanine and threonine substitutions for I98 strongly decrease fusion with no significant effect on either surface expression or receptor binding, consistent with the idea that the residue is involved in the fusion helper function of MV H, as originally proposed by Hummel and Bellini (20). However, while the I98A mutation disrupts fusion promotion at a stage prior to the triggering of MV F, it does not abolish the H-F interaction at the cell surface. On the contrary, it seems to stabilize it.
Indeed, we have performed an extensive site-directed mutational analysis of the role in fusion of several residues in the HR-like domain in the stalk of MV H. Our results show that, although many mutations in this region severely decrease, even eliminate, fusion, none of them results in a decrease in the amount of H that co-IPs with F commensurate with the decrease in fusion (Table 1). When the percentage of each mutated H protein that co-IPs with F is examined relative to its HAd and fusion-promoting activities (Table 1), a correlation between complete loss of fusion and increased co-IP becomes apparent. All three group 2 mutations that completely eliminate fusion (I84A, L92A, and I99A) stabilize the H-F complex at the cell surface. I84A- and L92A-mutated H co-IP almost 300% and 180% compared to wt levels, respectively. While the increased co-IP could be related to the reduced HAd activities of these proteins, I98A-mutated H (group 3), which fuses at <1% of the wt level but has HAd activity near that of the wt, co-IPs twofold better than the wt. These findings are consistent with previous results demonstrating that the extent of MV fusion is inversely related to the strength of the interaction between the H and F proteins (37).
However, this inverse relationship is really demonstrated only by those mutated proteins that exhibit <1% of wt fusion. Mutated proteins that fuse at 7.9 to 15.5% of the wt level, e.g., P94S, L95A, F96L, and K97A, co-IP about as well as the wt protein (93 to 125%). With the exception of P94S-mutated H, these all fall in group 3, comprised of proteins that exhibit markedly decreased fusion despite essentially wt HAd activity. It is also noteworthy that these mutations lie in a linear 4-amino-acid sequence in the region. Although they show markedly decreased fusion-promoting activity, they do not co-IP more efficiently than the wt protein. Thus, it seems more likely that these mutations could directly affect the interaction with the F protein and that the domain could directly contribute to it.
However, if this is the case, one must be able to rationalize the ability of these mutated proteins to still form the complex with F but not promote fusion. This can be accounted for in at least two ways. First, it is possible that residues 94 to 97 are part of the only domain in H that interacts with F and that mutation of a single amino acid in it is insufficient to prevent formation of the complex with F. Second, multiple regions of MV H may be involved in the formation of the H-F complex and the mutations in the linear domain spanning residues 94 to 97 do not disrupt its ability to interact with F, though they do impair fusion. In either case, the H-F complex could be of low enough avidity that it is incapable of resulting in fusion. In this regard, it should be noted that the co-IP assay is designed to maximize capture of the H-F complex and, as such, could detect a low-avidity association between the two proteins.
Whatever the basis for the co-IP findings with the MV H mutated proteins, one thing is clear. These findings are in direct contrast to the findings with NDV, where several analogous HN mutations, which result in significant though less drastic fusion deficiencies, reduce the HN-F complex to an undetectable level (28). The differential effects of mutations in the HR-like domain in MV H and NDV HN on the glycoprotein interaction are consistent with the fusion activation of the F protein in the two viruses being regulated by different mechanisms. This, in turn, is consistent with the idea, originally proposed by Plemper et al. (36), of a differential mechanism for regulation of the fusion activation of F in viruses that recognize the abundant sialic acid moiety, e.g., NDV, hPIV3, and simian virus 5, and those, such as MV, that recognize specific receptors.
Acknowledgments
We gratefully acknowledge the excellent technical assistance of Anne Mirza and helpful discussions and/or critical reading of the manuscript by Judith Alamares, Paul Mahon, Vanessa Melanson, Anne Mirza, and Matteo Porotto. We also thank Roberto Cattaneo, Bernard Moss, Michael Oldstone, Shmuel Rosenblatt, and Paul Rota for essential reagents.
This work was supported by grant AI-49268 from the National Institutes of Health.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Alkhatib, G., and D. J. Briedis. 1986. The predicted primary structure of the measles virus hemagglutinin. Virology 150:479-490. [DOI] [PubMed] [Google Scholar]
- 2.Bagai, S., and R. A. Lamb. 1995. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J. Virol. 69:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossart, K. N., L. F. Wang, M. N. Flora, K. B. Chua, S. K. Lam, B. T. Eaton, and C. C. Broder. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 76:11186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo, R., and J. K. Rose. 1993. Cell fusion by the envelope glycoproteins of persistent measles viruses which caused lethal human brain disease. J. Virol. 67:1493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corey, E. A., A. M. Mirza, E. Levandowsky, and R. M. Iorio. 2003. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 77:6913-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, R., A. M. Mirza, P. J. Mahon, and R. M. Iorio. 1997. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch. Virol. Suppl. 13:115-130. [DOI] [PubMed] [Google Scholar]
- 8.Deng, R., Z. Wang, P. J. Mahon, M. Marinello, A. Mirza, and R. M. Iorio. 1999. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253:43-54. [DOI] [PubMed] [Google Scholar]
- 9.Deng, R., Z. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 10.Deroo, S., K. C. El Kasmi, P. Fournier, D. Theisen, N. H. Brons, M. Herrmann, J. Desmet, and C. P. Muller. 1998. Enhanced antigenicity of a four-contact-residue epitope of the measles virus hemagglutinin protein by phage display libraries: evidence of a helical structure in the putative active site. Mol. Immunol. 35:435-443. [DOI] [PubMed] [Google Scholar]
- 11.Dorig, R. E., A. Marcil, A. Chopra, and C. D. Richardson. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295-305. [DOI] [PubMed] [Google Scholar]
- 12.Elango, N., J. E. Coligan, R. C. Jambou, and S. Venkatesan. 1986. Human parainfluenza type 3 virus hemagglutinin-neuraminidase glycoprotein: nucleotide sequence of mRNA and limited amino acid sequence of the purified protein. J. Virol. 57:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Kasmi, K. C., D. Theisen, N. H. Brons, and C. P. Muller. 1998. The molecular basis of virus crossreactivity and neutralisation after immunisation with optimised chimeric peptides mimicking a putative helical epitope of the measles virus hemagglutinin protein. Mol. Immunol. 35:905-918. [DOI] [PubMed] [Google Scholar]
- 14.Erlenhoefer, C., W. J. Wurzer, S. Loffler, S. Schneider-Schaulies, V. ter Meulen, and J. Schneider-Schaulies. 2001. CD150 (SLAM) is a receptor for measles virus but is not involved in viral contact-mediated proliferation inhibition. J. Virol. 75:4499-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, P., N. H. Brons, G. A. Berbers, K. H. Wiesmuller, B. T. Fleckenstein, F. Schneider, G. Jung, and C. P. Muller. 1997. Antibodies to a new linear site at the topographical or functional interface between the haemagglutinin and fusion proteins protect against measles encephalitis. J. Gen. Virol. 78:1295-1302. [DOI] [PubMed] [Google Scholar]
- 16.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin, D. E. 2001. Measles virus, p. 1401-1441. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 18.Hsu, E. C., R. E. Dorig, F. Sarangi, A. March, C. Iorio, and C. D. Richardson. 1997. Artificial mutations and natural variations in the CD46 molecules from human and monkey cells define regions important for measles virus binding. J. Virol. 71:6144-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, A., H. Sheshberadaran, E. Norrby, and J. Kovamees. 1993. Molecular characterization of epitopes on the measles virus hemagglutinin protein. Virology 192:351-354. [DOI] [PubMed] [Google Scholar]
- 20.Hummel, K. B., and W. J. Bellini. 1995. Localization of monoclonal antibody epitopes and functional domains in the hemagglutinin protein of measles virus. J. Virol. 69:1913-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovamees, J., M. Blixenkrone-Moller, and E. Norrby. 1991. The nucleotide and predicted amino acid sequence of the attachment protein of canine distemper virus. Virus Res. 19:223-233. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 689-724. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 24.Langedijk, J. P. M., F. J. Daus, and J. T. van Oirschot. 1997. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J. Virol. 71:6155-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maisner, A., B. Mrkic, G. Herrler, M. Moll, M. A. Billeter, R. Cattaneo, and H. D. Klenk. 2000. Recombinant measles virus requiring an exogenous protease for activation of infectivity. J. Gen. Virol. 81:441-449. [DOI] [PubMed] [Google Scholar]
- 26.McGinnes, L. W., A. Wilde, and T. G. Morrison. 1987. Nucleotide sequence of the gene encoding the Newcastle disease virus hemagglutinin-neuraminidase protein and comparisons of paramyxovirus hemagglutinin-neuraminidase protein sequences. Virus Res. 7:187-202. [DOI] [PubMed] [Google Scholar]
- 27.Melanson, V. R., and R. M. Iorio. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melanson, V. R., and R. M. Iorio. 2004. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J. Virol. 78:13053-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirza, A. M., R. Deng, and R. M. Iorio. 1994. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidase glycoprotein: effects on antigenic structure and function. J. Virol. 68:5093-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris, S. J., D. P. Sarkar, J. M. White, and R. Blumenthal. 1989. Kinetics of pH-dependent fusion between 3T3 fibroblasts expressing influenza hemagglutinin and red blood cells. Measurement by dequenching of fluorescence. J. Biol. Chem. 264:3972-3978. [PubMed] [Google Scholar]
- 31.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogura, H., I. Matsunaga, Y. Takano, X. Ning, M. Ayata, K. Tanaka, T. Seto, K. Furukawa, N. Ito, M. Shingai, T. Kimura, K. Ichihara, H. Kubo, and T. Murakawi. 2000. Cell surface expression of immature H glycoprotein in measles virus-infected cells. Virus Res. 66:187-196. [DOI] [PubMed] [Google Scholar]
- 33.Ogura, H., H. Sato, S. Kamiya, and S. Nakamura. 1991. Glycosylation of measles virus haemmaglutinin protein in infected cells. J. Gen. Virol. 72:2679-2684. [DOI] [PubMed] [Google Scholar]
- 34.Ono, N., H. Tatsuo, Y. Hidaka, T. Aoki, H. Minagawa, and Y. Yanagi. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plemper, R. K., and R. W. Compans. 2003. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J. Virol. 77:4181-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239-44246. [DOI] [PubMed] [Google Scholar]
- 37.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plemper, R. K., A. S. Lakdawala, K. M. Gernert, J. P. Snyder, and R. W. Compans. 2003. Structural features of paramyxovirus F protein required for fusion initiation. Biochemistry 42:6645-6655. [DOI] [PubMed] [Google Scholar]
- 39.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 77:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stern, L. B., M. Greenberg, J. M. Gershoni, and S. Rozenblatt. 1995. The hemagglutinin envelope protein of canine distemper virus (CDV) confers cell tropism as illustrated by CDV and measles virus complementation analysis. J. Virol. 69:1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stone-Hulslander, J., and T. G. Morrison. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73:3630-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsurudome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190-203. [DOI] [PubMed] [Google Scholar]
- 44.von Messling, V., G. Zimmer, G. Herrler, L. Haas, and R. Cattaneo. 2001. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 75:6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Z., and R. M. Iorio. 1999. Amino acid substitutions in a conserved region in the stalk of the Newcastle disease virus HN glycoprotein spike impair its neuraminidase activity in the globular domain. J. Gen. Virol. 80:749-753. [DOI] [PubMed] [Google Scholar]
- 46.Waxham, M. N., J. Aronowski, A. C. Server, J. S. Wolinsky, J. A. Smith, and H. M. Goodman. 1988. Sequence determination of the mumps virus HN gene. Virology 164:318-325. [DOI] [PubMed] [Google Scholar]
- 47.Wild, T. F., J. Fayolle, P. Beauverger, and R. Buckland. 1994. Measles virus fusion: role of the cysteine-rich region of the fusion glycoprotein. J. Virol. 68:7546-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild, T. F., E. Malvoisin, and R. Buckland. 1991. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J. Gen. Virol. 72:439-442. [DOI] [PubMed] [Google Scholar]
- 49.Yanagi, Y., M. Takeda, S. Ohno, and F. Seki. 2006. Measles virus receptors and tropism. Jpn. J. Infect. Dis. 59:1-5. [PubMed] [Google Scholar]
- 50.Yao, Q., X. Hu, and R. W. Compans. 1997. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 71:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]