Abstract
The 5′ nontranslated region of poliovirus RNA contains two highly structured regions, the cloverleaf (CL) and the internal ribosomal entry site (IRES). A cellular protein, the poly(rC) binding protein (PCBP), has been reported to interact with the CL either alone or in combination with viral protein 3CDpro. The formation of the ternary complex is essential for RNA replication and, hence, viral proliferation. PCBP also interacts with stem-loop IV of the IRES, an event critical for the initiation of cap-independent translation. Until recently, no special function was assigned to a spacer region (nucleotides [nt] 89 to 123) located between the CL and the IRES. However, on the basis of our discovery that this region strongly affects the neurovirulent phenotype of poliovirus, we have embarked upon genetic and biochemical analyses of the spacer region, focusing on two clusters of C residues (C93-95 and C98-100) that are highly conserved among entero- and rhinoviruses. Replacement of all six C residues with A residues had no effect on translation in vitro but abolished RNA replication, leading to a lethal growth phenotype of the virus in HeLa cells. Mutation of the first group of C residues (C93-95) resulted in slower viral growth, whereas the C98-100A change had no significant effect on viability. Genetic analyses of the C-rich region by extensive mutagenesis and analyses of revertants revealed that two consecutive C residues (C94-95) were sufficient to promote normal growth of the virus. However, there was a distinct position effect of the preferred C residues. A 142-nt-long 5′-terminal RNA fragment including the CL and spacer sequences efficiently bound PCBP, whereas no PCBP binding was observed with the CL (nt 1 to 88) alone. Binding of PCBP to the 142-nt fragment was completely ablated after the two C clusters in the spacer were mutated to A clusters. In contrast, the same mutations had no effect on the binding of 3CDpro to the 142-nt RNA fragment. Stepwise replacement of the C residues with A residues resulted in impaired replication that covaried with weaker binding of PCBP in vitro. We conclude that PCBP has little, if any, binding affinity for the CL itself (nt 1 to 88) but requires additional nucleotides downstream of the CL for its function as an essential cofactor in poliovirus RNA replication. These data reveal a new essential function of the spacer between the CL and the IRES in poliovirus proliferation.
After entry into the host cell, the plus strand RNA genome of poliovirus (PV) engages in numerous protein-RNA interactions that are required for successful completion of the life cycle. The formation of these complexes is required for three different processes, translation, RNA replication, and encapsidation. Because of the small size of the viral RNA and the limited number of viral proteins available for complex formation, the virus has evolved to use multiple host proteins to carry out a productive infection. The interaction of viral and cellular proteins with the RNA genome of PV during the translation and replication of the viral genome has been the subject of numerous studies during the past 2 decades. Although many important discoveries have been made in this field, the details of these processes are not yet understood.
One such cellular protein is poly(rC) binding protein 2 (PCBP2, also known as hnRNP E2 or αCP-2), an RNA binding protein with function in both translation and replication of PV RNA and possibly in RNA stability (6, 12, 13, 22, 24). PCBP2 has a strong preference for binding to poly(rC) sequences. The cellular function of PCBP2 is to form ribonucleoprotein (RNP) complexes with cellular mRNAs, which regulate mRNA stability and translation (15, 20, 23). PCBP2 contains three hnRNP K homology (KH) domains, the first and third of which mediate poly(rC) binding (9). The protein has also been shown to form homodimers (13) and to interact with other hnRNPs (4, 16).
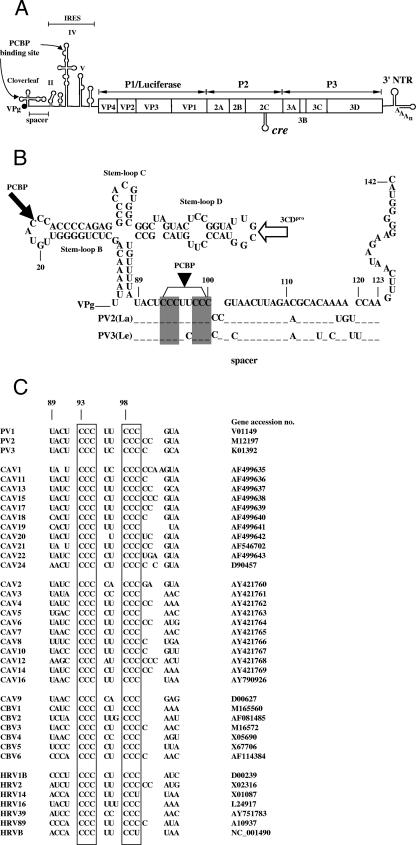
The 5′ nontranslated region (5′NTR) of PV RNA contains two highly structured regions, the cloverleaf (CL) and the internal ribosomal entry site (IRES) (Fig. 1A). The CL, which carries the 5′-terminal genome-linked protein VPg, consists of the 5′-terminal 88 nt containing stem-loops A to D (2, 13, 18, 24) (Fig. 1A and B). The C-rich region of stem-loop B has been reported to bind PCBP2 (1, 2, 13, 24) (Fig. 1A and B), while stem-loop D can specifically interact with the viral proteinase and RNA binding protein 3CDpro (1, 2, 13, 24, 28) (Fig. 1B). The formation of the ternary complex CL/PCBP/3CDpro at the 5′ end is essential for RNA replication and, hence, viral proliferation. Interestingly, viral protein 3AB also forms a complex with the CL and 3CDpro, which promotes viral RNA replication (14, 36).
FIG. 1.
Structure of PV genomic RNA and nucleotide sequence alignment of the region between the CL and the IRES. (A) Schematic diagram of full-length PV genomes [PV1(M)] and PV-Luc replicons. The single-stranded RNA is covalently linked to the virus-encoded protein VPg at the 5′ end of the 5′NTR. The 5′NTR consists of two highly structured cis-acting domains, the CL and the IRES, which are separated by a spacer region. The polyprotein (open box), consists of a structural (P1) region and two nonstructural domains (P2 and P3), specifying the replication proteins. In the replicons, the P1 domain of the polyprotein is replaced with the coding sequence of the firefly luciferase gene. A cis-replicating RNA element, cre, is located in the coding sequences of protein 2CATPase. (B) Nucleotide sequences of the PV1(M) CL and spacer and part of domain II (nt 1 to 142) that were included in the RNA probe used for the experiments. The spacer region of PV1(M) (nt 89 to 103), harboring the two C clusters (shaded areas), is compared with that of PV2(La) and PV3(Le). The closed arrows indicate the approximate PCBP binding sites, and the open arrow indicates the approximate binding site of 3CDpro. (C) Nucleotide sequence alignments of the 5′ terminus of the spacer regions (nt 89 to 103) of PVs, cluster C coxsackievirus A, cluster A coxsackievirus A, cluster B coxsackievirus B (plus coxsackievirus A9), and human rhinoviruses. The boxes indicate the C-rich region that is highly conserved among the enteroviruses. M, Mahoney; La, Lancing; Le, Leon.
PCBP2 interacts also with the IRES element specifically at domain IV (5, 6, 13) (Fig. 1A). This process is essential for IRES-regulated initiation of cap-independent translation of the polyprotein (10, 27, 31). In addition, the nucleocytoplasmic SR protein (SRp20) has been reported to bind to IRES-bound PCBP2 (4), an interaction that plays no role in genome replication but is important for translation initiation (4).
The CL and IRES elements are separated by a “spacer” sequence of 35 nt (Fig. 1A and B) to which, until recently, no special function(s) in viral proliferation had been assigned. This changed when we discovered an unexpected high degree of attenuation in CD155 transgenic mice of a PV [sPV(M)] that we had synthesized in the absence of a natural template (7). sPV(M) contained 27 mostly synonymous nucleotide exchanges designed as genetic markers (7). To our surprise, one of these genetic markers, a single nucleotide transition (A103G) mapping to the spacer region in the 5′NTR, was responsible for the strong attenuation phenotype of sPV(M) in the transgenic animals (8). Interestingly, a single A103G mutation in PV1(M) also triggered a robust temperature-sensitive phenotype in cells of neuronal origin (human SK-N-MC cells) (8). The molecular basis for the attenuation and temperature-sensitive phenotypes of the A103G virus is not yet known, but our results suggested that the defect is related to translation (8).
On the basis of these observations, we were curious about whether other signals are hidden in the spacer region that could play a role in viral proliferation, especially in RNA replication and translation. Accordingly, we have embarked upon a genetic and biochemical analysis of nucleotides in the spacer. When one compares the nucleotide sequences of the spacer regions from various entero- and rhinoviruses (Fig. 1C), two highly conserved clusters of C residues, each 3 nt long, stand out. Mutation of all six C residues to A residues had no effect on the translation of PV RNA in vitro but abolished RNA replication in HeLa cells, resulting in a dead growth phenotype. A more detailed analysis of the C-rich region revealed that a minimum of two consecutive C residues, located in the first cluster of C residues, or all three C residues from the second group, were required for virus growth. In addition, our studies showed that the efficient binding of PCBP2 to a 5′-terminal, 142-nt-long fragment of the PV RNA required the presence of at least two adjacent C residues from the C-rich region of the spacer. Interestingly, we have found that PCBP2 has little, if any, binding affinity for the CL per se (nt 1 to 88). For PCBP to function as an essential cofactor in PV genome replication, both the CL with its stem-loop B and additional C residues from the spacer region (nt 89 to 95) are required.
MATERIALS AND METHODS
Construction of plasmids.
pT7PV1(M) (32) refers to the full-length (nt 1 to 7525) PV1(M) cDNA (Fig. 1A). All mutations and final constructs were verified by sequencing with the ABI Prism DNA sequencing kit.
(i) Constructs with a substitution of nucleotides in the spacer region (nt 93 to 100).
For construction of the PV1(M) derivatives listed in Fig. 2, mutagenesis was performed with the site-directed mutagenesis kit (Stratagene) with pT7PV1(M) as the template. The mutagenic oligonucleotides are described in Table 1.
FIG. 2.
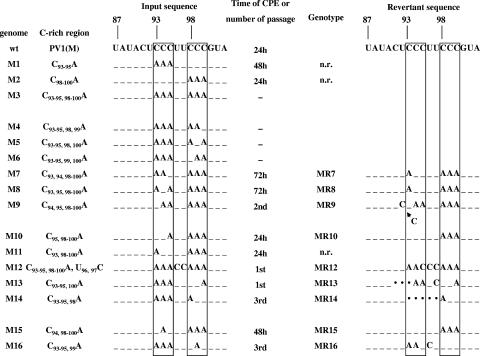
Characterization of mutants with nucleotide substitutions in the C-rich region of the spacer. Nucleotide sequences (nt 87 to 103) of the C-rich region in mutants used in these studies are shown. Dashes represent the nucleotides conserved between WT [PV1(M)] and the mutants. Boxed sequences indicate the C-rich regions in the spacer. RNA transcripts of the WT and mutant viruses were transfected into HeLa cells, and the time of CPE was determined (see Materials and Methods). Viruses that gave no CPE after transfection were passaged one or more times. RNAs derived from viruses that emerged after transfection were reverse transcribed, and their DNA sequences were determined. The arrow indicates the location of a C residue insertion in revertant MR9. Dots indicate deletions of nucleotides in the revertants. n.r., no revertant was obtained.
TABLE 1.
Oligonucleotides used for mutagenesis
| Primera | Sequence |
|---|---|
| Del. CL SL-b (F) | 5′-CAGCTCTGGGGTTGTACCCCAGAGGCCCAC-3′ |
| Del. CL SL-b (R) | 5′-CGTGGGCCTCTGGGGTACAACCCCAGAGC-3′ |
| C93-95A (F) | 5′-CGCCTGTTTTATACTAAATTCCCGTAAC-3′ |
| C93-95A (R) | 5′-CTAAGTTACGGGAATTTAGTATAAAACAGG-3′ |
| C98-100A (F) | 5′-CCTGTTTTATACTCCCTTAAAGTAACTTAGAC-3′ |
| C98-100A (R) | 5′-GCGTCTAAGTTACTTTAAGGGAGTATAAAAC-3′ |
| C93-95, 98-100A (F) | 5′-CGCCTGTTTTATACTAAATTAAAGTAACTTAGACG-3′ |
| C93-95, 98-100A (R) | 5′-GTGCGTCTAAGTTACTTTAATTTAGTATAAAACAGG-3′ |
| C93-95, 98, 99A (F) | 5′-CGCCTGTTTTATACTAAATTAACGTAACTTAGACGC-3′ |
| C93-95, 98, 99A (R) | 5′-GTGCGTCTAAGTTACGTTAATTTAGTATAAAACAGG-3′ |
| C93-95, 98, 100A (F) | 5′-CGCCTGTTTTATACTAAATTACAGTAACTTAG-3′ |
| C93-95, 98, 100A (R) | 5′-CGTCTAAGTTACTGTAATTTAGTATAAAACAG-3′ |
| C93-95, 99, 100A (F) | 5′-CGCCTGTTTTATACTAAATTCAAGTAACTTAG-3′ |
| C93-95, 99, 100A (R) | 5′-CGTCTAAGTTACTTGAATTTAGTATAAAACAG-3′ |
| C93, 94, 98-100A (F) | 5′-CGCCTGTTTTATACTAACTTAAAGTAACTTAGACGC-3′ |
| C93, 94, 98-100A (R) | 5′-GTGCGTCTAAGTTACTTTAAGTTAGTATAAAACAGG-3′ |
| C93, 95, 98-100A (F) | 5′-CGCCTGTTTTATACTACATTAAAGTAACTTAG-3′ |
| C93, 95, 98-100A (R) | 5′-CGTCTAAGTTACTTTAATGTAGTATAAAACAG-3′ |
| C94, 95, 98-100A (F) | 5′-CGCCTGTTTTATACTCAATTAAAGTAACTTAGACGC-3′ |
| C94, 95, 98-100A (R) | 5′-GTGCGTCTAAGTTACTTTAATTGAGTATAAAACAGG-3′ |
| C95, 98-100A (F) | 5′-CGCCTGTTTTATACTCCATTAAAGTAACTTAGACGC-3′ |
| C95, 98-100A (R) | 5′-GTGCGTCTAAGTTACTTTAATGGAGTATAAAACAGG-3′ |
| C93, 98-100A (F) | 5′-CGCCTGTTTTATACTACCTTAAAGTAACTTAGACGC-3′ |
| C93, 98-100A (R) | 5′-GTGCGTCTAAGTTACTTTAAGGTAGTATAAAACAGG-3′ |
| C93-95, 98-100A, U96, 97C (F) | 5′-CGCCTGTTTTATACTAAACCAAAGTAACTTAG-3′ |
| C93-95, 98-100A, U96, 97C (R) | 5′-CGTCTAAGTTACTTTGGTTTAGTATAAAACAG-3′ |
| C93-95, 100A (F) | 5′-CGCCTGTTTTATACTAAATTCCAGTAACTTAG-3′ |
| C93-95, 100A (R) | 5′-CGTCTAAGTTACTGGAATTTAGTATAAAACAG-3′ |
| C93-95, 98A (F) | 5′-CGCCTGTTTTATACTAAATTACCGTAACTTAGACGC-3′ |
| C93-95, 98A (R) | 5′-GTGCGTCTAAGTTACGGTAATTTAGTATAAAACAGG-3′ |
| C94, 98-100A (F) | 5′-CGCCTGTTTTATACTCACTTAAAGTAACTTAG-3′ |
| C94, 98-100A (R) | 5′-CGTCTAAGTTACTTTAAGTGAGTATAAAACAG-3′ |
| C93-95, 99A (F) | 5′-CGCCTGTTTTATACTAAATTCACGTAACTTAGACGC-3′ |
| C93-95, 99A (R) | 5′-GTGCGTCTAAGTTACGTGAATTTAGTATAAAACAGG-3′ |
F, forward; R, reverse.
(ii) Replicon constructs with a substitution of nucleotides in the spacer region (nt 93 to 100).
To test the effect of spacer mutations directly on RNA replication, we used a luciferase replicon (PV-Luc) which contains the firefly luciferase gene in place of the P1 coding region of the PV polyprotein. To introduce the mutations into PV-Luc (Fig. 1A) (19), mutant cDNAs C93-95A PV1(M), C98-100A PV1(M), and C93-100A PV1(M) (Fig. 2) were digested with AgeI and PmlI. The AgeI-PmlI fragments were cloned into similarly restricted PV-Luc.
(iii) pET21b/PCBP1.
Total RNA was isolated from HeLa cells with TRIzol (Invitrogen) according to the manufacturer's instructions. PCBP1 cDNA was obtained by reverse transcription (RT)-PCR with primers 5′-CTCGAGGCTGCACCCCATGCCCTTCTC-3′ and 5′-GAATTCTATGGATGCCGGTGTGACTG-3′. The PCR product was cloned into the pGEM-T Easy vector. The XhoI-EcoRI fragment of this plasmid was cloned into the XhoI/EcoRI-digested pET21b vector (Novagen).
Proteins.
Purified, His-tagged PCBP1, PCBP2, and 3CDpro recombinant proteins were expressed in Escherichia coli from plasmids pET21b/PCBP1, pET21b/PCBP2 (26), and pET21b/3CDpro(3Cpro/H40A) (26), respectively. The proteins were purified by nickel column chromatography (QIAGEN).
Antibodies.
Anti-PCBP2 polyclonal antiserum (33, 34) (a generous gift of Bert L. Semler, University of California, Irvine) and anti-3D monoclonal antibody (25) were used as primary antibodies for Western blot analysis.
In vitro transcription, translation, and RNA transfection.
Prior to transcription by T7 RNA polymerase, pT7PV1(M) and its derivatives were linearized with EcoRI, whereas PV-Luc and its derivatives were linearized with DraI. The RNA transcripts were transfected into monolayer cultures (35-mm-diameter dishes) of HeLa R19 cells by the DEAE-dextran method, as described previously (32). Transfected cells were incubated in Dulbecco's modified Eagle medium supplemented with 2% bovine calf serum at 37°C either until a complete cytopathic effect (CPE) was observed or for at least 3 days posttransfection. After three rounds of freezing and thawing, the lysate was clarified of cell debris by low-speed centrifugation. The supernatant, containing the virus, was used for further passaging to select PV variants capable of efficient replication in HeLa R19 cells. The RNAs extracted from the viral cell lysates served as templates for RT-PCR. Isolation of viral RNA, RT-PCR, purification of PCR products, and sequencing were carried out as described previously (7). In vitro RNA translations were performed with HeLa cell S10 cytoplasmic extracts at 34°C as described previously (21).
Luciferase assay.
After transfection with replicon RNA, HeLa R19 cells were incubated at 37°C in Dulbecco's modified Eagle medium (2% bovine calf serum). At 12 h posttransfection, the growth medium was removed from the dishes, and the cells were washed gently with 2 ml of phosphate-buffered saline. The HeLa cell dishes were overlaid with 300 μl of “passive” lysis buffer, supplied by Promega. The plates were rocked at room temperature for 15 min, and the lysate was transferred to a tube. Fifty microliters of luciferase assay reagent (Promega) was mixed with 20 μl of lysate, and the firefly luciferase activity was measured with an Optocomp I luminometer (MGM Instruments, Inc.).
RNA binding assay.
To test the binding of proteins to PV RNA fragments, RNA pull-down assays were used, similar to what was described before by Kim et al. (17). Truncated derivatives of PV CL-spacer RNA used in the RNA binding experiments were generated by PCR amplification with oligonucleotides described in Table 2. One microgram of gel-purified PCR products was used for the generation of biotinylated RNA probes. RNA transcripts were produced by T7 RNA polymerase (Stratagene) with nucleoside triphosphates in biotinylation buffer (1 mM ATP, 1 mM CTP, 1 mM GTP, 0.65 mM UTP, and 0.35 mM biotin-16-UTP [Roche]). After 2 h of incubation at 37°C, 5 U of RNase-free DNase (Roche) was added to remove the template DNA. The transcript RNAs were purified by phenol-chloroform extraction and ethanol precipitation. RNA pull-down experiments were performed with purified recombinant proteins [500 ng of PCBP1, 500 ng of PCBP2, and 250 ng of 3CDpro(3Cpro/H40A)] or 800 μg of HeLa cell S10 cytoplasmic extracts and 4 μg of biotinylated RNAs corresponding to PV1(M) RNAs (nt 1 to 91, 1 to 92, 1 to 93, 1 to 94, 1 to 95, 1 to 100, and 1 to 142) and to mutants RNAs (nt 1 to 142), as indicated. After incubation of the RNA-protein mixture in 1 ml of incubation buffer (10 mM HEPES [pH 7.4], 1.5 mM magnesium acetate, 90 mM potassium acetate, 2.5 mM dithiothreitol, 0.05% NP-40) for 30 min at 4°C, the samples were subjected to streptavidin-agarose resin (Pierce) adsorption and further incubated for 2 h. As nonspecific competitors, 20 μg of yeast tRNA (Roche) was added to the binding mixtures. After incubation, the resin was washed four times with incubation buffer, and then the resin-bound proteins were resolved by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis. Western blot analysis was performed with either anti-PCBP2 polyclonal antiserum or anti-3D monoclonal antibody.
TABLE 2.
Oligonucleotides used in making probes for RNA binding assays
| Primera | Sequence |
|---|---|
| T7-CL (F) | 5′-GCGAAATTAATACGACTCACTATAGGTTAAAAC-3′ |
| 91 (R) | 5′-GTATAAAACAGGCGTACAAG-3′ |
| 92 (R) | 5′-AGTATAAAACAGGCGTACAA-3′ |
| 93 (R) | 5′-GAGTATAAAACAGGCGTACA-3′ |
| 94 (R) | 5′-GGAGTATAAAACAGGCGTAC-3′ |
| 95 (R) | 5′-GGGAGTATAAAACAGGCGTAC-3′ |
| 100 (R) | 5′-GGGAAGGGAGTATAAAACAG-3′ |
| 142 (R) | 5′-GTACCCCCTTCTATTGAACTTGG-3′ |
F, forward; R, reverse.
RESULTS
A C-rich region between the CL and the spacer of PV RNA is required for viral growth.
Our previous studies have indicated that an A at position 103 in the spacer region of the PV 5′NTR between the CL and the IRES (Fig. 1B) is important for the replication of PV in the central nervous systems of PV receptor transgenic mice (CD155 transgenic mice), for replication in human neuronal cells at 39.5°C, and for the in vitro translation of the mutant RNA in SK-N-MC cell extracts (8). These results suggested the possibility that other nucleotides in the spacer region also have hitherto unknown functions either in translation or in replication of the viral genome. In the present study, we focused on a C-rich region in the spacer (nt 93 to 100) that consists of two clusters of three consecutive C residues (C93-95 [group I] and C98-100 [group II]) separated by two U residues (Fig. 1B). The two C clusters are almost fully conserved in the corresponding spacers of all of the enteroviruses and rhinoviruses that we analyzed (Fig. 1C). Entero- and rhinoviruses, whose genomes possess very similar 5′-terminal structures, make up two separate genera of the family Picornaviridae.
The experiments were initiated with mutant PV cDNAs containing either C93-95A or C98-100A substitutions in the spacer region. Mutant RNA transcripts derived from these cDNAs were transfected into HeLa cells, and the time was determined at which CPEs developed. Wild-type (WT) PV1(M) RNA transcripts were transfected into cells in parallel control experiments. As shown in Fig. 2, mutant M1, containing the C93-95A substitutions, developed a CPE at 48 h postinfection, 24 h after a CPE developed with WT transcripts. The replication properties of mutant M2 transcripts (C98-100A), on the other hand, were the same as that of the WT (Fig. 2). In contrast, the replacement of all six C residues in group I and group II with A residues (M3) resulted in no CPE, even after six blind passages of the cell supernatants, an observation suggesting a lethal viral phenotype (Fig. 2). These results indicated that three of the six C residues are sufficient for viral growth but that the three C residues of group I are more important in promoting growth than those C residues of group II.
We then analyzed in more detail the function of each C in group I and group II. Since the replacement of all six C residues in groups I and II (M3) yielded a dead phenotype, we chose to change back each one of these A residues to a C, hoping to recover replication and observe the possible emergence of revertants for further analyses.
When only one A residue was changed back in group II, the variants still expressed a lethal phenotype, regardless of the position of the A residue within the cluster (Fig. 2, M4 to M6). In contrast, a single A-to-C transversion in group I led to the recovery of replication (M7 to M9) even though the CPE was greatly delayed with mutants M7 and M8 (72 h). Mutant M9 was quasi-infectious, requiring two blind passages for virus to emerge. Sequence analyses of the emerging viral genomes revealed that, compared to the parental sequences (M7 to M9), all had undergone genetic variation, as expected. Whereas in MR7 and MR8 a single A residue in group I changed back to a C residue, the MR9 variant had inserted an extra C residue (Fig. 2). Thus, in all three revertants two consecutive C residues were regenerated, an observation indicating a minimum requirement for an adjacent C pair in group I for viral growth.
In the following experiments, we changed back one additional A to a C so that two adjacent C residues, flanked by an A residue, were located in either group I or group II. These experiments directly confirmed our previous conclusion that the presence of two consecutive C residues in group I (M10) is sufficient for the production of virus. M11 also replicated with the same phenotype as the WT virus (Fig. 2). Interestingly, the position of the two C residues within group I was less efficient in M10 than in M11, as M10 RNA yielded variant MR10, in which the A at nt 95 was changed back to a C and, hence, to a C triplet (Fig. 2). In contrast, two adjacent C residues, located in group II (M13, M14), resulted in a CPE only after blind passages (Fig. 2). The progeny contained either a deletion (MR14) or a transition and a deletion (MR13). Remarkably, these changes regenerated two adjacent C residues at positions 94 and 95 (MR13, MR14), the preferred site of the C doublet in group I (Fig. 2).
We were surprised to find that the lethal phenotype of the M3 mutant can be rescued by the genetically engineered replacement of U96-97 with C residues (M12) (Fig. 2). Although M12 is quasi-infectious, we were able to isolate a revertant that contained an A-to-C transversion at nt 95 (MR12). This indicates a stringent requirement for a C residue at this position. The last two mutants in this set contained three A residues in either group I or group II and another A in the center of the second group, disrupting the C triplet (M15, M16, Fig. 2). The MR15 variant derived from M15 emerged after a delayed CPE. It contained the reversion A94C (MR15) that reestablished the C triplet in group I. The M16 construct was quasi-infectious and required three blind passages to yield MR16. This variant had acquired two adjacent C residues at positions 95 and 96 (MR16) (Fig. 2).
The major conclusions of these mutational analyses are that the C-rich region of the spacer has a function in viral growth and that the C residues in group I are more important than those of group II. Two adjacent C residues (C94 and C95) in group I of the spacer are sufficient to support what appears to be normal WT viral growth under the conditions of the experiment, and the position of these two C residues is preferred over position C93 C94. On the other hand, the presence of such two C residues in group II results in highly impaired growth phenotypes.
Replacement of the six C residues with A residues in the spacer has no effect on translation, but it leads to a defect in RNA replication.
As we have discussed above, the replacement of the two triplets of C residues with A residues in the spacer (M3) abolishes virus growth. To determine at which stage of the viral life cycle these mutations exert their effect, we have carried out in vitro translations in HeLa cell extracts of full-length WT and mutant PV RNAs. As shown on Fig. 3A, translation of M3 mutant RNA and the processing of the resulting polyprotein are essentially the same (lane 4) as those of WT PV RNA (lane 1). Similarly, the A93-95 (M1) and A98-100 (M2) mutant RNAs exhibit the same translation and processing profiles as the WT RNA (Fig. 3A, lanes 2 and 3). In vitro translations in HeLa cell extract are excellent indicators of the quality of the viral RNA template, as well as of translation and of proteolytic processing (21).
FIG. 3.
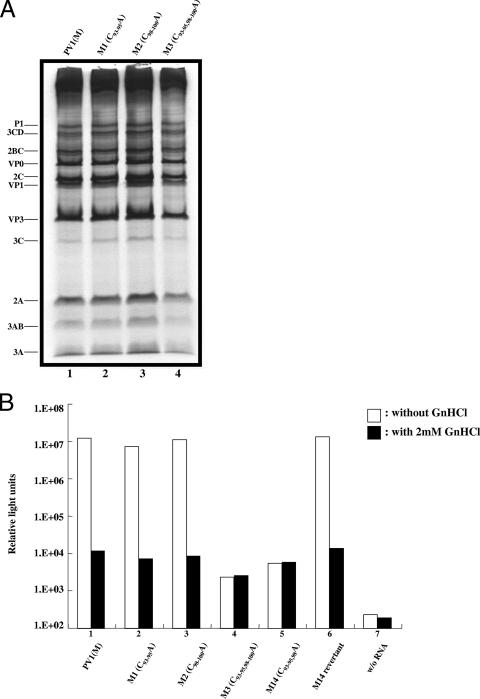
Analysis of translation and replication of WT and mutant PV RNAs. (A) In vitro translation of full-length transcript RNAs derived from PV1(M) and its derivates M1 (C93-95A), M2 (C98-100A), and M3 (C93-95, 98-100A). Reaction mixtures containing in vitro-transcribed mutant and WT PV RNAs (250 ng) were translated in a HeLa cell S10 cytoplasmic extract (see Materials and Methods). (B) Firefly luciferase activities of various PV-Luc replicons with or without (w/o) 2 mM GnHCl. Transfection of WT and mutant RNAs and the measurement of luciferase activities are described in Materials and Methods.
Since the group I and II C-to-A mutations in the spacer do not affect translation in vitro, we next examined their influence directly on RNA replication. We used luciferase replicons for this purpose in which the P1 domain of the polyprotein was replaced by the coding sequences of the firefly luciferase gene (19). Mutant RNAs were transfected into HeLa cells, and 12 h after transfection the luciferase activity was measured. The transfections were done both in the absence and in the presence of 2 mM guanidine hydrochloride (GnHCl), a potent inhibitor of PV RNA replication (35). The residual luciferase activity obtained in cultures with GnHCl represents translation of the input RNAs. As shown in Fig. 3B, with WT RNA there was a 1,000-fold increase in the luciferase signal when GnHCl was omitted from the culture, an observation indicating robust plus strand RNA synthesis under these conditions. In contrast, the M3 mutant RNA exhibited the same luciferase activity, both in the absence and in the presence of the drug, indicating a severe defect in RNA replication (Fig. 3B, lane 4). In contrast, the M1 and M2 mutant RNAs yielded luciferase signals comparable to that of WT PV RNA (lanes 2 and 3, respectively). The efficient replication of the M1 mutant under the conditions of the experiment was surprising considering the delay in a complete CPE reported in Fig. 2. However, the luciferase assays were performed at 12 h posttransfection. Apparently, at that time the mutant RNA had sufficient time to catch up with WT RNA in the single-cycle replication experiment. Indeed, when the luciferase activity was assayed at 6 h posttransfection, we did see a twofold lower signal with M1 RNA than with M2 or WT RNA, an observation suggesting a delayed replication cycle (data not shown). The phenotypes of M14 and MR14 will be discussed below. Together, these data indicate that the C-rich region is required for RNA replication.
Mutations in the C-rich region of the spacer affect the binding of PCBP to the 5′-terminal 142 nt of PV RNA.
It has been reported that the cellular RNA binding protein PCBP2 binds to the PV CL (1, 2, 13, 24), where the predominant site of contact is believed to be a triplet of three C residues in stem-loop B (C23-25) (13, 24; Fig. 1B). The CL structure itself comprises nt 1 to 88 (Fig. 1B). However, previous analyses of PCBP-CL binding have always involved 5′-terminal RNA fragments that were longer than 88 nt (usually 108 nt long) and included the C-rich region of the spacer analyzed here (2, 11, 13, 24, 34). Since our studies implicated the C-rich region contained in the spacer in RNA replication and viral growth and because PCBP2 has an affinity for C-rich sequences, we were interested in determining whether the group I and/or group II C residues participate in PCBP2 binding. To test this possibility, we used a pull-down assay described by Kim et al. (17), in which biotinylated RNA, after binding to protein, is adsorbed to streptavidin-agarose resin, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analyses of the RNA-protein complexes. The RNA probes were transcript RNAs containing sequences of the CL (nt 1 to 88), of the CL plus various lengths of the spacer, or of the CL plus the spacer and a short segment of domain II (nt 1 to 142). The source of proteins was either a HeLa cell extract or purified PCBP2 or PCBP1.
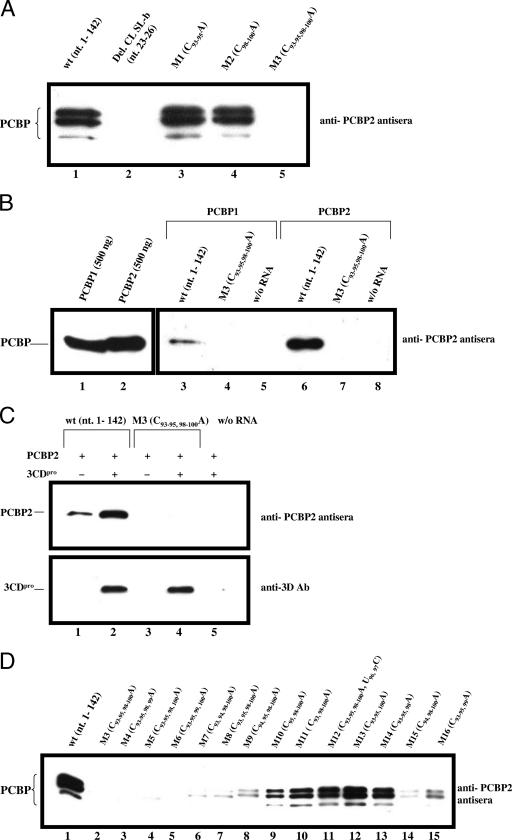
Figure 4A shows that in a pull-down assay with HeLa cell extracts and the WT RNA transcript (nt 1 to 142), two major bands and one minor band of PCBP were detected with anti-PCBP2 polyclonal antiserum (lane 1), an observation illustrating our probe's affinity for different PCBP subtypes. Walter et al. have previously shown that one of these PCBP bands is a doublet (33, 34). We obtained the same results with RNA probes containing either the M1 or the M2 mutation (Fig. 4A, lanes 3 and 4, respectively). In contrast, no PCBP binding was observed when all six C residues in group I and group II were replaced with A residues (M3), an observation indicating the importance of the C-rich region of the spacer in PCBP binding (Fig. 4A, lane 5), regardless of the presence of the C residues in stem-loop B of the CL. However, no PCBP binding was detected with the 142-nt RNA probe that contained a deletion of 4 nt in stem-loop B of the CL, which was previously shown to be required for the binding of this protein (Fig. 4A, lane 2) (13, 24). These results demonstrate that PCBP binding to the CL requires both stem-loop B of the CL and either the group I or group II C residues of the C-rich region from the spacer.
FIG. 4.
Analysis of PCBP binding to a 142-nt-long RNA fragment derived from the 5′ terminus of PV RNA. PCBP binding to WT and mutant RNA probes (nt 1 to 142) was analyzed with an RNA pull-down assay followed by Western analysis with anti-PCBP2 polyclonal antiserum (see Materials and Methods). (A) PCBP binding to WT and mutant RNA probes was measured with HeLa cell extracts. Nucleotides 23 to 26 (CCCA) were deleted from stem-loop B of the PV CL (Del. CL SL-b) (12, 13). (B) Binding of purified PCBP1 and PCBP2 to WT and mutant RNA probes (nt 1 to 142) (lanes 3 to 7). The binding of purified PCBP1 (500 ng) and PCBP2 (500 ng) to WT and mutant RNA probes was measured as described in Materials and Methods. Western blot analysis of 500 ng of PCBP1 and PCBP2 with anti-PCBP2 polyclonal antiserum (lanes 1 and 2). (C) Binding of PCBP2 to WT and mutant RNA probes in the absence and presence of 3CDpro. The binding of purified PCBP2 (500 ng) with or without (w/o) purified 3CDpro (250 ng) to WT and mutant RNA probes was measured (see Materials and Methods). Immunoblot analysis was performed by using either anti-PCBP2 polyclonal antiserum or anti-3D monoclonal antibody (Ab). Symbols: −, no protein added; +, protein added. (D) Binding of PCBP to WT and mutant RNA probes. The RNA binding experiments were performed with HeLa cell S10 cytoplasmic extracts and WT and mutant RNA probes (nt 1 to 142).
To confirm the identity of PCBP as the cellular protein that interacted with the WT probe (Fig. 4A), we repeated the RNA pull-down assay with purified PCBP1 and PCBP2. After individual binding of the purified polypeptides to the WT RNA probes, we observed a single band emerging with anti-PCBP2 polyclonal antiserum, which we conclude is either PCBP1 or PCBP2 (Fig. 4B). As expected, the M3 mutant RNA, containing a C-to-A replacement of all six C residues, did not exhibit any detectable interaction with purified PCBP1 (Fig. 4B, compare lane 3 with lane 4) or PCBP2 (Fig. 4B, compare lane 6 with lane 7). The anti-PCBP2 polyclonal antiserum used for Western blotting is equally efficient at detecting PCBP1 and PCBP2 (Fig. 4B, compare lane 1 with lane 2). Interestingly, the binding of PCBP1 to the WT RNA probe is not as strong as the binding of PCBP2 (Fig. 4B, compare lane 3 with lane 6).
It has been suggested previously that the interaction of PCBP2 with the extended CL (nt 1 to 108) in vitro is stimulated by the addition of viral proteinase 3CDpro, an RNA binding protein (11). The two proteins are believed to form a complex in which PCBP2 binds to stem-loop B and 3CDpro to stem-loop D of the CL (1, 2, 13, 24, 28). We have confirmed these results with the WT probe (nt 1 to 142), purified PCBP2, and purified 3CDpro (Fig. 4C, compare lane 1 with lane 2), where the presence of 3CDpro in the RNA-protein complex was demonstrated with anti-3Dpol antibodies (Fig. 4C, lane 2). In contrast, our M3 mutant probe did not bind PCBP2 either in the absence or in the presence of 3CDpro (Fig. 4C, compare lane 1 with lanes 3 and 4). Notably, 3CDpro alone did bind to the M3 probe (Fig. 4C, lane 4). These results indicate that the C-rich region of the spacer is required for the binding of PCBP2 but not of 3CDpro and that the addition of 3CDpro to PCBP2 does not rescue the effect of the M3 mutations.
To analyze in more detail the importance of the group I and group II C residues in the binding of PCBP, we tested all of our spacer mutants with the RNA pull-down assay and the same RNA probe (nt 1 to 142). Our results indicate that the mutants can be divided into two groups. The larger group (13 out of the 16 tested) shows a very good correlation between the growth phenotype of the mutants (Fig. 2) and their ability to bind PCBP (Fig. 4D). Specifically, the mutations leading to a lethal phenotype (M3 to M6) allow no detectable PCBP binding when present in the RNA probe (Fig. 4D, lanes 2 to 5). On the other hand, M11, which expresses a normal growth phenotype (Fig. 2), binds PCBP like the WT (Fig. 4D, compare lane 1 and lane 10). The mutants that possess a quasi-infectious growth phenotype (M7 to M10, M15, and M16) exhibit detectable but weaker-than-normal binding of the protein to the probe (Fig. 4D, compare lane 1 with lanes 6 to 9 and lanes 14 and 15).
Unexpectedly, a second and smaller group of mutants (M12 to M14) exhibited efficient binding of PCBP (Fig. 4D, compare lane 1 with lanes 11 to 13) but they were quasi-infectious, that is, severely impaired in growth (Fig. 2). Therefore, these results suggested the possibility that although these mutant RNAs were able to bind PCBP, this interaction did not promote efficient RNA replication. To test this hypothesis, we analyzed mutant M14 in the background of a luciferase replicon (Fig. 3B). As expected, the mutation reduced RNA replication to levels below detection by this assay (Fig. 3B, lane 5) but its variant MR14 that was isolated after three blind passages (Fig. 2) fully regained the ability to replicate (Fig. 3B, lane 6). The binding of PCBP to MR14 RNA was the same as that to the M14 RNA (data not shown). A closer examination of M14 RNA reveals the presence of two adjacent C residues located in the group II position. In the corresponding revertant, MR14, these two C residues have been moved into the group I position by deletion of 5 nt. It appears, therefore, that two adjacent C residues in group II at nt 99 and 100 (M14) do allow PCBP binding to the 5′-terminal fragment but this binding is inadequate for efficient RNA replication to occur. A very similar relationship exists between M13 and its variant MR13. Here, the virus introduced a C residue at position 97 by transition and then deleted 3 nt to move two C residues into the group I position (MR13, Fig. 2). A triplet of C residues is more favorable for PCBP binding, and hence, the position effect is less stringent. Therefore, the quasi-infectious M12 RNA introduced a C residue into nt 95 by transversion, thereby generating a C triplet (MR12, Fig. 2), which allows adequate RNA replication.
These data led us to conclude that there is an important functional difference between the C residues of groups I and II. Although two adjacent C residues in either group I or group II are sufficient for binding of PCBP, these same C residues in group II are not able to promote efficient RNA replication, hence the quasi-infectious nature of the viral mutant RNAs. In addition, data derived from our genetic analyses and biochemical PCBP binding studies suggest that, in vivo, the PCBP binding site that is preferred for RNA replication is located in group I, particularly at nucleotides C94 and C95. As we have noted above, however, a full triplet of C residues weakens the position effect. Accordingly, the single C triplet in group II (M1) is nearly as efficient in both PCBP binding and RNA replication as the single triplet of C residues in group I (M2). It is possible that long-term passage of M1 would regenerate adjacent C residues in the group I position. This experiment, however, has not been carried out.
The binding of PCBP2 to the CL requires nucleotides from the C-rich region of the spacer.
Since our results have indicated the importance of both stem-loop B of the CL and the C-rich region of the spacer in PCBP binding to the PV RNA 5′ terminus (nt 1 to 142), we were interested in determining the minimal length of the spacer RNA that is required for binding. Although the actual length of the CL is only 88 nt, previous studies, which showed PCBP binding to the CL, always used a 108-nt-long RNA segment (2, 11, 13, 24, 34). With HeLa cell extracts and 5′-terminal RNA fragments 91, 92, or 93 nt long, we observed no detectable binding of the protein to the probe (Fig. 5, lanes 1, 2 and 3, respectively). In contrast, normal PCBP binding was observed with RNAs 95 nt long or longer, which contained at least the first triplet of C residues (C93-95) (Fig. 5, lanes 5 and 6). This finding shows that 95 nt are required for optimal PCBP binding, although weak binding can already be detected with a 94-nt-long probe.
FIG. 5.
PCBP binding to the CL requires sequences from the spacer region. PCBP binding was analyzed with WT RNA probes of different lengths (nt 1 to 91 to nt 1 to 100) and HeLa cell extracts. Western blotting analysis was performed with anti-PCBP2 polyclonal antiserum (see Materials and Methods). w/o, without.
DISCUSSION
The experiments reported here were undertaken to determine whether the spacer (nt 89 to 123) between the CL and IRES of PV RNA is harboring hitherto unknown functions in either protein translation or RNA replication. Our previous studies have indicated that two nucleotide changes (A102G and A103G) in the spacer region of the synthetic PV sPV1(M) strongly attenuated neurovirulence in CD155 transgenic mice (7). The A102G and A103G mutations also caused a significant temperature-sensitive phenotype in human neuronal cells (SK-N-MC), and they reduced the efficiency of translation reactions in vitro in SK-N-MC cell extracts (8). Subsequent studies indicated that a single G103A reversion either in sPV1(M) or in an experimentally generated PV1(M) A102G/A103G variant was sufficient to restore all of the defective phenotypes to normal (8). These observations suggested the possibility that other regions of the spacer might also have functions at some stage of viral growth. A nucleotide alignment of the spacer region of many entero- and rhinoviruses has revealed two highly conserved triplets of C residues (C93-95 and C98-100) separated by two pyrimidines (U96-97) (Fig. 1C) that we have designated group I and group II triplets. We have selected these conspicuous genetic elements for our genetic and biochemical analyses.
Genetics of the C-rich domain in the spacer region.
The construction and analyses of 16 different genetic variants in the C-rich region of the spacer have allowed us to divide the mutant genomes into three phenotypes, replication competent, quasi-infectious, and dead. Specifically, our results suggest that the presence of three consecutive C residues in either group I (M2) or group II (M1) of the spacer is sufficient for the production of virus with growth properties similar, but not identical, to those of the WT virus. A comparison of the growth phenotypes of M1 and M2 hinted at the possibility of a position effect of the C triplet in replication (Fig. 2).
RNA genomes with only a CC duplet in the group I position (M10, M11) replicated either immediately with WT kinetics (M11) or proceeded to rapidly exchange an A residue for a C residue in group I, thereby creating a C triplet (MR10). In stark contrast, replication of genomes M13 and M14 with two consecutive C residues in group II is severely obstructed (quasi-infectious). Significantly, variants isolated from M13 or M14 RNA-infected cells managed to establish two adjacent C residues in group I (MR13 and M14, respectively). This position effect is also apparent in mutants that were constructed to contain only a single C residue in either group I or group II (M4 to M9). Whereas mutant genomes M4 to M6 expressed a dead phenotype, quasi-infectious genomes M7 to M9 allowed sufficient replication for the selection of variants with a C duplet in group I (MR7 to MR9).
These data support a rule that, if presented with a choice, the virus strongly prefers C triplets or C duplets in the group I position rather than in the group II position. The rule is slightly violated in MR12 and MR16, where the duplet is moved to the 95-96 position, 1 nt out of group I (Fig. 2). We have not yet performed long-term passage experiments with the variants listed in Fig. 2. But we predict that eventually new variants would emerge from cells infected with MR12 or MR16 carrying a CC duplet in the group I position.
It should be noted that drastic sequence changes in the spacer downstream of the C clusters do not appear to significantly influence PV replication. For example, the PV cre element (63 nt) has been inserted into the spacer at nt 103, where it can rescue a lethal mutation in endogenous 2C(cre) (Fig. 1A) (37). PV with cre in the spacer (which is temperature sensitive, just like the A103G virus) has been found to be highly attenuated in CD155 transgenic mice; it has recently been successfully used to cure immunocompetent CD155 transgenic mice carrying a lethal neuroblastoma (30).
Effect of spacer mutagenesis on translation and RNA replication.
To determine at which stage of viral growth the mutations exert their effect, we have first compared the translation of WT and mutant PV RNAs in HeLa cell extracts. There were no significant differences in either the translation efficiency or the processing of the polypeptides between the WT and mutant RNAs (Fig. 3A), an observation suggesting that the translation of these viral constructs that we consider representative of all mutants, is normal. We next examined the effect of the mutations directly on RNA replication in HeLa cells by using a luciferase replicon (19). Our results clearly indicated that the mutations in the C-rich region of the spacer affect RNA replication. The lethal M3 mutation caused a total block in RNA replication, and the M1 mutation caused a retardation of RNA replication when the luciferase signal was assayed at 6 h posttransfection. On the other hand, the viable M2 mutant exhibited luciferase activities comparable to those of the WT.
Binding of the CL-spacer sequence to PCBP.
The discovery of PCBP as a cellular RNA binding protein involved in two major steps in PV proliferation has advanced our understanding of the interplay between PV genetic elements and host factors. PCBP plays a decisive role in the regulation of IRES-mediated, cap-independent initiation of translation of the virus genome (5, 6, 13, 33). In this process, PCBP binds to domain IV of the IRES (Fig. 1A) and, as recently reported, it collaborates in translation regulation with SRp20, another cellular protein and a member of the SR protein family (4).
In addition to IRES binding, PCBP also has an affinity for the C-rich region in stem-loop B of the 5′-terminal CL (1, 2, 13, 34). However, we report here that this affinity is not sufficient to mediate binding of the cellular protein to the PV genome in this region. Rather, an additional C-rich site, located in the spacer region just downstream of the CL, is required for PCBP binding, regardless of the isotype of the cellular RNA binding protein (PCBP1 or PCBP2). Binding of PCBP to this second C-rich site is essential for viral proliferation.
Specifically, we have tested spacer mutants described above to determine the effects of different nucleotide changes (C to A) on PCBP binding. These experiments revealed that the presence of only a single C in group I and group II resulted in either poor binding of PCBP or no detectable binding, respectively. In contrast, the presence of two or three consecutive C residues in either group I or group II was found to be sufficient for efficient PCBP binding. When the PCBP binding and viral proliferation abilities of the mutants were compared, they could be divided into two groups. With the larger group of mutants (M1 to -11, M15, and M16), there was a very good correlation between PCBP binding and viral growth. A smaller group of mutants (M12 to M14), on the other hand, interacted with PCBP very efficiently but exhibited a highly impaired growth phenotype. These results suggested that the PCBP/RNA complex formed with these mutants functioned only poorly in RNA replication. We confirmed this hypothesis by using one of these mutants (M14) with a luciferase replicon. The mutant was found to be defective in RNA replication as assayed by the luciferase signal (the mutant is quasi-infectious, and it required three blind passages for recovery of the viable variant MR14; Fig. 2), while its progeny virus MR14 regained the ability to replicate.
As mentioned, it has been previously shown by different investigators that PCBP2 binds to stem-loop B of the CL, most likely to the triplet of C residues at nt 23 to 25 (13, 24). Although the CL consists of only 88 nt, the RNA probes used in the previous studies of PCBP binding to CL were always 108 nt long (nt 1 to 108) (2, 11, 13, 24, 34). However, when we tested the CL structure alone (nt 1 to 91), no PCBP binding was observed (Fig. 5, lane 5). For efficient binding to occur, a minimum length of 95 nt from the 5′ terminus is required (Fig. 5, lane 2). On the other hand, our probe of 142 nt failed to bind PCBP when the binding site in stem-loop B of the CL was deleted (Fig. 4A, lane 2). These data strongly suggest that the binding sites in both the CL and the spacer are required for PCBP binding and that they collaborate together.
PCBP, a polypeptide of 36 kDa, contains three hnRNP KH RNA binding domains. Experimental evidence suggests that only KH1 is essential for RNA replication (and translation), while the KH2 and KH3 domains are dispensable for initiation of RNA synthesis to occur (29, 34). The requirement for two intact PCBP binding sites in the CL and the adjacent spacer and the robust “position effect” of the PCBP binding site downstream of the CL indicate a spacing restriction for the protein-RNA interaction. For this reason, we consider it unlikely that this spacer function could be transposed to another locus in the genome (e.g., into the spacer between the IRES and the open reading frame) while the original sequence downstream of the CL would be inactivated for PCBP binding. However, such experiments remain to be carried out.
Since it was previously demonstrated that PCBP forms an RNP complex with the CL and protein 3CDpro (1, 2, 24), we examined the effect of the spacer mutations on the binding of 3CDpro. As expected, we observed an enhancement of PCBP binding in the presence of 3CDpro. However, a spacer mutation that ablated PCBP binding had no effect on the interaction between 3CDpro and the 142-nt-long RNA. The three-dimensional structure of the enterovirus CL is unknown. It is likely, however, that an interaction of PCBP, of 3CDpro, or of PCBP/3CDpro changes the three-dimensional structure of the CL in a specific manner. The details of the interactions among proteins, the CL, and the spacer (stoichiometry, succession of binding) remain to be elucidated.
Recently, Bailey and Tapprich have presented a refined secondary structure of the 5′-terminal sequence (CL-spacer-IRES) of coxsackievirus B3 (3). Although a long-range pairing interaction between sequences of domain II and domain V has been uncovered, the spacer region between the CL and the first base-paired region, containing the C region, remained available for PCBP binding.
PV, just like all RNA viruses, replicates under conditions of genetic austerity because it has chosen to exist without proofreading and editing functions during RNA replication, and hence, its genome is small (35). Since the PV genome is densely packed with genetic information, we should not be surprised that the sequence between the CL and the IRES has an essential function(s) in viral proliferation. In this regard, the term “spacer” for this sequence is a misnomer.
Acknowledgments
We thank Bert Semler for the clone of PCBP2 and also for anti-PCBP2 polyclonal antiserum.
Hidemi Toyoda was the recipient of a scholarship from the Pediatric Oncology Research Foundation (Japan). This work was supported by NIH grant AI5122.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, J. M., and W. E. Tapprich. 2007. Structure of the 5′ nontranslated region of the coxsackievirus B3 genome: chemical modification and comparative sequence analysis. J. Virol. 81:650-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedard, K. M., S. Daijogo, and B. L. Semler. 2007. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 26:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cello, J., A. V. Paul, and E. Wimmer. 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297:1016-1018. [DOI] [PubMed] [Google Scholar]
- 8.De Jesus, N., D. Franco, A. Paul, E. Wimmer, and J. Cello. 2005. Mutation of a single conserved nucleotide between the cloverleaf and internal ribosome entry site attenuates poliovirus neurovirulence. J. Virol. 79:14235-14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejgaard, K., and H. Leffers. 1996. Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur. J. Biochem. 241:425-431. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenfeld, E., and N. L. Teterina 2002. Initiation of translation of picornavirus RNAs: structure and function of the internal ribosome entry site, p. 159-169. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 11.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 14.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 15.Holcik, M., and S. A. Liebhaber. 1997. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. USA 94:2410-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, J. H., B. Hahm, Y. K. Kim, M. Choi, and S. K. Jang. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298:395-405. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. H., K. Y. Paek, K. Choi, T. D. Kim, B. Hahm, K. T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23:708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen, G. R., B. L. Semler, and E. Wimmer. 1981. Stable hairpin structure within the 5′-terminal 85 nucleotides of poliovirus RNA. J. Virol. 37:328-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X., H. H. Lu, S. Mueller, and E. Wimmer. 2001. The C-terminal residues of poliovirus proteinase 2A(pro) are critical for viral RNA replication but not for cis- or trans-proteolytic cleavage. J. Gen. Virol. 82:397-408. [DOI] [PubMed] [Google Scholar]
- 20.Makeyev, A. V., and S. A. Liebhaber. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 22.Murray, K. E., A. W. Roberts, and D. J. Barton. 2001. Poly(rC) binding proteins mediate poliovirus mRNA stability. RNA 7:1126-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostareck-Lederer, A., D. H. Ostareck, and M. W. Hentze. 1998. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 23:409-411. [DOI] [PubMed] [Google Scholar]
- 24.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 25.Paul, A. V., J. Mugavero, A. Molla, and E. Wimmer. 1998. Internal ribosomal entry site scanning of the poliovirus polyprotein: implications for proteolytic processing. Virology 250:241-253. [DOI] [PubMed] [Google Scholar]
- 26.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 8:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rieder, E., W. Xiang, A. Paul, and E. Wimmer. 2003. Analysis of the cloverleaf element in a human rhinovirus type 14/poliovirus chimera: correlation of subdomain D structure, ternary protein complex formation and virus replication. J. Gen. Virol. 84:2203-2216. [DOI] [PubMed] [Google Scholar]
- 29.Silvera, D., A. V. Gamarnik, and R. Andino. 1999. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem. 274:38163-38170. [DOI] [PubMed] [Google Scholar]
- 30.Toyoda, H., J. Yin, S. Mueller, E. Wimmer, and J. Cello. 2007. Oncolytic treatment and cure of neuroblastoma by a novel attenuated poliovirus in a novel poliovirus-susceptible animal model. Cancer Res. 67:2857-2864. [DOI] [PubMed] [Google Scholar]
- 31.Trono, D., J. Pelletier, N. Sonenberg, and D. Baltimore. 1988. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science 241:445-448. [DOI] [PubMed] [Google Scholar]
- 32.van der Werf, S., J. Bradley, E. Wimmer, F. W. Studier, and J. J. Dunn. 1986. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter, B. L., J. H. Nguyen, E. Ehrenfeld, and B. L. Semler. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 36.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin, J., A. V. Paul, E. Wimmer, and E. Rieder. 2003. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: analysis of single- and dual cre viral genomes and proteins that bind specifically to PV-cre RNA. J. Virol. 77:5152-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]