Abstract
The phosphatidylinositol 3-kinase (PI3K)-protein kinase B (Akt) signaling pathway is one of the major oncogenic pathways and is activated in many types of human cancers, including hepatocellular carcinoma. It can also be activated by the hepatitis C virus (HCV) nonstructural 5A (NS5A) protein. In the present study, we set out to determine the regulatory effects of this pathway on the replication of hepatitis B virus (HBV). Our results demonstrate that the expression of a constitutively active Akt1 profoundly inhibited HBV RNA transcription and consequently reduced HBV DNA replication in HepG2 cells. This suppression of HBV gene transcription was apparently mediated by the activation of mTOR, as it was abolished by the mTOR inhibitor rapamycin. Moreover, treatment of HBV-expressing HepG2.2.15 cells with inhibitors of PI3K, Akt, and mTOR increased the transcription of 3.5-kb and 2.4-kb viral RNA as well as the replication of HBV DNA. This observation implies that the basal level activation of this pathway in HepG2 cells regulated HBV replication. Consistent with previous reports showing that the HCV NS5A protein could bind to the p85 subunit of PI3K and activate the PI3K-Akt signal transduction pathway, our results showed that expression of this protein could inhibit HBV RNA transcription and reduce HBV DNA replication in HepG2 cells. Taken together, our results suggest that the activation of the PI3K-Akt pathway during liver oncogenesis may be at least partially responsible for the elimination of HBV replication from tumor cells and may also provide an explanation for the observed suppression of HBV replication by HCV coinfection.
Hepatitis B virus (HBV) infection is a significant public health problem affecting an estimated 400 million individuals worldwide (28). Patients who are chronically infected with HBV have an increased risk of morbidity and mortality from cirrhosis and primary hepatocellular carcinoma (HCC) (32). The treatments currently approved for chronic hepatitis B patients, including alpha interferon and four nucleoside analogues that inhibit viral DNA polymerase, are limited by low rates of sustained response, side effects, and the emergence of drug resistance (30).
HBV is the prototype virus of the family Hepadnaviridae and infects primarily hepatocytes. It is a small-DNA virus that contains a relaxed circular (rc) partially double-stranded DNA genome (49). Unlike other mammalian DNA viruses, HBV replicates via reverse transcription of its pregenomic (pg) RNA (48). In marked contrast to retroviruses, HBV genomic DNA integration into host cellular chromosomes is not an essential step in its life cycle. Instead, upon infection, incoming viral rcDNA is transported into the nucleus of the hepatocyte and converted into episomal covalently closed circular (ccc)DNA, which serves as the template for the transcription of viral RNAs. The viral pgRNA is translated to produce both the core protein and the reverse transcriptase (RT) (48). The RT protein binds to the epsilon sequence within the pgRNA to prime viral DNA synthesis and initiate nucleocapsid assembly (52, 53). Subsequently, the viral polymerase converts the pgRNA into rcDNA. The nucleocapsids mature as rcDNA is formed and can either be enveloped and secreted out of cells or deliver their rcDNA into the nucleus to amplify nuclear cccDNA (14, 39, 56).
HBV replication is regulated by many extra- and intracellular factors. For example, it has been known for a long time that HBV replication is cell density and/or cell cycle dependent (34, 40, 59). Specific hormones and inflammatory cytokines have been shown to modulate the virus replication in cultured cells and in vivo (15, 17, 20, 21, 35). Furthermore, HBV replication also appears to be influenced by certain pathological conditions. For example, coinfections with other hepatitis viruses, such as hepatitis A virus, hepatitis C virus (HCV), and hepatitis D virus, suppress HBV replication (41, 50, 55). Interestingly, despite the existence of integrated HBV DNA in the majority of HBV-related hepatocellular carcinomas, the replicative forms of HBV DNA are usually negative in most tumor cells. Consistent with this observation, most HCC-derived cell lines do not support HBV replication upon transfection of a wild-type HBV genome (42, 47, 54). Moreover, studies done with woodchuck hepatitis virus-infected woodchucks demonstrated that woodchuck hepatitis virus replication was largely eliminated in precancerous nodules (58). Those observations suggest that the activation of certain cellular oncogenic pathways can inhibit HBV replication. However, the molecular mechanisms by which these extracellular and intracellular factors regulate HBV replication remain largely unknown. A detailed understanding of HBV replication and its regulation by these factors would advance our knowledge of viral pathogenesis and could provide clues for the development of novel therapeutics.
The phosphatidylinositol 3-kinase(PI3K)-protein kinase B (Akt) signal transduction pathway is a crucial regulator of a number of cellular processes, including proliferation, differentiation, and cell survival. Activation of this pathway has been documented as a frequent occurrence in many types of human cancer (22, 51). A recent comprehensive microarray study with a large number of human hepatocellular carcinomas revealed that the activation of Akt1 is one of the most consistent features of HBV-induced HCC (7). Moreover, the PI3K-Akt pathway is activated during infection by many viruses, including HCV (45, 46). It is generally believed that the activation of the PI3K-Akt pathway by viruses inhibits apoptosis and promotes the survival of infected cells to favor viral replication (11). It is, therefore, interesting to know if the PI3K-Akt pathway regulates HBV replication. Our results demonstrate that the activation of this pathway inhibits HBV RNA transcription and consequently reduces viral DNA replication. Those observations suggest a molecular basis for the observed inhibition of HBV replication during HCC oncogenesis and HCV coinfection.
MATERIALS AND METHODS
Cell culture and kinase inhibitors.
HepG2 cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium-F12 (DMEM/F12) medium (Invitrogen) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. HepG2.2.15 and HepAD38 cells were kindly provided by George Acs (Mt. Sinai Medical College, New York, NY) and Christoph Seeger (Fox Chase Cancer Center, Philadelphia, PA), respectively. These two HBV-expressing stable cell lines were cultured in the same way as HepG2, except for the addition of G418 at 400 μg/ml (1). Where needed, tetracycline was routinely added at 1 μg/ml to suppress HBV pgRNA transcription in HepAD38 cells (26). Huh7.5 cells were kindly provided by Charles M. Rice (Rockefeller University, New York, NY) and maintained as previously described (4). Protein kinase inhibitor LY294002, Akti-1 and Akti-2 (Akti1/2), and rapamycin were purchased from Calbiochem.
Plasmid construction.
To construct the HBV plasmid pHBV1.3, HBV EcoRI monomer was PCR amplified with the primer pairs 5′-TTG TGG GTC TTT TGG GTT TTG CTG-3′ (nucleotides 1000 to 1023; GenBank accession no. V01460) and 5′-TCT CAT TAA CTG TGA GTG GGC CTA-3′ (nucleotides 2618 to 2595). The PCR fragment was then digested with BspEI. The resulting 1.3-kb HBV DNA was gel purified and inserted into the SmaI- and BspEI-restricted pCMVHBV vector. The plasmids expressing the wild-type Akt1 (pcDNA3Akt1), the dominant negative form of Akt1 (pCMVAkt1aa), and the constitutively active form of Akt1 (pCMVmyr-Akt1) have been described previously (57). Plasmids expressing the HCV nonstructural 5A (NS5A) protein were obtained from Christoph Seeger (Fox Chase Cancer Center, Philadelphia, PA).
Cell transfection.
HepG2 cells were seeded into 35-mm-diameter dishes at a density of 1.2 × 106 cells per dish in antibiotic-free complete DMEM/F12 medium. After 6 h, each well was transfected with a total of 4 μg of plasmid with Lipofectamine 2000 (Invitrogen) by following the manufacturer's directions. Transfected cells were harvested at the indicated times. The levels of HBV mRNA and core associated DNA were determined by Northern and Southern blot hybridization, respectively.
Nucleic acid analysis.
Intracellular viral core DNA was extracted as described previously (16). Briefly, cells from one 35-mm-diameter dish were lysed with 0.5 ml of lysis buffer containing 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% NP-40, and 2% sucrose at 37°C for 10 min. Cell debris and nuclei were removed by centrifugation, and the supernatant was mixed with 130 μl of 35% PEG-8000 containing 1.5 M NaCl. After 1 h of incubation in ice, viral nucleocapsids were pelleted by centrifugation at 12,000 × g for 10 min at 4°C, followed by 1 h of digestion at 37°C in 400 μl of digestion buffer containing 0.5 mg/ml pronase (Calbiochem), 0.5% sodium dodecyl sulfate (SDS), 150 mM NaCl, 25 mM Tris-HCl (pH 8.0), and 10 mM EDTA. The digestion mixture was extracted twice with phenol, and DNA was precipitated with ethanol, dissolved in Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). One-third of the DNA sample from each plate was resolved by electrophoresis into a 1.5% agarose gel. The gel was then subjected to denaturation in a solution containing 0.5 M NaOH and 1.5 M NaCl, followed by neutralization in a buffer containing 1 M Tris-HCl (pH 7.4) and 1.5 M NaCl. DNA was then blotted onto a Hybond-XL membrane (GE Health care) in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Total cellular RNA was extracted with TRIzol reagents (Invitrogen) by following the instructions of the manufacturer. Ten micrograms of total RNA was resolved in 1.5% agarose gel containing 2.2 M formaldehyde and transferred onto a Hybond-XL membrane in 20× SSC buffer.
Extraction of cccDNA was carried out by using a modified Hirt extraction procedure (23). Briefly, cells from one 35-mm-diameter dish were lysed in 3 ml of 10 mM Tris-HCl (pH 7.5), 10 mM EDTA, and 0.7% SDS. After 30 min of incubation at room temperature, the lysate was transferred into a 15-ml tube, followed by the addition of 0.8 ml of 5 M NaCl and incubation at 4°C overnight. The lysate was then clarified by centrifugation at 12,000 × g for 30 min at 4°C and extracted twice with phenol and once with phenol-chloroform. DNA was precipitated with 2 volumes of ethanol overnight at room temperature and dissolved in Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). One-third of the cccDNA sample was then denatured at 85°C for 5 min, separated in a 1.2% agarose gel, and transferred onto a Hybond-XL membrane.
For the detection of HBV DNA and RNA, membranes were probed with a full-length minus- or plus-strand-specific HBV riboprobe labeled with [α-32P]UTP (800 Ci/mmol; Perkin Elmer). Hybridization was carried out in 5 ml of EKONO hybridization buffer (Genotech) with 1 h of prehybridization at 65°C and overnight hybridization at 65°C, followed by a 1-h wash with 0.1× SSC and 0.1% SDS at 65°C. The membrane was exposed to a phosphorimager screen, and hybridization signals were quantified with QuantityOne software (Bio-Rad).
Western blotting analysis.
Transfected and/or drug-treated cells were washed once with phosphate-buffered saline (PBS) buffer, scraped, and suspended in PBS buffer containing 5 mM EDTA. Cells were pelleted by centrifugation at 3,000 × g for 5 min, and the pellets were suspended in a lysis buffer containing 10 mM Tris-HCl (pH 7.5), 10% glycerol and 1% NP-40, 1 mM sodium orthovanadate, and 1× protease inhibitor cocktail (Roche). Cell lysates were centrifuged at 10,000 × g for 10 min at 4°C. The supernatants were collected, and protein concentrations were determined by using a Bradford assay (Bio-Rad). One hundred micrograms of total protein per sample was electrophoresed on SDS-10% polyacrylamide gels and transferred onto polyvinylidene difluoride membrane (Bio-Rad). Membranes were blocked with PBS containing 0.1% Tween 20 and 5% nonfat dry milk and probed with antibodies against Akt1 (catalog no. 9272; Cell Signaling Technology), Ser473-phospho-Akt1 (catalog no. 9271; Cell Signaling Technology), HCV NS5A (a gift from Chen Liu, Florida State University, Jacksonville, FL), and β-actin (Chemicon International). Bound antibodies were revealed by horseradish peroxidase-labeled secondary antibodies and visualized with an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech) according to the protocol of the manufacturer.
Reporter assay.
HepG2 cells were seeded into 24-well plates at a density of 2 × 105 cells per well in antibiotic-free complete DMEM/F12 medium. After 6 h, each well was transfected with a total of 0.8 μg of plasmid containing 0.4 μg of pCMVRLuc (Promega), which expresses Renilla sp. luciferase under the control of a cytomegalovirus immediate-early (CMV IE) promoter, and 0.4 μg of the control plasmid pUC19 or a plasmid that expresses the wild-type, dominant negative or constitutively active forms of Akt1, respectively, with Lipofectamine 2000 (Invitrogen) by following the manufacturer's directions. Cells were lysed at 24 h after transfection, and luciferase activity levels in the cell lysates were determined with a luciferase assay kit (Promega).
RESULTS
Expression of constitutively active Akt1 inhibits HBV gene expression and replication.
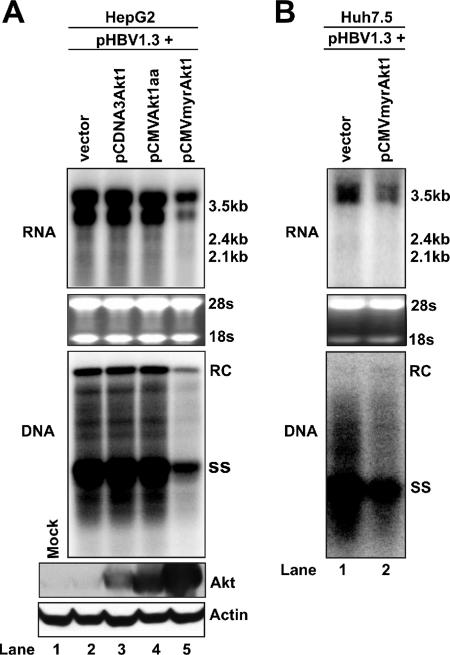
To investigate the possible regulatory effects of the PI3K-Akt signal transduction pathway on HBV replication, we cotransfected a plasmid containing a 1.3-fold-overlength HBV genome (pHBV1.3) with a plasmid that expressed the wild-type Akt1 (pcDNA3Akt1), the dominant negative Akt1 (pCMVAkt1aa), or the constitutively active Akt1 (pCMVmyrAkt1) form into HepG2 cells. The dominant negative Akt1 (Aktaa) form was created by replacing Thr-308 and Ser-473 with Ala residues and, therefore, cannot be activated by phosphorylation (57). The constitutively active Akt1 (myrAkt1) form was created by placing a myristoylation signal sequence at the N terminus of Akt1 protein. The resulting protein is myristoylated and therefore anchored to the plasma membrane to be constitutively active. Five days after transfection, total cellular RNA and cytoplasmic HBV core DNA were extracted, and viral RNA transcription and DNA replication were analyzed by Northern and Southern blot hybridizations, respectively. As shown in Fig. 1A, while expression of the wild-type Akt1 did not apparently affect the levels of HBV mRNA and HBV DNA replicative intermediates, the expression of myrAkt1 significantly reduced the levels of HBV mRNA and viral DNA.
FIG. 1.
Effects of Akt1 activation on HBV RNA transcription and DNA replication in hepatocyte-derived cell lines. (A) A plasmid that carries pgRNA of wild-type HBV (pHBV1.3) was cotransfected with the control plasmid pUC19 (lane 2) or plasmids that express the wild-type (lane 3), dominant negative (lane 4), and constitutively active Akt1 (lane 5) forms into HepG2 cells. In these transfection experiments, 2 μg of pHBV1.3 was cotransfected with 2 μg of vector DNA or Akt1 expression plasmids into a 35-mm dish of cells. Cells were harvested at day 5 after transfection and viral RNA transcription, and DNA replication was determined by Northern (upper panel) and Southern (middle panel) blot hybridization analyses, respectively. For RNA analysis, each lane was loaded with 10 μg of total RNA. Ribosomal RNAs (28S and 18S) were presented as loading controls. The positions of HBV 3.5-kb, 2.4-kb, and 2.1-kb RNAs are indicated. For DNA analysis, HBV core DNA was probed with a genome-length minus strand-specific HBV riboprobe. The positions of relaxed circular (RC) and single-stranded (SS) DNA are indicated. Expression of Akt1 by transfected plasmids was determined by Western blot analysis, and the levels of β-actin served as loading controls (lower panel). (B) Huh7.5 cells were cotransfected with 2 μg of pHBV1.3 and 2 μg of pUC19 or pCMVmyr-Akt1. Cells were harvested at day 5 after transfection, and viral RNA transcription and DNA replication were determined by Northern (upper panel) and Southern (lower panel) blot hybridization analyses as described above.
The expression of Akt1 protein in transfected cells was confirmed by Western blot analysis (Fig. 1A, lower panel). Interestingly, compared to the other forms of Akt proteins, myrAkt1 seemed to accumulate to a higher level in transfected cells, indicating that the mutant protein might have a longer half-life in cells. The overexpression of all three forms of Akt1 proteins was well tolerated; as determined by microscopy observation and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, no evidence of cytotoxicity was observed for any experimental set (data not shown).
To further determine the effect of constitutively active Akt1 expression on HBV RNA transcription and DNA replication in another hepatocyte-derived cell line, Huh7.5 cells were cotransfected with plasmids pHBV1.3 and pCMVmyrAkt1 or control plasmid pUC19, respectively. Five days after transfection, total cellular RNA and cytoplasmic HBV core DNA were extracted, and viral RNA transcription and DNA replication were analyzed as described above. As observed for HepG2 cells, the expression of constitutively active Akt1 significantly reduces the levels of both HBV RNA and capsid-associated HBV DNA in Huh7.5 cells (Fig. 1B).
To rule out the possibility that overexpression of constitutively active Akt1 nonspecifically decreases transcription, the plasmid pCMVRLuc, which expresses Renilla luciferase under the control of the CMV IE promoter, was cotransfected into HepG2 cells with the control plasmid pUC19 or a plasmid expressing the wild-type, the dominant negative, or the constitutively active form of Akt1. Twenty-four hours after transfection, cells were lysed and luciferase activity levels in the cell lysates were determined. The results demonstrated that the expression of all three forms of Akt1 did not significantly affect the CMV IE promoter-driven luciferase gene expression (data not shown).
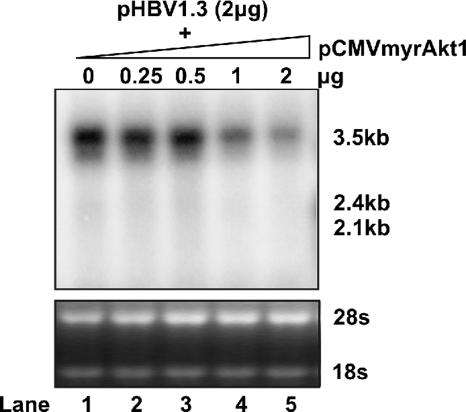
According to the replication cycle of HBV, our results suggest that the primary target of Akt1 activation in HBV replication is most likely the viral RNA transcription. The reduction of HBV DNA replication should be a secondary effect. To further evaluate the potency of myrAkt on HBV RNA transcription, we performed a dose-response experiment. HepG2 cells were cotransfected with 2 μg of plasmid pHBV1.3 and various amounts of plasmid pCMVmyrAkt1. Five days after transfection, total cellular RNA was extracted, and viral RNA levels were determined by Northern blot analysis. The results showed that the inhibition of HBV RNA transcription by myrAkt1 was dose dependent. The cotransfection of 1 and 2 μg of plasmid expressing myrAkt1 resulted in a four- and sixfold reduction of HBV pgRNA, respectively (Fig. 2).
FIG. 2.
Dose response of constitutively active Akt1 on HBV RNA transcription. The HBV DNA genome (2 μg) was cotransfected into a 35-mm dish of HepG2 cells with either 2 μg of the control vector pUC19 (lane 1) or 0.5 to 2 μg (lanes 2 to 5) of pCMVmyrAkt1. When less than 2 μg of pCMVmyrAkt1 was used for cotransfection, the control vector pUC19 was added to bring the amount of cotransfected DNA up to 2 μg. Total RNA was extracted at day 5 posttransfection and analyzed by Northern blot hybridization. rRNA served as the loading control.
The inhibition of HBV gene transcription by constitutively active Akt1 could be abolished by rapamycin treatment.
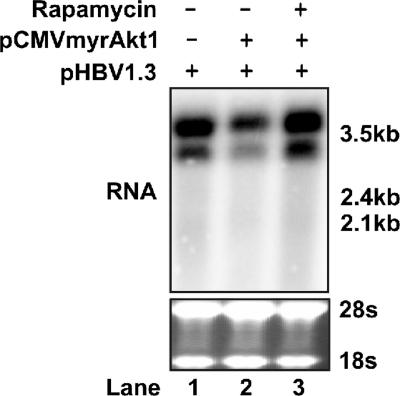
Activated Akt1 can mediate many cellular signaling events by potentially phosphorylating over 9,000 proteins in mammalian cells (27). The mammalian target of rapamycin (mTOR), an unconventional serine/threonine kinase, is one of the well-studied Akt downstream targets. Akt activates mTOR via the direct phosphorylation of TSC2 and by the inhibition of AMPK, thereby resulting in activation of the mTOR complex 1 (mTORC1) that contains mTOR, raptor, and mLST8. Rapamycin works through a gain-of-function mechanism in which it binds to FKBP12 to generate a drug receptor complex that then binds and inhibits the kinase activity of mTORC1 (18). To investigate whether mTOR is responsible for the observed suppression of HBV gene transcription by the activation of Akt1, HepG2 cells were cotransfected with pHBV1.3 and pCMVmyrAkt1. Twelve hours after transfection, cells were either left untreated or treated with the mTOR inhibitor rapamycin for another 4 days. HBV RNA levels were determined by Northern blot assay. As shown in Fig. 3, in agreement with the previous results (Fig. 1 and 2), expression of myrAkt1 substantially suppressed HBV RNA transcription. However, in the presence of 10 ng/ml rapamycin, this inhibitory effect of myrAkt1 was entirely abolished.
FIG. 3.
Rapamycin rescues the inhibition of HBV transcription by Akt1 activation. The HBV DNA genome (2 μg) was cotransfected into HepG2 cells in a 35-mm dish with either 2 μg of the control plasmid (lane 1) or pCMVmyrAkt1 (lanes 2 and 3). Twelve hours after transfection, rapamycin was added to the culture medium to a final concentration of 10 ng/ml (lane 3); DMSO concentration in all experiment groups was normalized at 0.01%. Culture media and the inhibitor were replaced every other day, and cells were harvested at day 5 after transfection. Total RNA was extracted and analyzed by Northern blot hybridization. rRNA served as the loading control.
Taken together, the results presented in the above two sections clearly demonstrate that the activation of Akt1 can inhibit HBV gene transcription, most probably through the activation of mTOR.
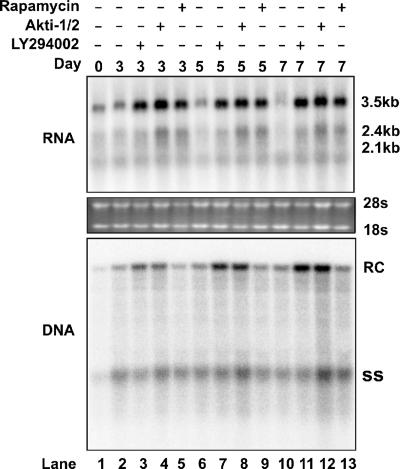
Inhibition of the PI3K-Akt pathway promotes HBV RNA transcription and DNA replication in HepG2.2.15 cells.
The results presented in Fig. 1A show that the overexpression of the dominant negative Akt1 does not affect HBV gene transcription and DNA replication. This result suggests that the basal level activation of cellular Akt1 might not regulate HBV replication in HepG2 cells. However, due to the existence of other isoforms of Akt proteins, such as Akt2 and Akt3, it is possible that inhibited Akt1 activity can be compensated for by those other Akt proteins. Moreover, to further explore the upstream signaling components in Akt-mTOR-mediated HBV gene transcription regulation, we tested the effects of the PI3K, Akt, and mTOR inhibitors on HBV gene transcription and replication in a HepG2-derived stable cell line that supports constitutive HBV replication (1). HepG2.2.15 cells were treated with the control solvent DMSO (dimethyl sulfoxide), 10 μM LY294002, 5 μM Akti-1/2, and 10 ng/ml rapamycin. Cells were harvested at days 3, 5, and 7 of treatment. Viral RNA and DNA replication intermediates were determined by Northern and Southern blot hybridizations. As shown in Fig. 4, the treatment of HepG2.2.15 cells with any of the three kinase inhibitors significantly increased the levels of HBV mRNA at all three time points examined. Interestingly, the 3.5-kb pgRNA and 2.4-kb mRNA were more profoundly affected by the inhibitors. Consistent with the increase of HBV pgRNA, HBV DNA replication is also increased in the presence of the PI3K and Akt inhibitors. However, curiously, HBV DNA levels in rapamycin-treated cells do not increase concurrently. This result seems to indicate that in addition to rapamycin's positive effect on HBV RNA transcription, it inhibits HBV DNA replication. A possible explanation for this observation is that activation of the PI3K-Akt-mTOR pathway negatively regulates HBV RNA transcription, but rapamycin could also act on a certain cellular target(s), other than mTOR, in HepG2.2.15 cells to regulate HBV DNA replication.
FIG. 4.
Effects of kinase inhibitors on HBV replication in HepG2.2.15 cells. HepG2.2.15 cells cultured in 35-mm dishes were left untreated or treated with LY294002 (10 μM), Akti-1/2 (5 μM), and rapamycin (10 ng/ml). The DMSO concentration for all experimental groups was normalized at 0.1%. Cells were harvested at days 3, 5, and 7 of treatment. Total RNA and HBV DNA replicative intermediates were extracted and analyzed by Northern blot and Southern blot hybridization, respectively.
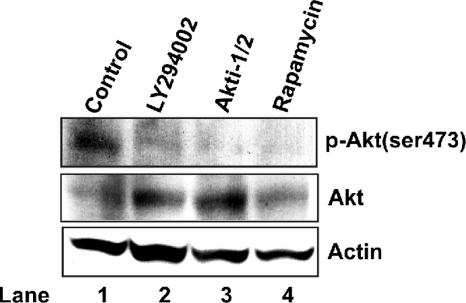
To ensure that the kinase inhibitors had the intended affect on Akt activity under these experimental conditions, we measured the effects of the inhibitors on the levels of cellular total Akt1 and Ser-473-phosphorylated Akt1 by using Western blot analysis (Fig. 5). As expected, inhibition of the Akt1 upstream kinase PI3K by LY294002 reduced the level of Ser-473-phosphorylated Akt1 in treated cells. Isoenzyme-specific Akt inhibitor Akti-1/2 selectively inhibits both Akt1 and Akt2 kinase activity and also blocks the phosphorylation and activation of the corresponding Akt isoforms by phosphoinositide-dependent kinase 1 (PDK1) (2). In agreement with its proposed mechanism of action, Akti-1/2 treatment decreased the level of phosphorylated Akt1 in HepG2.2.15 cells. To our surprise, the mTOR inhibitor rapamycin also reduced the level of phosphorylated Akt1 in treated cells. In searching for an explanation of this result, we noticed that a recent report from Sarbassov and colleagues showed that prolonged treatment of certain types of tumor cells with rapamycin inhibits the assembly of a distinct mTORC2. The mTORC2 can phosphorylate and activate Akt. The prolonged rapamycin treatment reduces the mTORC2 level and, consequently, Akt phosphorylation is inhibited (37).
FIG. 5.
Suppression of Akt1 activation by PI3K, Akt, and mTOR inhibitors. HepG2.2.15 cells cultured in 35-mm dishes were untreated (lane 1) or treated with LY294002 (10 μM), Akti-1/2 (5 μM), or rapamycin (10 ng/ml). The DMSO concentration in all experimental groups was normalized at 0.1%. Three days after treatment, cells were harvested, and total levels of Akt1 and Ser-473-phosphorylated Akt1 [p-Akt(ser473)] were determined by Western blot analysis as described in Materials and Methods. β-Actin served as a loading control.
In summary, these results demonstrated that the basal level activity of the PI3K-Akt-mTOR pathway regulates HBV gene transcription in HepG2.2.15 cells.
Inhibition of the PI3K-Akt pathway enhances HBV cccDNA-dependent transcription.
Thus far, all the experiments presented above utilized either pHBV1.3 transient transfection to initiate HBV gene transcription and DNA replication or the HBV-producing stable cell line HepG2.2.15. It had been reported previously that in those cells, HBV RNA was transcribed mainly from plasmid DNA or integrated viral transgene. The formation of cccDNA in those cells is very inefficient, and viral transcripts from cccDNA templates comprise less than 10% of total viral RNA in those cells (10, 17, 61).
As mentioned above, cccDNA is the authentic transcription template in HBV-infected hepatocytes. It was speculated previously that the regulation of cccDNA-dependent transcription may be substantially different from that of plasmid DNA or integrated HBV genome (3). Moreover, we observed that in pHBV1.3-transfected HepG2 and Huh7.5 cells, the precore/core mRNA showed as doublet bands and the abundance of two envelope protein mRNAs (2.1 and 2.4 kb) was lower than those observed for HepG2.2.15 cells (compare Fig. 1 and Fig. 4). It is clear that the transfection of this plasmid efficiently initiates HBV DNA replication as shown in Fig. 1 and secretes virion particles (data not shown), indicating that the functional pgRNA and the envelope RNA are produced by the plasmid. Interestingly, the doublet bands of the 3.5-kb precore/core mRNA in HepG2 cells transfected with a plasmid containing 1.3-mer HBV genome had been previously observed by others (33). Hence, explanations for those observations could be that one of the doublet bands of 3.5-kb mRNA is derived from an alternative initiation of HBV precore/core mRNA transcription with the plasmid template and the transcription regulation on integrated HBV DNA and plasmid DNA templates and also, possibly, that cccDNA might be different. It is important, therefore, to confirm whether the PI3K-Akt-mTOR pathway could regulate cccDNA-dependent transcription.
To this end, we took advantage of a HepG2-derived cell line (HepAD38) that supports inducible HBV replication (26). Upon removal of tetracycline from the culture medium, HBV RNA synthesis, DNA replication, and cccDNA formation sequentially occur. Ten days after tetracycline removal, cccDNA accumulated to approximately 20 copies per cell (61). At this time point, tetracycline was added back to the culture medium to shut off pgRNA transcription from the transgene. The HBV DNA polymerase inhibitor lamivudine was also added to arrest viral DNA synthesis and prevent further cccDNA formation. Due to its short half-life (approximately 6 to 10 h), the pgRNA transcribed from the transgene was almost completely degraded after 3 days of tetracycline treatment (17, 61). The remaining low level of pgRNA was newly transcribed from cccDNA.
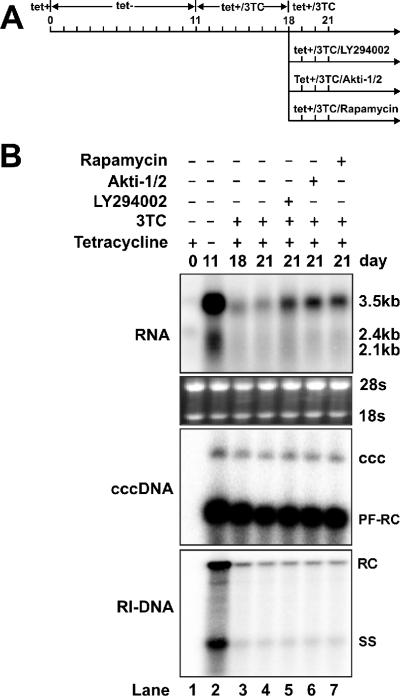
To test the effects of PI3K, Akt, and mTOR inhibitors on cccDNA-dependent transcription, HepAD38 cells were cultured until confluent, and tetracycline was then removed from the culture medium to induce HBV replication and cccDNA formation. After 11 days, tetracycline and lamivudine were added into the culture medium and cultured for an additional 7 days. Cells were then treated with the control solvent DMSO, 10 μM LY294002, 5 μM Akti-1/2, or 10 ng/ml rapamycin in the presence of tetracycline and lamivudine for 3 days (Fig. 6A). The intracellular viral RNA, cccDNA, and viral DNA replicative intermediates were determined by Northern blot and Southern blot hybridizations.
FIG. 6.
The PI3K-Akt signaling pathway regulates HBV cccDNA-dependent transcription. (A) Experimental procedure. HepAD38 cells were cultured in 35-mm dishes until confluent, and tetracycline was then removed from the culture medium to induce HBV replication and cccDNA formation. After 11 days, tetracycline and lamivudine (3TC) were added to the culture medium to shut off the viral pgRNA transcription from the integrated HBV genome and to arrest viral DNA replication. Seven days later, one set of cells (lane 4) continued to be cultured with medium containing tetracycline and 3TC, while another three sets of cells were treated with 10 μM LY294002 (lane 5), 5 μM Akti-1/2 (lane 6), and 10 ng/ml rapamycin (lane 7) in the presence of tetracycline and 3TC for 3 days. The DMSO concentration in all experiment groups was normalized at 0.1%. (B) Cells were harvested at the indicated time points, and the levels of viral RNA and viral DNA replicative intermediates (RI-DNA) were determined by Northern blot and Southern blot assays, as described in Materials and Methods. As shown in the middle panel, HBV cccDNA was extracted with a modified Hirt procedure, and samples were denatured at 85°C for 5 min before loading to increase the sensitivity of cccDNA detection. Under this condition, cccDNA is not denatured, but protein-free rcDNA (PF-RC) species are denatured into single-stranded DNA (61).
Consistent with our previous report, HBV cccDNA was stably maintained after tetracycline and lamivudine treatment (Fig. 6B, lanes 2 to 7). Also in agreement with our previous results, cccDNA transcription was very inefficient and contributed less than 10% of total pgRNA, observed at day 11 after tetracycline removal (Fig. 6B, compare lanes 2 and 3) (61). The levels of pgRNA transcribed from cccDNA remained from days 18 to 21 (Fig. 6B, compare lanes 3 and 4). However, during the same period of time, the treatment of cells with PI3K, Akt, and mTOR inhibitors significantly increased pgRNA levels, indicating that the PI3K-Akt-mTOR signal transduction pathway regulates cccDNA-dependent viral RNA transcription.
HCV NS5A protein activates Akt and inhibits HBV gene expression.
Because HBV and HCV share similar transmission routes, coinfection of the two viruses is frequently diagnosed (13). Previous clinical studies revealed that patients with chronic HCV infection frequently had occult HBV infection (8), suggesting that HCV infection might suppress HBV replication. This notion has been further supported by the observation that HCV superinfection of chronic HBV carriers inhibited HBV replication (41). This inhibition could be due to either the immune response induced by HCV infection or a direct effect of HCV proteins. Using double-fluorescent in situ hybridization with liver biopsy samples obtained from chronic HCV-infected patients with occult HBV infection, Rodriguez-Inigo and colleagues demonstrated that HBV DNA levels in hepatocytes coinfected with the two viruses are consistently lower than those in cells that are infected with HBV alone (36). This result suggests that the observed inhibition of HBV replication by HCV is most likely due to an HCV-induced intracellular mechanism.
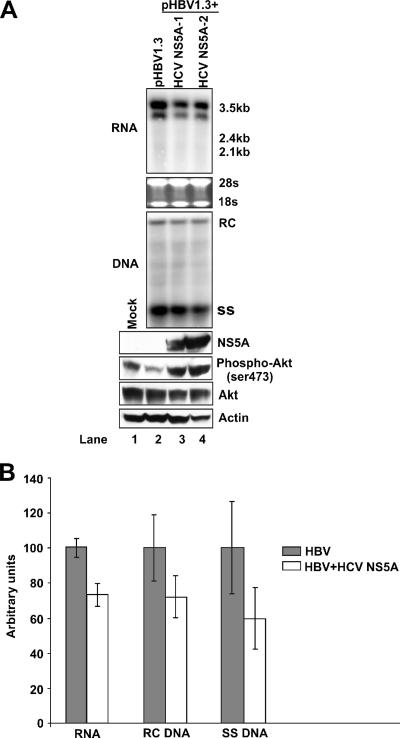
It has been reported by several laboratories that HCV NS5A protein interacts with the p85 subunit of PI3K and activates the PI3K-Akt signal transduction pathway (19, 31, 45, 46). In the context of our findings presented above, we imagined that HCV NS5A protein might be involved in the inhibitory effect of HCV upon HBV replication in hepatocytes coinfected with the two viruses. To test this hypothesis, HepG2 cells were cotransfected with a wild-type HBV genome (pHBV1.3) and the control vector pUC19 or with plasmids that expressed two forms of HCV NS5A proteins cloned from interferon-sensitive (HCV NS5A-1) and interferon-resistant (HCV NS5A-2) patient sera. Western blot analysis of transfected cell lysates confirmed that NS5A proteins were expressed. In agreement with previous reports, HCV NS5A protein expression in HepG2 cells activates Akt1 (Fig. 7A, lower panel). Viral RNA and DNA analyses consistently demonstrate modest reductions of the levels of HBV RNA and DNA replicative intermediates in cells transfected with plasmids that express HCV NS5A protein (Fig. 7A and B).
FIG. 7.
HCV NS5A activates Akt1 and inhibits HBV replication. (A). The wild-type HBV genome (pHBV1.3) was cotransfected into HepG2 cells with control vector pUC19 (lane 2), wild-type HCV NS5A-expressing plasmid (HCV NS5A-1) (lane 3), or interferon-resistant HCV NS5A-expressing plasmid (HCV NS5A-2) (lane 4). In these transfection experiments, 2 μg of HBV DNA was cotransfected with 2 μg of vector DNA or HCV NS5A expression plasmid into a 35-mm dish of cells. Five days after transfection, cells were harvested, and total RNA, HBV DNA replicative intermediates, and protein were extracted and analyzed by Northern blotting, Southern blotting, and immunoblotting assays, as described in Materials and Methods. (B). HepG2 cells were cotransfected with plasmids pHBV1.3 and pUC19 or a plasmid that expresses HCV NS5A-1 and were harvested at day 4 posttransfection. Intracellular HBV RNA and DNA were assayed as described above and quantified using Bio-Rad QuantityOne software. Results from triplicate experiments are presented.
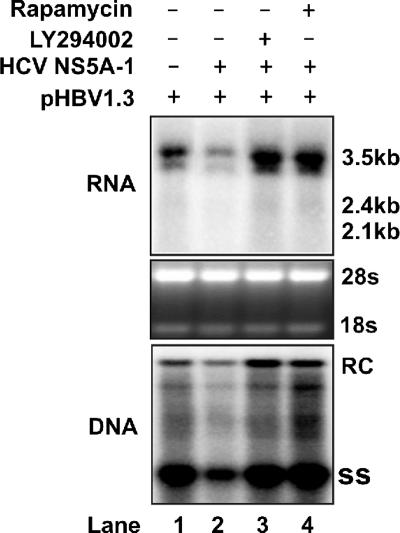
To determine if the observed suppression of HBV RNA transcription by HCV NS5A proteins is mediated by the activation of the PI3K-Akt signal transduction pathway, HepG2 cells were cotransfected with pHBV1.3 and HCV NS5A-1 expression plasmids and treated with the control solvent DMSO, PI3K, and mTOR inhibitors, respectively, for 5 days. Viral RNA and DNA analysis results demonstrated that HCV NS5A-mediated inhibition of HBV RNA and DNA synthesis was completely abolished by treatment with the inhibitors (Fig. 8).
FIG. 8.
Inhibition of HBV replication by HCV NS5A can be rescued by treatment with Akt and mTOR inhibitors. The wild-type HBV genome was cotransfected into HepG2 cells with the control vector pUC19 (lane 1) or the wild-type HCV NS5A-expressing plasmid (HCV NS5A-1) (lanes 2 to 4). In these transfection experiments, 2 μg of HBV DNA was cotransfected with 2 μg of vector DNA or HCV NS5A expression plasmid into a 35-mm dish of cells. Twelve hours after transfection, LY294002 (lane 3) or rapamycin (lane 4) was added to the culture medium at concentrations described above. Cells were harvested at day 5 after transfection. The viral RNA and DNA levels were determined by Northern blotting and Southern blotting assays as described in Materials and Methods.
In summary, the results presented in this section clearly demonstrate that HCV NS5A protein inhibits HBV replication by activation of the PI3K-Akt signal transduction pathway.
DISCUSSION
HBV replication is regulated by many intracellular factors and extracellular environmental cues. However, the molecular pathways involved are largely unknown. Previous studies have revealed that several intracellular signaling pathways could regulate HBV replication. For example, it has been shown that activation of the Ras-mitogen-activated protein kinase pathway inhibits HBV gene transcription (60). Because this pathway is linked to many external stimuli, such as growth factors and hormones, it implies that certain factors may regulate HBV replication through this pathway. The HBV protein X (HBx) has been shown to enhance HBV replication in HepG2 cells, and a series of studies reported by Schneider and colleagues demonstrated that HBx protein does so by triggering Ca2+ release and thereby activating Src kinase, which promotes HBV DNA replication via an unknown mechanism (5, 6).
In this report, we demonstrate that activation of the PI3K-Akt pathway profoundly suppresses the transcription of HBV pgRNA and 2.4-kb mRNA, but not 2.1-kb mRNA (Fig. 4, upper panel). This suggests that the observed inhibition is possibly mediated by transcriptional regulatory proteins that recognize precore/core and L, but not M/S, promoters. The molecular mechanism by which the PI3K-Akt-mTOR pathway regulates HBV gene transcription remains to be determined.
Activation of the PI3K-Akt pathway has been demonstrated to occur during the early phase of infection by a variety of cytopathic viruses, such as human immunodeficiency virus, respiratory syncytial virus, rubella virus, Coxsackie B virus, herpes simplex virus, flaviviruses, influenza virus, adenovirus, and severe acute respiratory syndrome-associated coronavirus, and to prolong short-term survival of infected cells and hence promote virus replication (see reference 11 for a recent review). However, the interplay between noncytopathic viruses, such as HBV and HCV, and this signaling pathway has caught attention only recently. It had been shown that HCV infection activates the PI3K-Akt pathway to limit virus replication and promote cell survival (31). A previous study showed that HBx protein, when overexpressed alone, associated with the catalytic subunit of PI3K and increased phosphorylation of the p85 adaptor subunit, thereby activating PI3K (29). But in the context of HBV replication as described in this study, the activation of Akt1, the downstream target of PI3K, was not observed (Fig. 7, lower panel, compare lanes 1 and 2), indicating that the activation of this pathway in HBV-infected cells might not occur.
Our findings that the activation of PI3K-Akt pathway suppresses HBV gene transcription and that the basal level activity of this pathway regulates virus replication may have several important biological and clinical implications. First, as mentioned previously, Akt activation has been reported to be one of the most consistent features of HBV-induced HCC (7). Currently, it is not yet known when and how this occurs during HCC development. We reason that the activation of Akt during the oncogenic transformation of hepatocytes, along with other cellular changes, may be responsible for the elimination of HBV replication from tumor and even precancerous cells.
Second, the PI3K-Akt signal transduction pathway can be activated by several growth factor receptor tyrosine kinases (25). We have also demonstrated that, as with PI3K and Akt inhibitors, the treatment of HepG2.2.15 cells with the receptor tyrosine kinase inhibitor imatinib mesylate (Gleevec) increased HBV RNA transcription and DNA replication (data not shown). Consistent with this finding, Ikeda and colleagues reported that imatinib mesylate treatment for a patient with chronic myeloid leukemia reactivated an occult HBV infection and led to fulminant hepatitis (24). Those observations imply that HBV replication might normally be controlled in cells by the activation of tyrosine kinase-mediated signaling pathways and possibly by the PI3K-Akt pathway. Therefore, in the clinical trials of the compounds that inhibit receptor tyrosine kinase and the PI3K-Akt-mTOR signal transduction pathway, HBV replication and liver functions should always be carefully monitored.
Finally, previous work suggests that HCV-mediated inhibition of HBV replication in patients coinfected with the two viruses most likely occurs through an HCV protein-mediated intracellular pathway (36). Studies in search of the HCV proteins that are responsible for the inhibition have reported that HCV core and NS2 proteins inhibited HBV replication in cultured cells, but so far, the underlined molecular mechanism(s) is not clear (9, 12, 38, 43, 44). Here we demonstrate that the HCV NS5A protein inhibits HBV gene transcription and DNA replication via the activation of the PI3K-Akt pathway and, thus may be, at least in part, responsible for the observed inhibition of HBV replication during HCV coinfection.
In conclusion, our work presented here demonstrates that the PI3K-Akt-mTOR signal transduction pathway regulates HBV replication under certain physiological and pathological conditions and, thus, may play important roles in viral pathogenesis.
Acknowledgments
We thank Anand Mehta and Pamela Norton for critical reading of the manuscript and helpful suggestions.
This work was supported by an award from the Commonwealth of Pennsylvania through the Hepatitis B Foundation.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Acs, G., M. A. Sells, R. H. Purcell, P. Price, R. Engle, M. Shapiro, and H. Popper. 1987. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc. Natl. Acad. Sci. USA 84:4641-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, S. F., D. Defeo-Jones, S. Fu, P. J. Hancock, K. M. Haskell, R. E. Jones, J. A. Kahana, A. M. Kral, K. Leander, L. L. Lee, J. Malinowski, E. M. McAvoy, D. D. Nahas, R. G. Robinson, and H. E. Huber. 2005. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem. J. 385:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckel-Mitchener, A., and J. Summers. 1997. A novel transcriptional element in circular DNA monomers of the duck hepatitis B virus. J. Virol. 71:7917-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchard, M. J., R. J. Puro, L. Wang, and R. J. Schneider. 2003. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J. Virol. 77:7713-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 7.Boyault, S., D. S. Rickman, A. de Reynies, C. Balabaud, S. Rebouissou, E. Jeannot, A. Herault, J. Saric, J. Belghiti, D. Franco, P. Bioulac-Sage, P. Laurent-Puig, and J. Zucman-Rossi. 2007. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 45:42-52. [DOI] [PubMed] [Google Scholar]
- 8.Cacciola, I., T. Pollicino, G. Squadrito, G. Cerenzia, M. E. Orlando, and G. Raimondo. 1999. Occult hepatitis B virus infection in patients with chronic hepatitis C liver disease. N. Engl. J. Med. 341:22-26. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S. Y., C. F. Kao, C. M. Chen, C. M. Shih, M. J. Hsu, C. H. Chao, S. H. Wang, L. R. You, and Y. H. Lee. 2003. Mechanisms for inhibition of hepatitis B virus gene expression and replication by hepatitis C virus core protein. J. Biol. Chem. 278:591-607. [DOI] [PubMed] [Google Scholar]
- 10.Chou, Y.-C., K.-S. Jeng, M.-L. Chen, H.-H. Liu, T.-L. Liu, Y.-L. Chen, Y.-C. Liu, C.-P. Hu, and C. Chang. 2005. Evaluation of transcriptional efficiency of hepatitis B virus covalently closed circular DNA by reverse transcription-PCR combined with the restriction enzyme digestion method. J. Virol. 79:1813-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooray, S. 2004. The pivotal role of phosphatidylinositol 3-kinase-Akt signal transduction in virus survival. J. Gen. Virol. 85:1065-1076. [DOI] [PubMed] [Google Scholar]
- 12.Dumoulin, F. L., A. von dem Bussche, J. Li, L. Khamzina, J. R. Wands, T. Sauerbruch, and U. Spengler. 2003. Hepatitis C virus NS2 protein inhibits gene expression from different cellular and viral promoters in hepatic and nonhepatic cell lines. Virology 305:260-266. [DOI] [PubMed] [Google Scholar]
- 13.Fong, T. L., A. M. Di Bisceglie, J. G. Waggoner, S. M. Banks, and J. H. Hoofnagle. 1991. The significance of antibody to hepatitis C virus in patients with chronic hepatitis B. Hepatology 14:64-67. [DOI] [PubMed] [Google Scholar]
- 14.Ganem, D., and H. E. Varmus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 15.Gripon, P., C. Diot, A. Corlu, and C. Guguen-Guillouzo. 1989. Regulation by dimethylsulfoxide, insulin, and corticosteroids of hepatitis B virus replication in a transfected human hepatoma cell line. J. Med. Virol. 28:193-199. [DOI] [PubMed] [Google Scholar]
- 16.Guo, H., W. S. Mason, C. E. Aldrich, J. R. Saputelli, D. S. Miller, A. R. Jilbert, and J. E. Newbold. 2005. Identification and characterization of avihepadnaviruses isolated from exotic anseriformes maintained in captivity. J. Virol. 79:2729-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, J. T., M. Pryce, X. Wang, M. I. Barrasa, J. Hu, and C. Seeger. 2003. Conditional replication of duck hepatitis B virus in hepatoma cells. J. Virol. 77:1885-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay, N. 2005. The Akt-mTOR tango and its relevance to cancer. Cancer Cell 8:179-183. [DOI] [PubMed] [Google Scholar]
- 19.He, Y., H. Nakao, S. L. Tan, S. J. Polyak, P. Neddermann, S. Vijaysri, B. L. Jacobs, and M. G. Katze. 2002. Subversion of cell signaling pathways by hepatitis C virus nonstructural 5A protein via interaction with Grb2 and P85 phosphatidylinositol 3-kinase. J. Virol. 76:9207-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hertogs, K., E. Depla, T. Crabbé, W. De Bruin, W. Leenders, H. Moshage, and S. H. Yap. 1994. Spontaneous development of anti-hepatitis B virus envelope (anti-idiotypic) antibodies in animals immunized with human liver endonexin II or with the F(ab′)2 fragment of anti-human liver endonexin II immunoglobulin G: evidence for a receptor-ligand-like relationship between small hepatitis B surface antigen and endonexin II. J. Virology 68:1516-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hild, M., O. Weber, and H. Schaller. 1998. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J. Virol. 72:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, M. M., and B. A. Hemmings. 2002. Inhibition of protein kinase B/Akt. implications for cancer therapy. Pharmacol. Ther. 93:243-251. [DOI] [PubMed] [Google Scholar]
- 23.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, K., Y. Shiga, A. Takahashi, T. Kai, H. Kimura, K. Takeyama, H. Noji, K. Ogawa, A. Nakamura, H. Ohira, Y. Sato, and Y. Maruyama. 2006. Fatal hepatitis B virus reactivation in a chronic myeloid leukemia patient during imatinib mesylate treatment. Leuk. Lymphoma 47:155-157. [DOI] [PubMed] [Google Scholar]
- 25.Kharas, M. G., and D. A. Fruman. 2005. ABL oncogenes and phosphoinositide 3-kinase: mechanism of activation and downstream effectors. Cancer Res. 65:2047-2053. [DOI] [PubMed] [Google Scholar]
- 26.Ladner, S. K., M. J. Otto, C. S. Barker, K. Zaifert, G. H. Wang, J. T. Guo, C. Seeger, and R. W. King. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41:1715-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawlor, M. A., and D. R. Alessi. 2001. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 114:2903-2910. [DOI] [PubMed] [Google Scholar]
- 28.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 29.Lee, Y. I., S. Kang-Park, S. I. Do, and Y. I. Lee. 2001. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 276:16969-16977. [DOI] [PubMed] [Google Scholar]
- 30.Locarnini, S. 2005. Molecular virology and the development of resistant mutants: implications for therapy. Semin. Liver Dis. 25(Suppl. 1):9-19. [DOI] [PubMed] [Google Scholar]
- 31.Mannova, P., and L. Beretta. 2005. Activation of the N-Ras-PI3K-Akt-mTOR pathway by hepatitis C virus: control of cell survival and viral replication. J. Virol. 79:8742-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahon, B. J. 2005. Epidemiology and natural history of hepatitis B. Semin. Liver Dis. 25(Suppl. 1):3-8. [DOI] [PubMed] [Google Scholar]
- 33.Oropeza, C. E., and A. McLachlan. 2007. Complementarity between epsilon and phi sequences in pregenomic RNA influences hepatitis B virus replication efficiency. Virology 359:371-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozer, A., V. I. Khaoustov, M. Mearns, D. E. Lewis, R. M. Genta, G. J. Darlington, and B. Yoffe. 1996. Effect of hepatocyte proliferation and cellular DNA synthesis on hepatitis B virus replication. Gastroenterology 110:1519-1528. [DOI] [PubMed] [Google Scholar]
- 35.Pasquetto, V., S. F. Wieland, S. L. Uprichard, M. Tripodi, and F. V. Chisari. 2002. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J. Virol. 76:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Inigo, E., J. Bartolome, N. Ortiz-Movilla, C. Platero, J. M. Lopez-Alcorocho, M. Pardo, I. Castillo, and V. Carreno. 2005. Hepatitis C virus (HCV) and hepatitis B virus (HBV) can coinfect the same hepatocyte in the liver of patients with chronic HCV and occult HBV infection. J. Virol. 79:15578-15581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarbassov, D. D., S. M. Ali, S. Sengupta, J. H. Sheen, P. P. Hsu, A. F. Bagley, A. L. Markhard, and D. M. Sabatini. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22:159-168. [DOI] [PubMed] [Google Scholar]
- 38.Schuttler, C. G., N. Fiedler, K. Schmidt, R. Repp, W. H. Gerlich, and S. Schaefer. 2002. Suppression of hepatitis B virus enhancer 1 and 2 by hepatitis C virus core protein. J. Hepatol. 37:855-862. [DOI] [PubMed] [Google Scholar]
- 39.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sells, M. A., A. Z. Zelent, M. Shvartsman, and G. Acs. 1988. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J. Virol. 62:2836-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheen, I. S., Y. F. Liaw, C. M. Chu, and C. C. Pao. 1992. Role of hepatitis C virus infection in spontaneous hepatitis B surface antigen clearance during chronic hepatitis B virus infection. J. Infect. Dis. 165:831-834. [DOI] [PubMed] [Google Scholar]
- 42.Shih, C. H., L. S. Li, S. Roychoudhury, and M. H. Ho. 1989. In vitro propagation of human hepatitis B virus in a rat hepatoma cell line. Proc. Natl. Acad. Sci. USA 86:6323-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shih, C. M., C. M. Chen, S. Y. Chen, and Y. H. Lee. 1995. Modulation of the trans-suppression activity of hepatitis C virus core protein by phosphorylation. J. Virol. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih, C. M., S. J. Lo, T. Miyamura, S. Y. Chen, and Y. H. Lee. 1993. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J. Virol. 67:5823-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Street, A., A. Macdonald, K. Crowder, and M. Harris. 2004. The hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 279:12232-12241. [DOI] [PubMed] [Google Scholar]
- 46.Street, A., A. Macdonald, C. McCormick, and M. Harris. 2005. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular β-catenin and stimulation of β-catenin-responsive transcription. J. Virol. 79:5006-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suk, F. M., M. H. Lin, M. Newman, S. Pan, S. H. Chen, J. D. Liu, and C. Shih. 2002. Replication advantage and host factor-independent phenotypes attributable to a common naturally occurring capsid mutation (I97L) in human hepatitis B virus. J. Virol. 76:12069-12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 49.Summers, J., A. O'Connell, and I. Millman. 1975. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc. Natl. Acad. Sci. USA 72:4597-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Nunen, A. B., O. Pontesilli, F. Uytdehaag, A. D. Osterhaus, and R. A. de Man. 2001. Suppression of hepatitis B virus replication mediated by hepatitis A-induced cytokine production. Liver 21:45-49. [DOI] [PubMed] [Google Scholar]
- 51.Vogt, P. K., A. G. Bader, and S. Kang. 2006. Phosphoinositide 3-kinase: from viral oncoprotein to drug target. Virology 344:131-138. [DOI] [PubMed] [Google Scholar]
- 52.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, G. H., and C. Seeger. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663-670. [DOI] [PubMed] [Google Scholar]
- 54.Wang, R. Y., C. N. Shen, M. H. Lin, D. Tosh, and C. Shih. 2005. Hepatocyte-like cells transdifferentiated from a pancreatic origin can support replication of hepatitis B virus. J. Virol. 79:13116-13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, J.-C., P.-J. Chen, M. Y. Kuo, S.-D. Lee, D.-S. Chen, and L.-P. Ting. 1991. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J. Virol. 65:1099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, T. T., L. Coates, C. E. Aldrich, J. Summers, and W. S. Mason. 1990. In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology 175:255-261. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, G. H., M. Jeffers, A. Bellacosa, Y. Mitsuuchi, G. F. Vande Woude, and J. R. Testa. 2001. Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc. Natl. Acad. Sci. USA 98:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, C., T. Yamamoto, T. Zhou, C. E. Aldrich, K. Frank, J. M. Cullen, A. R. Jilbert, and W. S. Mason. 2006. The liver of woodchucks chronically infected with the woodchuck hepatitis virus contains foci of virus core antigen-negative hepatocytes with both altered and normal morphology. Virology 359:283-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh, C. T., H. T. Chiu, C. M. Chu, and Y. F. Liaw. 1998. G1 phase dependent nuclear localization of relaxed-circular hepatitis B virus DNA and aphidicolin-induced accumulation of covalently closed circular DNA. J. Med. Virol. 55:42-50. [DOI] [PubMed] [Google Scholar]
- 60.Zheng, Y., J. Li, D. L. Johnson, and J. H. Ou. 2003. Regulation of hepatitis B virus replication by the Ras-mitogen-activated protein kinase signaling pathway. J. Virol. 77:7707-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, T., H. Guo, J. T. Guo, A. Cuconati, A. Mehta, and T. M. Block. 2006. Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral Res. 72:116-124. [DOI] [PubMed] [Google Scholar]