Abstract
The host innate immune response is an important deterrent of severe viral infection in humans and animals. Nuclear import factors function as key gatekeepers that regulate the transport of innate immune regulatory cargo to the nucleus of cells to activate the antiviral response. Using severe acute respiratory syndrome coronavirus (SARS-CoV) as a model, we demonstrate that SARS-COV ORF6 protein is localized to the endoplasmic reticulum (ER)/Golgi membrane in infected cells, where it binds to and disrupts nuclear import complex formation by tethering karyopherin alpha 2 and karyopherin beta 1 to the membrane. Retention of import factors at the ER/Golgi membrane leads to a loss of STAT1 transport into the nucleus in response to interferon signaling, thus blocking the expression of STAT1-activated genes that establish an antiviral state. We mapped the region of ORF6, which binds karyopherin alpha 2, to the C terminus of ORF6 and show that mutations in the C terminus no longer bind karyopherin alpha 2 or block the nuclear import of STAT1. We also show that N-terminal deletions of karyopherin alpha 2 that no longer bind to karyopherin beta 1 still retain ORF6 binding activity but no longer block STAT1 nuclear import. Recombinant SARS-CoV lacking ORF6 did not tether karyopherin alpha 2 to the ER/Golgi membrane and allowed the import of the STAT1 complex into the nucleus. We discuss the likely implications of these data on SARS-CoV replication and pathogenesis.
Severe acute respiratory syndrome (SARS) is a potentially fatal disease that likely originated in the Guandong Province of China in the fall of 2002 (14, 26). The global outbreak infected more than 8,000 people and resulted in about 800 deaths, with economic losses in the tens of billions of dollars (www.who.int/csr/sars/en/). The disease is caused by a new human coronavirus (CoV), designated SARS-CoV, that likely evolved from closely related strains in civet cats, raccoon dogs, and bats (28, 45). Low-level seropositivity in archived human sera is suggestive of multiple introductions into human populations prior to 2002 (63). The underlying mechanisms governing the high mortality rates during SARS-CoV infection remain largely unknown.
SARS-CoV is a single-stranded positive-polarity enveloped virus. The genome is approximately 29.7 kb long with the 5′ two-thirds of the genome encoding proteins important for replicase function and the 3′ one-third of the genome encoding structural proteins and group-specific (accessory) proteins. The accessory open reading frames (ORFs) are unique to each different strain of CoV and are predicted to encode functions important in pathogenesis (11). Among the SARS-CoV accessory ORFs, ORF3a, ORF6, ORF7a, and ORF7b are not essential for in vitro or in vivo replication or release of infectious viruses (62), although each one is reported to be incorporated into virions (22, 23, 50, 54).
An important host defense against invading pathogens is the innate immune system. The innate immune system consists of secreted cytokines, intracellular signaling pathways, and the expression of type I interferon (IFN) that signals to neighboring cells to induce an antiviral state (reviewed in reference 16). SARS-CoV has been shown to affect IFN regulatory factor 3 (IRF3) trafficking and activation during infection (55). In murine models, SARS-CoV infection is cleared normally from infected CD1−/− and RAG1−/− mice but not STAT1−/− mice, suggesting that innate immunity coordinates an effective antiviral response, and protection in these models does not require adaptive immunity (17, 21).
Interactions between the adaptive immune system and CoVs have been studied with great detail, yet the role of innate immunity and the virus-host interactions that modulate the innate immune response during infection are less well characterized (5, 7, 27). Taguchi and Siddell showed that different strains of mouse hepatitis virus (MHV) were differentially susceptible to type I IFN and hypothesized that this difference influenced pathogenic outcomes (56). Ye et al. showed previously that the MHV nucleocapsid (N) protein interacts with protein kinase R and 2′,5′-oligoadenylate synthase machinery to inhibit viral RNA degradation (60). Kamitani et al. demonstrated that SARS nsp1 protein expression led to the degradation of cellular RNA messages by an unknown mechanism, including IFN-β mRNA, suggesting a possible role in antagonizing type I IFN induction (24). In addition, Kopecky-Bromberg et al. demonstrated that different arms of the type I IFN induction pathway can be blocked by the expression of SARS-CoV ORF3b, ORF6, and N protein in vitro (25).
As type I interferon (IFN-α/β) is secreted from a cell, IFN-α/β can bind to the IFN-α/β receptor on the surface of neighboring cells or the same cell in a paracrine or autocrine fashion. Binding leads to the activation of the receptor, recruitment of JAK kinase, and phosphorylation of STAT1 via JAK. Phosphorylation induces STAT1 to form a complex with STAT2 and IRF9, forming the interferon-stimulated gene factor 3 (ISGF3) complex. ISGF3 binds import factor karyopherin alpha 1 (KPNA1) and recruits karyopherin beta 1 (KPNB1) for transport into the nucleus (35), where STAT1 acts as a transcription factor for many IFN-induced genes that establish the antiviral state (34, 36). The mechanism by which ORF6 blocks STAT1 transport into the nucleus is not known, nor has the function of ORF6 been established in the context of virus infection.
Multiple levels of regulation occur at the level of protein import of ISGF3 to the nucleus. After being appropriately phosphorylated, STAT1 interacts with the ISGF3 complex, exposing an unusual nuclear localization signal (NLS) on its surface for the recruitment of a specific karyopherin, KPNA1 (34, 51). Each of the karyopherins (designated KPNA1 to KPNA4) recognizes NLS sites in distinct yet sometimes overlapping sets of protein cargo destined for transport into the nucleus (reviewed in reference 8). After binding cargo, an N-terminal repeat domain termed the importin beta binding site in each KPNA engages KPNB1, which chaperones each KPNA/cargo complex through the nuclear pore (18, 38). Ebola virus VP24 has been shown to affect STAT1 binding to KPNA1 by blocking the binding site on KPNA1 and thus blocking STAT1 nuclear import and signaling (47).
Previous studies by our group indicated that ORF6 expression from plasmids blocked STAT1 translocation to the nucleus (25). In this paper, we demonstrate that ORF6 sequesters host nuclear import factors into the rough endoplasmic reticulum (ER) (rER)/Golgi membrane, thus blocking the nuclear import of STAT1, and functions as an IFN antagonist in the context of SARS-CoV infection.
MATERIALS AND METHODS
SARS infections.
Vero, MA104, 293, and Caco2 cells were infected with an infectious clone of SARS CoV strain Urbani (icSARS) (61), SARS strain Urbani (GenBank accession number AY2787421), or Sendai virus (SeV) (ATCC VR-907) for 18 h at a multiplicity of infection (MOI) of 5. Vero cells (ATCC CRL-1586) (African green monkey kidney cells), MA104 cells (ATCC CRL-2378.1) (African green monkey kidney cells), HEK293 cells (ATCC CRL-11268) (human embryonic kidney cells), and Caco2 cells (ATCC CRL-2102) (human intestinal epithelial cells) were grown in standard growth medium according to ATCC recommendations. Growth medium was harvested for an IFN bioassay (see below), cells were treated with TRIzol (Invitrogen, Carlsbad, CA), and RNA was extracted according to the manufacturer's directions.
Plasmids.
SARS-CoV ORF6 was cloned using icSARS as a template for PCR primers. The ORF was amplified using forward primer 1 (5′-ACGTCCCGGGTGGATAATCTAACTCCA-3′) and reverse primer 5′-GGCCGAATTCACCATGTTTCATCTAGTTGACTT-3′. PCR products were digested with EcoRI and XmaI and cloned into pCAGGS-HA containing a hemagglutinin (HA) tag at the XmaI site. For ORF6 59 to 63 Ala constructs, forward primer 1 was used with reverse primers 5′-CCCGGGTTCATCATCTAACTCCGAATAATTCTTCTT-3′ and 5′-ACGTCCCGGGTGCCGCTGCAGCTGCCATAGGTTCTTCATC-3′, respectively. For ORF6 54 to 58 Ala, two rounds of PCR were used. The first step involved forward primer 1 as described above and reverse primer 5′-TGCCGCTGCAGCTGCATCTAACTCCGAATAATTCT-3′. The PCR product was then reamplified using the forward primer 1 and reverse primer 5′-ACGTCCCGGGTGGATAATCTAACTCTGCCGCTGCAGC-3′. For ORF6 49 to 53 Ala, two rounds of PCR were used. The first step used forward primer 1 and reverse primer 5′-TTCATCTGCCGCTGCAGCTGCATTCTTCTTAGTTA-3′. The PCR product was then reamplified using forward primer 1 and reverse primer 5′-ACGTCCCGGGTGGATAATCTAACTCCATAGGTTCTTCATCTGCCGCTGC-3′. KPNA2ΔN was cloned using forward primer 5′-CGATAAGGCGGCCGCGATGATTGTCAAAGGCATAAATAGC-3′ and reverse primer 5′-CATCAATCACAACCTGAGTCTGTTCATCTG-3′. After PCR amplification, the amplicon was cloned into parent vector pCAGGS-KPNA2/FLAG using NotI and PstI.
IFN bioassay and IFN-β reverse transcription (RT)-PCR.
MA104 cells were infected with icSARS or SeV at an MOI of 5. Medium was removed from the cells for the bioassay, and cells were treated with TRIzol for RNA extraction. The bioassay was described previously for use with human type I IFN and A549 cells (52). Briefly, the pH of the medium was lowered to 2 with 2 N HCl and incubated at 4°C overnight. The pH of the medium was then adjusted to pH 7 with NaOH. One hundred microliters of the medium was then added to A549 cells plated in a 96-well plate and incubated for 24 h. After Encephalomyocarditis virus infection for 1 h, the cultures were incubated for 24 h before the plates were scored for cytopathic effect.
Isolated RNA was converted to cDNA with Superscript II (Invitrogen, Carlsbad, CA) and random hexamers and used for a PCR for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and IFN-β. GAPDH-specific primers were 5′-GTCTTCACCACCATGGAGAAGGCTGGGGCT-3′ and 5′-ACAGCCTTGGCAGCGCCAGTAGAGGCAGGG-3′, while IFN-β primers were 5′-GACGCCGCATTGACCATCTA-3′ and 5′-CCTTAGGATTTCCACTCTGACT-3′.
STAT1 localization.
The same procedure was used for transfections of both 293 and Vero cells in culture. Cells were grown on glass coverslips in 24-well plates. Five hundred nanograms of STAT1/green fluorescent protein (GFP) plasmid, 500 ng of ORF6 plasmid, or various derivative constructs and Fugene 6 (Roche, Indianapolis, IN) were used for transfections according to the manufacturer's directions. At 24 h after transfection, cells were treated with 100 IU/ml of IFN-β (Calbiochem, San Diego, CA) for 1 h. Cells were fixed with 4% paraformaldehyde (PFA) and imaged in the Microscopy Service Laboratory (University of North Carolina at Chapel Hill) using a Zeiss LSM5 Pa confocal laser scanning microscope.
Immunofluorescence.
HA-tagged ORF6, Flag-tagged KPNA (a gift of Megan Shaw, Mount Sinai Medical Center), and Flag-tagged KPNB1 (a gift of Nancy Reich, Stony Brook University, Stony Brook, NY) plasmids were transfected as described above, and cells were fixed with 4% PFA. Fixed cells were incubated with rabbit anti-HA antibody (Sigma-Aldrich, St. Louis, MO) and mouse anti-Flag M2 antibody (Sigma-Aldrich, St. Louis, MO). Coverslips were incubated with secondary antibodies: Alexa Fluor 488-, 546-, or 633-conjugated goat anti-rabbit antibody (Molecular Probes) and Alexa Fluor 488-, 546-, or 633-conjugated goat anti-mouse antibody (Molecular Probes). Confocal imaging was performed in the Microscopy Service Laboratory (University of North Carolina at Chapel Hill) using a Zeiss LSM5 Pa confocal laser scanning microscope.
Flag immunoprecipitations.
Flag-tagged karyopherins were transfected as described above. After 24 h of expression, cells were lysed in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40), the extract was centrifuged for 10 min at 4°C, and the supernatant was removed. Twenty-five microliters of washed EZ View Red Anti-Flag M2 Affinity Gel beads (catalog number F2426; Sigma, St. Louis, MO) was added to each extract and rotated overnight at 4°C. The extract was then washed three times with lysis buffer and resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer before boiling and electrophoresis.
Split YFP localization.
KPNA2 and ORF6 were cloned into a “Venus” split yellow fluorescent protein (YFP) plasmid by PCR amplification and the restriction enzymes BspEI/Xba1 and NotI/ClaI, respectively (plasmids were a gift of Stephen Michnick, University of Montreal). Primers used for the amplification of KPNA2 cDNA were 5′-ACTGTCCGGAATGTCCACCAACGAGAATGC-3′ and 5′-GTCAAGATCTCTAAAAGTTAAAGGTCCCAGGAGCCCCATC-3′, while primers used for the amplification of ORF6 cDNA were 5′-ACTGGCGGCCGCACCATGTTTCATCTTGTTGACTT-3′ and 5′-CAGTATCGATTGGATAATCTAACTCCA-3′. Plasmids were transfected as described above and visualized using a Zeiss Confocal microscope and a YFP-specific filter.
RESULTS
SARS does not induce IFN in infected cells.
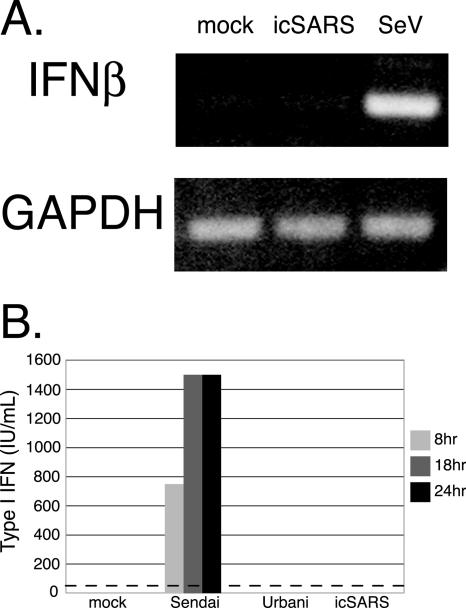
Cultures of cells were infected with icSARS (infectious clone of SARS strain Urbani), SARS strain Urbani, or SeV at an MOI of 5 and evaluated for IFN induction by the detection of IFN-β transcripts and secretion of type I IFN into the medium. We found robust levels of IFN-β RNA transcripts in MA104 cells following SeV infection, but not SARS-CoV infection, using RT-PCR (Fig. 1A). Media from infected MA104 cells were analyzed for the presence of secreted IFN-β protein by IFN bioassay. Type I IFNs were not detected in the medium from SARS infections of MA104 cells (Fig. 1B). While similar results were also seen in SARS-CoV-infected 293 cells, human airway epithelial cells, and Caco2 cells, robust levels of type I interferon were secreted in SeV-infected cultures of each cell line (data not shown).
FIG. 1.
SARS-CoV encodes an IFN antagonist. (A) MA104 cells were infected with SARS and SeV or mock infected for 12 h. Extracted RNA was analyzed by RT-PCR for the induction of IFN-β mRNA. RT-PCR-amplified GAPDH transcripts are shown as a loading control. (B) Media from icSARS-, Urbani virus-, and SeV-infected MA104 cells were analyzed for secreted type I IFN across a time course of infection. An IFN bioassay (as described in Materials and Methods) was used to analyze the amount of type I IFN secreted from infected cells. The dotted line is the minimal level of detection for the assay.
SARSΔORF6 does not block STAT1 nuclear translocation.
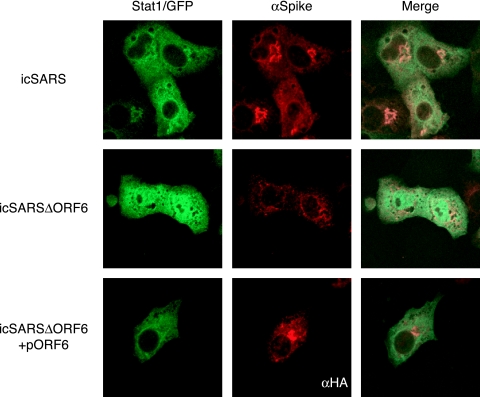
We previously showed that ORF6 expressed from a plasmid blocks the nuclear translocation of STAT1 after IFN-β treatment (25). To determine if STAT1 transport was blocked in the context of virus infection, cultures of Vero cells were transfected with STAT1/GFP and then infected with wild-type icSARS or icSARS which lacks ORF6 (icSARSΔORF6) (62). Following virus infection, STAT1 was seen localized in the cytoplasm at 12 h postinfection (data not shown), in agreement with our data showing that SARS-CoV does not induce IFN and thus does not induce STAT1 nuclear import. After treatment of uninfected cultures with IFN-β, STAT1 was translocated to the nucleus within 60 min. Following IFN-β treatment of a wild-type icSARS-infected culture, STAT1 was not translocated into the nucleus (Fig. 2A, top). Under identical conditions, STAT1/GFP was successfully transported into the nucleus of icSARSΔORF6-infected cells (Fig. 2A, middle), demonstrating that ORF6 expression may be responsible for the nuclear trafficking block of STAT1 during virus infection. In agreement with these findings, the icSARSΔORF6 deletion phenotype was reversed by providing ORF6 in trans via an expression plasmid (pORF6) prior to infection and IFN-β treatment (Fig. 2A, bottom). As a control, recombinant icSARS viruses lacking either ORF3a or ORF7ab prevented the translocation of STAT1/GFP to the nucleus after IFN treatment, as seen for the wild-type icSARS virus (data not shown).
FIG. 2.
STAT1 localization during SARS infection. STAT1/GFP plasmid was transfected into Vero cells, and its localization was assayed after SARS infection. At 24 h posttransfection, cells were infected with either icSARS (top) or icSARSΔORF6 (middle) at an MOI of 3 for 12 h. Before fixation in 4% PFA, cells were treated with 100 IU of IFN-β for 1 h. The cells in the bottom panels were cotransfected with STAT1/GFP and HA-ORF6 prior to infection with icSARSΔORF6. Cells were then labeled with anti-SARS spike antibody to visualize the SARS-infected cells or anti-HA antibody (to visualize the transfected cells) and an Alexa 546-conjugated secondary antibody. Cells were visualized by using a confocal microscope.
KPNA1 interacts with ORF6.
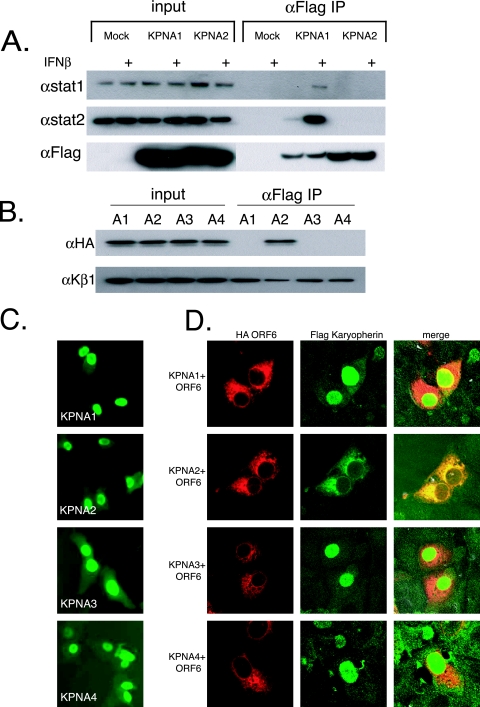
Previous studies indicated that ORF6 does not prevent STAT1 phosphorylation or dimerization (25) or degrade IFN-α/β receptors in infected cells (data not shown), suggesting a novel mechanism of action. Activated STAT1 (and the ISGF3 complex) is normally translocated into the nucleus by binding KPNA1, which then recruits and binds KPNB1. Together, the STAT1:KPNA1:KPNB1 complex is imported into the nucleus (34). As Ebola virus VP24 was shown to bind KPNA1 and block its interaction with STAT1 (47), it is possible that ORF6 may block STAT1's nuclear import by interacting with nuclear import factors. To determine if karyopherin interactions were altered in the STAT1 signaling pathway, we transfected Flag-tagged KPNA1, KPNA2, KPNA3, and KPNA4 into 293 cells and assayed their interactions with STAT1 and STAT2 in the presence or absence of IFN-β treatment. Transfected 293 cells were either mock treated or treated with IFN-β for 30 min. Extracts were immunoprecipitated with anti-Flag antibody and run on SDS-PAGE gels for analysis. We found that STAT1 and STAT2 interacted with KPNA1 and not KPNA2, KPNA3, or KPNA4 specifically after IFN-β treatment, confirming previously described work (Fig. 3A) (34).
FIG. 3.
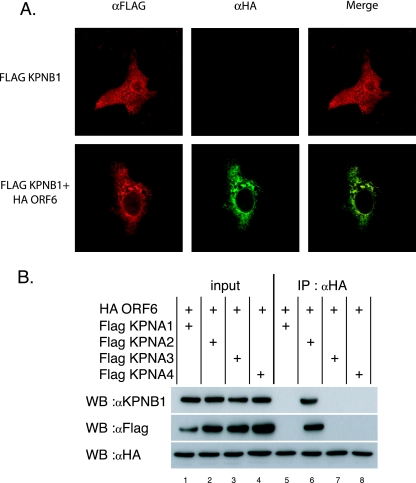
ORF6 affects localization of KPNA2. (A) 293 cells were transfected with Flag-tagged karyopherin plasmids. At 24 h posttransfection, half of the cells were treated with 100 IU of IFN-β for 30 min, and protein was harvested as described in Materials and Methods. Proteins were then immunoprecipitated (IP) with anti-Flag (αFlag) M2 antibodies and run on an SDS-PAGE gel before being transferred for Western blotting. Blots were probed with STAT1, STAT2, or Flag antibodies. + indicates treatment with 100 IU/ml of IFN-β. (B) Immunoprecipitations were performed on 293 cells cotransfected with each Flag-tagged karyopherin and HA-tagged ORF6. Lysates were prepared and immunoprecipitated as described in Materials and Methods. The top panel was probed with anti-HA antibody to visualize ORF6, and the bottom panel was probed with anti-KPNB1. A1, KPNA1; A2, KPNA2; A3, KPNA3; A4, KPNA4. (C) Vero cells were transfected with Flag-tagged KPNA plasmids. At 24 h posttransfection, cells were fixed and labeled with anti-Flag antibody. (D) Vero cells were cotransfected with Flag-tagged KPNA plasmids and HA-ORF6 plasmid. At 24 h posttransfection, cells were fixed and labeled with anti-Flag antibody and anti-HA antibody. Anti-Flag antibody was visualized with Alexa Fluor 488-conjugated secondary antibody, and anti-HA antibody was visualized with Alexa Fluor 546-conjugated secondary antibody. Note the overlapping localization of ORF6 and KPNA2.
To test the hypothesis that ORF6 may interact with a KPNA, coimmunoprecipitations were performed following single transfections and cotransfections of 293 cells. At 24 h after transfection of HA-tagged ORF6 with each Flag-tagged KPNA, proteins were coimmunoprecipitated with anti-Flag antibody and separated on an SDS-PAGE gel. We found that ORF6 was specifically coimmunoprecipitated with KPNA2 but not with KPNA1, KPNA3, or KPNA4 (Fig. 3B, top). Interestingly, STAT1 normally interacts with KPNA1 during normal IFN-induced nuclear transport and not with KPNA2.
Since ORF6 is an ER/Golgi membrane protein and interacts with KPNA2 by immunoprecipitation, we hypothesized that KPNA2, which is normally distributed in the cytoplasm, may become membrane associated, resulting in a loss of nuclear localization. To confirm the biochemical association, we cotransfected Flag-tagged KPNA1, KPNA2, KPNA3, and KPNA4 into Vero cells with HA-tagged ORF6 and determined the localization of each protein. By confocal microscopy, the expression of KPNA1, KPNA2, KPNA3, and KPNA4 resulted in a predominantly nuclear localization, a phenotype typically observed following expression from plasmids (Fig. 3C) (53). Following cotransfection with ORF6, KPNA1, KPNA3, and KPNA4 were still localized to the nucleus (Fig. 3D). In agreement with our immunoprecipitation data, KPNA2 was colocalized with ORF6 on the ER/Golgi membrane, further supporting an interaction between ORF6 and KPNA2.
KPNA2 and ORF6 interact in vivo.
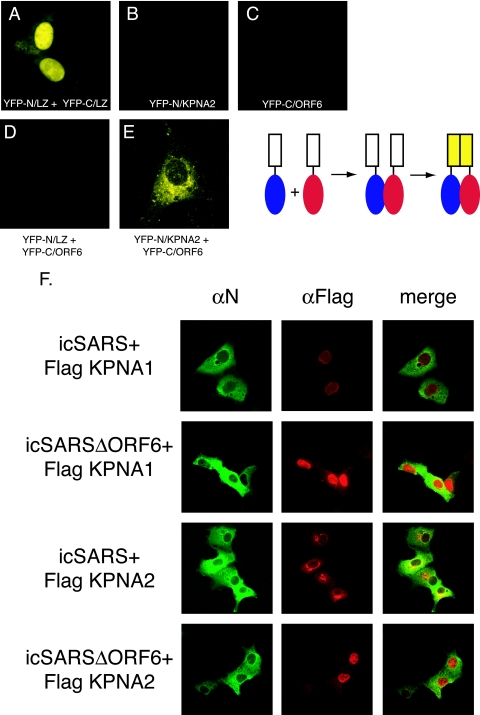
To further demonstrate an intimate in vivo interaction between ORF6 and KPNA2, we utilized the enhanced Venus split YFP system (40). In this system, the N- and C-terminal halves of YFP are independently fused to two putative interacting proteins. If these proteins interact, that interaction event brings the two YFP halves in intimate proximity, which folds and reconstitutes the YFP protein, leading to fluorescence. The N terminus of YFP was fused in frame to KPNA2, and the C terminus of YFP was fused in frame to ORF6. Each construct was transfected into Vero cells for 24 h prior to visualization. Positive-control leucine zipper plasmids successfully reconstituted YFP fluorescence as reported previously (40) (Fig. 4A). Transfection of the KPNA2 or ORF6 construct independently did not result in YFP fluorescence (Fig. 4B and C). As a control, KPNA2 and ORF6 fusions were transfected with their complementary leucine zipper/YFP plasmids, and no fluorescence was seen, demonstrating that interacting partners were needed for fluorescence (Fig. 4D). Cotransfection of KPNA2/YFP-N and ORF6/YFP-C resulted in robust YFP fluorescence localized to the ER/Golgi membrane (Fig. 4E), confirming an in vivo interaction between KPNA2 and ORF6.
FIG. 4.
Interaction between KPNA2 and ORF6 in vivo. The top panel shows a schematic of the split YFP assay. The cargo is fused to each half of YFP. When the cargo interacts, the YFP halves are brought together to re-form and fluoresce. Vero cells were transfected with YFP plasmids, and 24 h after transfection, YFP fluorescence was visualized on a confocal microscope using a YFP filter. (A) YFP-N/leucine zipper (LZ) and YFP-C/leucine zipper (positive controls). (B) YFP-N/KPNA2. (C) YFP-C/ORF6. (D) YFP-N/leucine zipper plus YFP-C/ORF6. (E) YFP-N/KPNA2 and YFP-C/ORF6. (F) Karyopherin localization during SARS infection. Vero cells were transfected with Flag-tagged KPNA1 or KPNA2 24 h prior to infection with either icSARS or icSARSΔORF6. Cells were fixed in 4% PFA prior to antibody staining. SARS mouse anti-N antibody (αN) was used to localize SARS-infected cells, and rabbit anti-Flag antibody was used to localize karyopherins. Mouse anti-N antibody was visualized with Alexa Fluor 488-conjugated secondary antibody, and anti-Flag antibody was visualized with Alexa Fluor 546-conjugated secondary antibody.
SARS-CoV, but not SARSΔORF6, retains KPNA2 at the ER/Golgi membrane.
Since KPNA2 nuclear localization was altered in ORF6-transfected cells, we next determined whether KPNA2 localization was also altered following SARS-CoV infection. Utilizing wild-type icSARS and SARSΔORF6 recombinant viruses, we infected Vero cells that had been previously transfected with Flag-tagged KPNA1 or KPNA2. Eight hours postinfection, cells were fixed and analyzed by confocal microscopy for the localization of KPNA1 and KPNA2 in SARS-infected cells. Using immunofluorescence for the SARS N protein as a marker to identify icSARS-CoV- and icSARSΔORF6-infected cells, KPNA1 in control and virus-infected cells remains localized predominantly in the nucleus, supporting data from previous studies showing that KPNA1 is not interacting directly with ORF6 (Fig. 4F). In contrast, a clear difference in KPNA2 localization was seen in wild-type- and icSARSΔORF6-infected cells. In wild-type virus-infected cells expressing ORF6, KPNA2 was localized to the rER/Golgi. In cultures infected with recombinant virus lacking ORF6, KPNA2 was localized predominantly to the nucleus (Fig. 4F). Taken together, these data support a direct interaction between KPNA2 and ORF6 in the context of virus infection.
KPNB1 relocalizes to the ER/Golgi membrane when ORF6 is present.
Although ORF6 causes a relocalization of KPNA2 from the cytosol to the ER/Golgi membrane, KPNA2 is not directly involved in the translocation of the STAT1:STAT2:IRF9 (ISGF3) complex into the nucleus; rather, KPNA1 interacts with KPNB1 to initiate ISGF3's nuclear localization. If KPNA2 interacted directly with KPNB1, tethering it to the ER/Golgi complex, this association might subsequently limit KPNB1's interaction with the STAT1 complex via KPNA1. To test this hypothesis, 293 cells were transfected with the karyopherin plasmids for 24 h, and coimmunoprecipitations were performed with antibodies directed against the Flag-tagged KPNAs. Western blots were probed with anti-KPNB1 antibody to determine if KPNB1 was coimmunoprecipitated with each KPNA (Fig. 3B, bottom). We find that KPNB1 interacts with each of the four KPNAs tested, confirming data from previously reported studies (43).
We have shown that ORF6 interacts with KPNA2 on the ER/Golgi membrane. If this interaction is similar to KPNA2 binding an NLS on a protein destined for the nucleus, then following KPNA2's binding to an NLS, a conformational change occurs in KPNA2, revealing the N-terminal importin beta binding domain that binds KPNB1 for nuclear import (9). As ORF6 is membrane bound, KPNA2 binding to ORF6 may present the importin beta binding domain for KPNB1 binding and allow KPNB1 to be tethered to the ER/Golgi membrane as well.
To determine whether KPNB1 subcellular localization was altered in ORF6-transfected cells, HA-tagged ORF6 and Flag-tagged KPNB1 were transfected into Vero cells, and the tagged proteins were localized. We find that the KPNB1 distribution was diffusely cytoplasmic and nuclear in the absence of ORF6, consistent with data from previous reports in the literature (33) (Fig. 5A). However, in ORF6-cotransfected cultures, KPNB1 colocalized with ORF6 to the ER/Golgi membrane and was localized mostly to the cytoplasm and not the nucleus.
FIG. 5.
ORF6 interacts with KPNB1. (A) ORF6 colocalization with KPNB1. Vero cells were transfected with Flag-tagged KPNB1 and HA-ORF6. At 24 h posttransfection, cells were fixed and stained. Cells were incubated with mouse anti-HA (αHA) and rabbit anti-Flag antibodies. Anti-Flag antibody was visualized with Alexa Fluor 546-conjugated secondary antibody, and anti-HA antibody was visualized with Alexa Fluor 488-conjugated secondary antibody. Coverslips were visualized with a Zeiss confocal microscope. (B) 293 cells were transfected with HA-tagged ORF6 and Flag-tagged KPNA1, KPNA2, KPNA3, or KPNA4. Cells were lysed 24 h posttransfection and immunoprecipitated (IP) with anti-HA antibody. Proteins were separated on a 4 to 12% SDS PAGE gel and analyzed by Western blot (WB) probing with anti-HA, anti-Flag, and anti-KPNB1 antibody.
To further prove an interaction in vivo, we transfected ORF6 and each KPNA into 293 cells and evaluated whether KPNB1 was coimmunoprecipitated with the ORF6:KPNA2 complex by immunoprecipitation with anti-HA antibody. We found that KPNB1 was no longer associated with each of the four KPNAs as seen in Fig. 3B but rather was coimmunoprecipitated only with the KPNA2:ORF6 complex (Fig. 5B, lane 6). These data support the hypothesis that ORF6 binds KPNA2 as cargo, activating KPNA2's KPNB1 binding domain and tethering KPNB1 to the ORF6:KPNA2 complex on the rER/Golgi membrane.
A KPNA2 mutant binds ORF6 but not KPNB1.
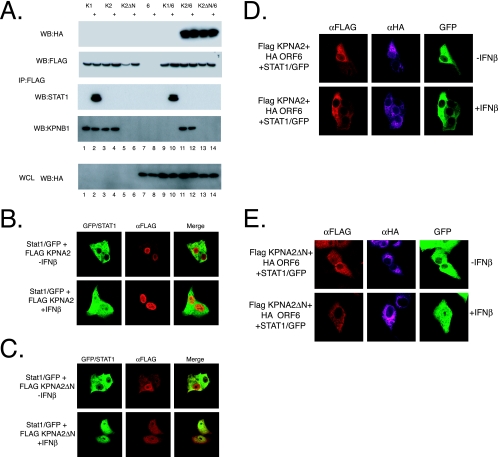
As described above, KPNAs bind to KPNB1 via their N-terminal domain, called the importin beta binding site. If the KPNA2:ORF6 complex recruits KPNB1, then KPNA2 with a deletion of its importin beta binding site would no longer be able to bind KPNB1 in either the presence or absence of ORF6. To test this hypothesis, we produced an N-terminal deletion of KPNA2, deleting amino acids 2 to 80 (KPNA2ΔN). 293 cells were transfected with either Flag-tagged wild-type KPNA1, KPNA2, or KPNA2ΔN in addition to HA-tagged ORF6. At 24 h posttransfection, cells were treated with IFN-β for 60 min and then lysed, and anti-Flag immunoprecipitations were performed. As shown in Fig. 3A, KPNA1 binds to STAT1 only after treatment with IFN-β (Fig. 6A, lane 2). Consistent with previous findings, this interaction is independent of ORF6 expression (Fig. 6A, lanes 9 and 10), since in the presence of ORF6, KPNA1 still binds STAT1 after IFN-β treatment. Moreover, Western blotting of the Flag immunoprecipitates for KPNB1 shows that in the absence of ORF6, KPNB1 associates with KPNA1 and KPNA2 but not KPNA2ΔN, confirming that the importin beta binding site of KPNA2 is necessary for KPNB1 binding. In the presence of ORF6, KPNA1 no longer binds KPNB1; however, under identical conditions, KPNA2 binds KPNB1 efficiently (Fig. 6A, lanes 9 and 11). As predicted, KPNA2ΔN binds ORF6 but no longer interacts with KPNB1 (Fig. 6A, lane 13). Together, these results indicate that ORF6 binds KPNA2 as well as KPNB1; however, this binding does not reduce the KPNA1:STAT1 complex in the cell, and it reduces only the KPNA1:STAT1:KPNB1 complex.
FIG. 6.
KPNA2 interactions with ORF6 and KPNB1. (A) 293 cells were transfected with either Flag-tagged KPNA1, KPNA2, or KPNA2ΔN in combination with HA-tagged ORF6. At 24 h after transfection, cells were treated with 100 IU/ml IFN-β for 30 min, and lysates were collected. Proteins were immunoprecipitated (IP) with anti-Flag antibodies as described in Materials and Methods. Immunoprecipitated extracts were then separated on a 4 to 12% SDS-PAGE gel and analyzed by Western blot (WB) probed with anti-HA, anti-KPNB1, anti-STAT1, and anti-Flag antibodies. Whole-cell extracts (WCL) were also blotted for ORF6 transfection with anti-HA antibodies. (B and C) Vero cells were transfected with STAT1/GFP and either Flag-tagged KPNA2 or KPNA2ΔN. At 24 h posttransfection, cells were treated with 100 IU/ml IFN-β for 60 min before fixation. Proteins were visualized with anti-Flag (αFLAG) antibodies and for the presence of GFP using confocal microscopy. (D and E) Vero cells were transfected as described above (B) except for the addition of HA-tagged ORF6. Cells were stained with anti-Flag and anti-HA antibodies and visualized using confocal microscopy for Flag (Alexa Fluor 546)-, HA (Alexa Fluor 633)-, and GFP-tagged proteins.
While immunoprecipitations provide strong biochemical support for in vitro interactions, we next examined how these interactions would affect STAT1 localization in a cell. To that end, we transfected STAT1/GFP with the wild-type and mutant KPNA constructs in the presence or absence of ORF6 to determine how STAT1 localization was affected following IFN-β treatment. First, we tested the effects of KPNA2 and KPNA2ΔN expression on STAT1/GFP localization to control for potential nuclear import effects associated with plasmid expression of a karyopherin. Flag-tagged KPNA2 and STAT1/GFP were localized in Vero cells before and after the addition of IFN-β. Prior to IFN treatment, STAT1 is localized to the cytoplasm, and KPNA2 is located predominately in the nucleus (Fig. 6B). After IFN treatment, STAT1 efficiently localizes to the nucleus in the presence or absence of Flag-tagged KPNA2 (Fig. 6B). Transfection of Flag-tagged KPNA2ΔN and STAT1/GFP indicated that STAT1 was localized in the cytoplasm before IFN-β treatment; however, KPNA2ΔN was localized to both the cytoplasm and, to a lesser extent, the nucleus (Fig. 6C, D, and E). This agrees with biochemical studies suggesting that KPNA2ΔN is impaired for KPNB1 binding and less efficiently imported to the nucleus. After IFN-β treatment, STAT1 efficiently localizes to the nucleus as before.
As shown by immunoprecipitation, KPNA2ΔN binds ORF6 but no longer binds KPNB1. If KPNB1 recruitment to the ER/Golgi membrane limits STAT1 nuclear localization, then KPNA2ΔN expression in the presence of ORF6 should not block STAT1 nuclear localization in the cell. We find that the cotransfection of STAT1/GFP, wild-type KPNA2, and ORF6 blocks STAT1 nuclear localization after IFN-β treatment (Fig. 6D). Interestingly, the cotransfection of STAT1/GFP, KPNA2ΔN, and ORF6 did not block STAT1 nuclear localization after IFN-β treatment (Fig. 6E). STAT1 was efficiently imported into the nucleus in those cells. These data demonstrate that KPNB1:KPNA2:ORF6 complex formation was necessary to inhibit STAT1 nuclear transport.
The C-terminal tail of ORF6 binds to KPNA2.
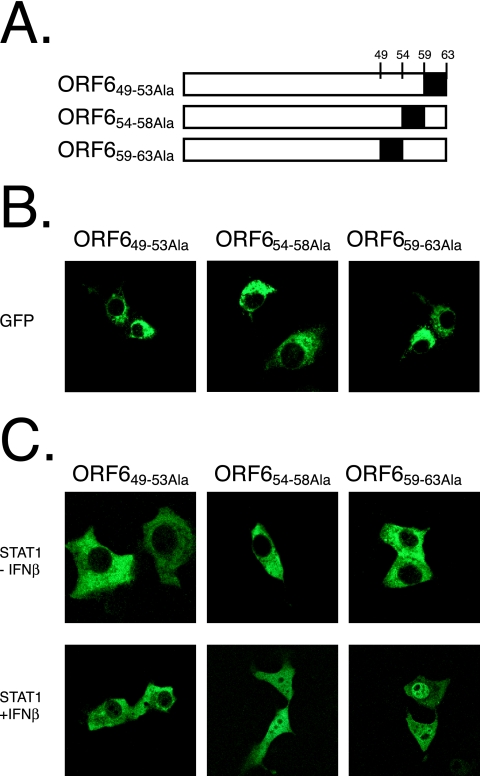
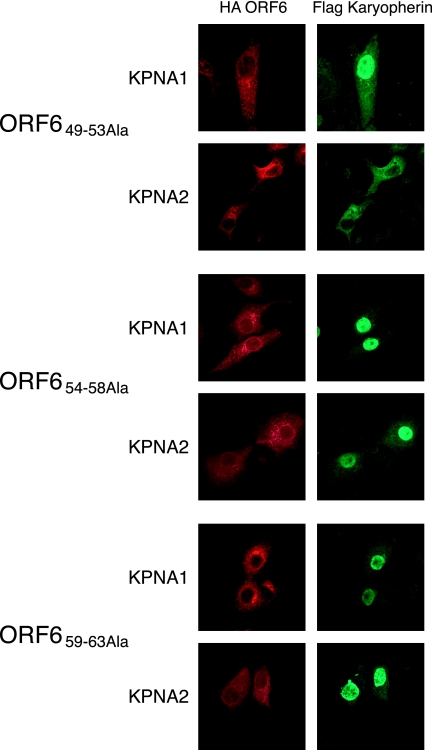
Based on structural predictions, the C terminus of ORF6 was predicted to protrude from the ER/Golgi membrane into the cytoplasm (structure algorithm from http://bp.nuap.nagoya-u.ac.jp/sosui/). We hypothesized that the C terminus of ORF6 may be interacting with the karyopherins and constructed a series of alanine mutations in the C terminus of ORF6 to more precisely map the ORF6 region necessary for antagonist activity. Amino acids 49 to 53, 54 to 58, and 59 to 63 of ORF6 were independently mutated to alanine (Fig. 7A). When fused to GFP, each product localized to the ER/Golgi membrane similarly to wild-type ORF6 (Fig. 7B). After cotransfection with STAT1-GFP into Vero cells, ORF6 with amino acids 49 to 53 mutated to Ala (ORF649-53Ala), but not ORF654-58Ala or ORF659-63Ala, was able to block STAT1 nuclear transport after the addition of IFN-β (Fig. 7C). We further characterized the alanine mutants by determining whether KPNA2 would be retained in the ER/Golgi membrane. Under conditions in which ORF654-58Ala and ORF659-63Ala did not retain KPNA2, the ORF649-53Ala product still retained KPNA2 at the ER/Golgi membrane by confocal microscopy (Fig. 8). These data demonstrate that the C-terminal 10 amino acids of ORF6 are critical for KPNA2 binding and STAT1 blockage.
FIG. 7.
C-terminal mutations of ORF6 affect STAT1 localization. (A) Schematic of ORF6 alanine mutants expressed in B to D. Numbers correspond to amino acids in ORF6. The black box indicates the amino acids that are changed to alanine in the protein (B) Each alanine mutant was fused to a C-terminal GFP and expressed in Vero cells. Localization is shown via a GFP filter on a confocal microscope. (C) Each HA-tagged alanine mutant was cotransfected with STAT1/GFP. At 24 h posttransfection, cells were treated with 100 IU/ml of IFN-β for 60 min before fixation in 4% PFA. STAT1/GFP was visualized on a confocal microscope.
FIG. 8.
C-terminal mutations of ORF6 affect karyopherin localization. (A) HA-tagged alanine scanning mutants of ORF6 (as described in the text) were cotransfected with either Flag-tagged KPNA1 or KPNA2. At 24 h posttransfection, cells were fixed and labeled with anti-HA and anti-Flag antibodies. Cells were visualized using a confocal microscope for Alexa Fluor 488 and Alexa Fluor 546 fluorescence.
DISCUSSION
Viruses have evolved diverse mechanisms to evade host innate and adaptive immunity (1, 2, 4, 6, 16, 19, 20, 30-32, 37, 41, 49). SARS-CoV infection does not induce type I IFN in most cells in culture and likely encodes several proteins that disrupt IFN production. In support of data from previous work (25), we show that SARS-CoV encoding a functional ORF6 protein was able to block STAT1-mediated signaling, while deletion mutants lacking ORF6 could not block STAT1 translocation into the nucleus after IFN treatment. SARS infection also retains KPNA2 at the ER/Golgi membrane; however, in SARSΔORF6-infected cells, KPNA2 resides in the nucleus. ORF6 also retained KPNB1 at the ER/Golgi membrane in complex with KPNA2. Deletion of the N terminus of KPNA2, which binds KPNB1, no longer retained KPNB1 at the ER/Golgi membrane in the presence of ORF6 and did not antagonize STAT1 nuclear import in response to IFN-β.
Under normal conditions, the NLS:KPNA2 interaction reveals an importin beta binding motif at the N terminus of KPNA2 which binds KPNB1. This complex is then imported into the nucleus, where Ran-GTP induces the release of KPNA2 and KPNB1 from the cargo, and it is recycled back out into the cytoplasm and able to bind to new cargo. Our working model is that the cytoplasmic tail of ORF6, acting as a mock NLS, functions to bind and retain KPNA2 on the ER/Golgi membrane (Fig. 9). Since membrane-bound ORF6 is unable to be imported into the nucleus, KPNA2 is retained in a bound state with a functionally exposed importin beta binding domain at its N terminus. Although the importin beta binding domain recruits and binds KPNB1, the complex cannot localize to the nucleus and remains tethered in a bound state. Since the levels of Ran-GTP are very low in the cytoplasm, KPNB1 and KPNA2 cannot release the bound ORF6 cargo and are functionally retained at the ER/Golgi membrane.
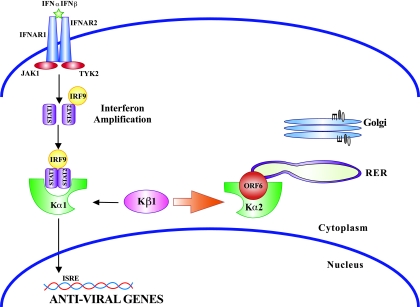
FIG. 9.
Model for ORF6 function. Upon IFN-α or -β stimulation of the IFN receptor on the surface of a cell, the STAT1:STAT2:IRF9 complex is formed. The NLS formed from STAT1 and STAT2 is recognized by KPNA1 (Kα1) for import into the nucleus. SARS-CoV-infected cells that are ER/Golgi membrane localized (shown here only on the ER membrane for clarity) bind to KPNA2, which recruits KPNB1 to the membrane complex as well. This creates a concentration-dependent competition between the ORF6:KPNA2 and ISGF3:KPNA1 complexes for the free unbound KPNB1 in the cytoplasm. This depletion of free KPNB1 in the cytoplasm produces a block in import of the ISGF3 complex in response to both IFN-β and IFN-γ and leaves KPNB1 bound to KPNA2 and ORF6 on the ER/Golgi membrane.
ORF6 and innate immunity.
Unlike other IFN antagonists that have cytoplasmic localizations (for example, Ebola virus VP24 [47] and VP35 [2], Nipah virus V [53], and hepatitis C virus [19]), ORF6 is uniquely localized to the ER/Golgi membrane. ORF6 binds to KPNA2, retargeting its localization from the cytoplasm to the ER/Golgi membrane. Although KPNA2 does not participate directly in STAT1 import, the retention of KPNA2 on the ER/Golgi membrane and the subsequent recruitment of KPNB1 to the ER/Golgi membrane as well probably deplete KPNB1 concentrations in the cytoplasm, retarding the translocation of the STAT1:KPNA1 complex from entering the nucleus. In virus-infected but not uninfected cells, ORF6 expression should block STAT1 signaling in the context of either autocrine or paracrine signaling, suggesting that the SARS-CoV-infected cell would be highly resistant to IFN and provide a protected environment for efficient SARS-CoV replication. Experiments examining the dynamics of STAT1 localization in icSARS-CoV and icSARSΔORF6 during in vivo infection in mice are in progress. The ORF6:KPNA2:KPNB1 interaction also has the potential to simultaneously antagonize several innate immune pathways in infected cells. Reductions in the concentration of free cytoplasmic KPNB1 pools would reduce the rate of KPNA- and KPNB-mediated transport as well as their cargo involved in innate immune responses to viral infection (e.g., AP-1, IRF3, IRF7, and STAT1) (8). KPNA2 also imports other cargo into the nucleus of cells, a function that is likely antagonized by ORF6. For example, NF-κB uses KPNA2 for nuclear transport (10), and we have shown that NF-κB-inducible genes are poorly induced during SARS infection in vitro (15).
ORF6 and nucleocytoplasmic shuttling.
Cytoplasmic protein levels of KPNB1 have been shown to be important for the rapid kinetics of NLS-containing cargo. Changes in the levels of injected KPNB1 in cells can drastically modify the nuclear import kinetics of a protein (59). Yang and Musser found that under conditions of low KPNB1 protein concentration, only ∼50% of the nuclear pore-associated cargos are imported into the nucleus, yet with high concentrations of KPNB1, ∼80% of the cargo is imported into the nucleus and with a sevenfold-faster transit (59). Alterations of the free KPNB1 concentration in the cytoplasm by ORF6 may mimic the lower concentration of KPNB1 seen in those experiments and explain the diminished STAT1 nuclear translocation.
Our data predict that the relative concentrations of ORF6, KPNB1, and KPNA2 within an infected cell will influence the efficiency of the ORF6-STAT1 antagonistic activity. SARS-CoV infection has been reported in bronchial epithelium, type I and II pneumocytes, T lymphocytes, macrophages/monocytes, fibroblasts, vascular endothelial cells, epithelial mucosa of the intestine, and neurons. Karyopherins, especially KPNA2, are differentially expressed in different tissues and cell types in vivo (39). The antagonistic activity of SARS-CoV ORF6 may be ineffective in some virus-infected tissues due to lower KPNA2 expression levels. One possible explanation for the finding that SARS infection of plasmacytoid (pDC), but not conventional dendritic cells, induced large amounts of IFN (5) may be related to the levels of karyopherins. If there are low levels of KPNA2 in a cell, the effect of ORF6 to block STAT1 will be diminished. Alternatively, various levels of ORF6 and other SARS IFN antagonists may be expressed in different cell types. For example, SARS-CoV infection in macrophages and dendritic cells has been characterized by low levels of viral gene expression and the release of few viral progeny (5). If the SARS-CoV IFN antagonists are not expressed at high enough levels in those cell types, their ability to block the IFN sensing or signaling pathways would be diminished, leading to increased IFN production.
SARS-CoV infection is most severe in the elderly (3, 13, 29, 42). Karyopherin expression varies with the age of the cell as well (46). In fact, nuclear import/export-associated KPNA2, Ran binding protein 1, and cellular apoptosis susceptibility (an exportin) mRNAs are found to be down-regulated in fibroblasts from old donors compared to fibroblasts from young donors (46). In this study, old human fibroblasts further showed reduced import rates of a reporter protein compared to those of young donor fibroblasts, suggesting that older cells and, by analogy, older individuals may be more susceptible to SARS due to altered nuclear import/export machinery levels. Recently, it has been shown that SARS infection of senescent mice increases pathogenesis and weight loss in comparison to young mice (12, 58). Senescent mice show increased viral titers and lung pathology, suggesting that altered nuclear import mechanics may contribute to a less developed and robust innate and adaptive immune response.
ORF6 and pathogenesis.
Previous work has shown that the insertion of ORF6 into a nonvirulent strain of MHV resulted in disease enhancement and death in a mouse model of MHV (44). Tangudu et al. further showed that ORF6 accelerates the replication of MHV, a phenotype that may contribute to increased lethality in mice (57). Those data, in agreement with our findings, support the hypothesis that ORF6 is an important virulence protein during in vivo infection. Although the mechanism of action in the MHV backbone is unknown, it seems plausible that ORF6 antagonism of the KPNA2 import machinery is a likely possibility. In those studies, however, ORF6 was expressed from a relatively weak mRNA 4 transcription regulatory sequence, so it will be very important to carefully evaluate and compare levels of ORF6 expression in heterologous (MHV) and homologous (SARS-CoV) genome backbones because the abundance of product correlates with STAT1 antagonism. We are currently investigating the role of ORF6 in in vivo pathogenesis using our deletion virus and the mouse-adapted SARS-CoV model (48).
Consonant with the findings reported in this paper, STAT1 knockout mice are unable to clear SARS-CoV infection. Virus titers remain high, and infection rapidly disseminates from the lungs to the liver and spleen, demonstrating the importance of the STAT1 pathway in regulating disease severity in vivo (21). Although the removal of STAT1 leads to increased SARS-CoV-induced disease, this does not preclude virus-mediated antagonism of STAT1. ORF6 mediated the antagonism of STAT1, which, unlike the knockout animal, is unlikely to be 100% efficient and probably functions to attenuate signaling by type I and type II IFNs in infected cells, thereby promoting viral replication and spread. An ORF6 block in type I and type II IFN amplification in infected tissue will greatly diminish the amplification of the IFN signal to surrounding cells. Therefore, additional studies to assess the contribution of ORF6 to the in vivo pathogenesis of SARS-CoV will be extremely important.
The mechanism by which ORF6 antagonizes the IFN signaling pathway via nuclear import machinery may well be an unexplored commonality of many diverse viruses, with each virus having its own mechanism of inhibiting nuclear import (for example, Ebola virus VP24 versus SARS-COV ORF6). Compared with antagonists that target individual cargo and signaling cascades, the perturbation of nuclear import machinery may affect a broader range of host signaling networks and contribute significantly to increased pathogenesis. The identification of other viral proteins inhibiting similar pathways will contribute to our understanding of the mechanisms responsible for the pathogenesis of SARS-CoV, identify novel viral drug targets, and provide essential tools to investigate nuclear import.
Acknowledgments
We thank Robert Bagnell of the UNC Microscopy Center, the UNC Histology Core, the UNC Sequencing Core, and the UNC Nucleic Acids Facility for their assistance. We also thank Megan Shaw (Mount Sinai Medical Center, NY), Nancy Reich (University of Stony Brook, NY), and Stephen Mishnick (University of Montreal) for reagents. We also thank Richard Frieman for illustration assistance.
M.F. is supported by an NIH NRSA postdoctoral fellowship (F32-AI066542); R.S.B., M.H., and P.P. are supported by NIH P01-AI05944302; and P.P. is a Senior Fellow of the Ellison Medical Foundation.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Ahn, K., T. H. Meyer, S. Uebel, P. Sempe, H. Djaballah, Y. Yang, P. A. Peterson, K. Fruh, and R. Tampe. 1996. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. 15:3247-3255. [PMC free article] [PubMed] [Google Scholar]
- 2.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booth, C. M., L. M. Matukas, G. A. Tomlinson, A. R. Rachlis, D. B. Rose, H. A. Dwosh, S. L. Walmsley, T. Mazzulli, M. Avendano, P. Derkach, I. E. Ephtimios, I. Kitai, B. D. Mederski, S. B. Shadowitz, W. L. Gold, L. A. Hawryluck, E. Rea, J. S. Chenkin, D. W. Cescon, S. M. Poutanen, and A. S. Detsky. 2003. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 289:2801-2809. [DOI] [PubMed] [Google Scholar]
- 4.Cardenas, W. B., Y. M. Loo, M. Gale, Jr., A. L. Hartman, C. R. Kimberlin, L. Martinez-Sobrido, E. O. Saphire, and C. F. Basler. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cervantes-Barragan, L., R. Zust, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2006. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Changeux, S. P., and P. Palese (ed.). 2005. Modulation of host gene expression and innate immunity by viruses. Springer, Heidelberg, Germany.
- 7.Charley, B., and H. Laude. 1988. Induction of alpha interferon by transmissible gastroenteritis coronavirus: role of transmembrane glycoprotein E1. J. Virol. 62:8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chook, Y. M., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703-715. [DOI] [PubMed] [Google Scholar]
- 9.Cingolani, G., C. Petosa, K. Weis, and C. W. Muller. 1999. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399:221-229. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham, M. D., J. Cleaveland, and S. G. Nadler. 2003. An intracellular targeted NLS peptide inhibitor of karyopherin alpha:NF-kappa B interactions. Biochem. Biophys. Res. Commun. 300:403-407. [DOI] [PubMed] [Google Scholar]
- 11.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deming, D., T. Sheahan, M. Heise, B. Yount, N. Davis, A. Sims, M. Suthar, J. Harkema, A. Whitmore, R. Pickles, A. West, E. Donaldson, K. Curtis, R. Johnston, and R. Baric. 2006. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 3:e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly, C. A., A. C. Ghani, G. M. Leung, A. J. Hedley, C. Fraser, S. Riley, L. J. Abu-Raddad, L. M. Ho, T. Q. Thach, P. Chau, K. P. Chan, T. H. Lam, L. Y. Tse, T. Tsang, S. H. Liu, J. H. Kong, E. M. Lau, N. M. Ferguson, and R. M. Anderson. 2003. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 361:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 15.Frieman, M., M. Heise, and R. Baric. 20 April 2007, posting date. SARS coronavirus and innate immunity. Virus Res. [Epub ahead of print.] doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed]
- 16.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 17.Glass, W. G., K. Subbarao, B. Murphy, and P. M. Murphy. 2004. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 173:4030-4039. [DOI] [PubMed] [Google Scholar]
- 18.Gorlich, D., P. Henklein, R. A. Laskey, and E. Hartmann. 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810-1817. [PMC free article] [PubMed] [Google Scholar]
- 19.Hiscott, J., J. Lacoste, and R. Lin. 2006. Recruitment of an interferon molecular signaling complex to the mitochondrial membrane: disruption by hepatitis C virus NS3-4A protease. Biochem. Pharmacol. 72:1477-1484. [DOI] [PubMed] [Google Scholar]
- 20.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53-56. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, R. J., G. Gao, T. Rowe, P. Bell, D. Flieder, J. Paragas, G. P. Kobinger, N. A. Wivel, R. G. Crystal, J. Boyer, H. Feldmann, T. G. Voss, and J. M. Wilson. 2004. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 78:11416-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C., N. Ito, C. T. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 80:7287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, C., C. J. Peters, and S. Makino. 2007. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 81:5423-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamitani, W., K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 103:12885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopecky-Bromberg, S. A., L. Martínez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 27.La Bonnardiere, C., and H. Laude. 1983. Interferon induction in rotavirus and coronavirus infections: a review of recent results. Ann. Rech. Vet. 14:507-511. (In French.) [PubMed] [Google Scholar]
- 28.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 30.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKɛ molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo, H. W., M. Ali-Seyed, Y. Wu, G. Bartholomeusz, S. C. Hsu, and M. C. Hung. 2006. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J. Cell. Biochem. 98:1570-1583. [DOI] [PubMed] [Google Scholar]
- 34.McBride, K. M., G. Banninger, C. McDonald, and N. C. Reich. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 21:1754-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride, K. M., C. McDonald, and N. C. Reich. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1transcription factor. EMBO J. 19:6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride, K. M., and N. C. Reich. 2003. The ins and outs of STAT1 nuclear transport. Sci. STKE 2003:RE13. [DOI] [PubMed] [Google Scholar]
- 37.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 38.Moroianu, J., G. Blobel, and A. Radu. 1996. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc. Natl. Acad. Sci. USA 93:6572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadler, S. G., D. Tritschler, O. K. Haffar, J. Blake, A. G. Bruce, and J. S. Cleaveland. 1997. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 272:4310-4315. [DOI] [PubMed] [Google Scholar]
- 40.Nyfeler, B., S. W. Michnick, and H. P. Hauri. 2005. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. USA 102:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 42.Peiris, J. S., K. Y. Yuen, A. D. Osterhaus, and K. Stohr. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 43.Pemberton, L. F., G. Blobel, and J. S. Rosenblum. 1998. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 10:392-399. [DOI] [PubMed] [Google Scholar]
- 44.Pewe, L., H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 79:11335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon, L. L., D. K. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. Leung, S. K. Lau, P. C. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pujol, G., H. Soderqvist, and A. Radu. 2002. Age-associated reduction of nuclear protein import in human fibroblasts. Biochem. Biophys. Res. Commun. 294:354-358. [DOI] [PubMed] [Google Scholar]
- 47.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaecher, S. R., J. M. Mackenzie, and A. Pekosz. 2007. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 81:718-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shabman, R. S., T. E. Morrison, C. Moore, L. White, M. S. Suthar, L. Hueston, N. Rulli, B. Lidbury, J. P. Ting, S. Mahalingam, and M. T. Heise. 2007. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J. Virol. 81:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw, M. L., A. Garcia-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen, S., P. S. Lin, Y. C. Chao, A. Zhang, X. Yang, S. G. Lim, W. Hong, and Y. J. Tan. 2005. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 330:286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taguchi, F., and S. G. Siddell. 1985. Difference in sensitivity to interferon among mouse hepatitis viruses with high and low virulence for mice. Virology 147:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tangudu, C., H. Olivares, J. Netland, S. Perlman, and T. Gallagher. 2007. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 81:1220-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel, L. N., A. Roberts, C. D. Paddock, G. L. Genrich, E. W. Lamirande, S. U. Kapadia, J. K. Rose, S. R. Zaki, and K. Subbarao. 2007. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV). Vaccine 25:2173-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, W., and S. M. Musser. 2006. Nuclear import time and transport efficiency depend on importin beta concentration. J. Cell Biol. 174:951-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 81:2554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng, B. J., K. H. Wong, J. Zhou, K. L. Wong, B. W. Young, L. W. Lu, and S. S. Lee. 2004. SARS-related virus predating SARS outbreak, Hong Kong. Emerg. Infect. Dis. 10:176-178. [DOI] [PMC free article] [PubMed] [Google Scholar]