Abstract
The ataxia telangiectasia-mutated (ATM) protein, a member of the related phosphatidylinositol 3-like kinase family encoded by a gene responsible for the human genetic disorder ataxia telangiectasia, regulates cellular responses to DNA damage and viral infection. It has been previously reported that herpes simplex virus type 1 (HSV-1) infection induces activation of protein kinase activity of ATM and hyperphosphorylation of transcription factor, Sp1. We show that ATM is intimately involved in Sp1 hyperphosphorylation during HSV-1 infection rather than individual HSV-1-encoded protein kinases. In ATM-deficient cells or cells silenced for ATM expression by short hairpin RNA targeting, hyperphosphorylation of Sp1 was prevented even as HSV-1 infection progressed. Mutational analysis of putative ATM phosphorylation sites on Sp1 and immunoblot analysis with phosphopeptide-specific Sp1 antibodies clarified that at least Ser-56 and Ser-101 residues on Sp1 became phosphorylated upon HSV-1 infection. Serine-to-alanine mutations at both sites on Sp1 considerably abolished hyperphosphorylation of Sp1 upon infection. Although ATM phosphorylated Ser-101 but not Ser-56 on Sp1 in vitro, phosphorylation of Sp1 at both sites was not detected at all upon infection in ATM-deficient cells, suggesting that cellular kinase(s) activated by ATM could be involved in phosphorylation at Ser-56. Upon viral infection, Sp1-dependent transcription in ATM expression-silenced cells was almost the same as that in ATM-intact cells, suggesting that ATM-dependent phosphorylation of Sp1 might hardly affect its transcriptional activity during the HSV-1 infection. ATM-dependent Sp1 phosphorylation appears to be a global response to various DNA damage stress including viral DNA replication.
Transcription factor Sp1 is a 95- to 105-kDa protein that binds to GC-rich recognition elements (GC-boxes) through C-terminal zinc finger motifs (30). Sp1 functions as a transactivator of gene expression, and its recognition elements are distributed widely in various promoters of cellular and viral genes (15, 16, 19). As with many other transcription factors, the transcription activity of Sp1 is regulated in part by posttranslational modifications, which include phosphorylation, glycosylation, acetylation, and sumoylation (5, 9, 26, 54). It has been recently demonstrated that phosphorylation of Sp1 alters its transcription activity in a wide variety of physiological processes, including cell cycle progression, terminal differentiation, and viral infection (2, 9, 17, 23, 28, 36, 41). Also, relationships between the phosphorylation site(s) on Sp1 and specific kinases have been clarified, with documentation of targeting of Ser-59 by cyclin A-CDK, Ser-220 (quoted as Ser-131 in the original study) by DNA-dependent protein kinase (DNA-PK), Thr-453 and Thr-739 by p42/p44 mitogen-activated protein kinase (MAPK), and Thr-668 (quoted as Thr-579 in the original study) by casein kinase II (2, 9, 11, 17, 41). Cyclin A-CDK-mediated phosphorylation increases DNA-binding activity of Sp1 (17, 23). Inactivation of Thr-453 and Thr-739 phosphorylation sites by p42/p44 MAPK decreases by half the transcriptional activity (41). Mutation of the Thr-668 phosphorylation site which is in the Zinc finger motif of Sp1 increases its DNA-binding activity, further suggesting that casein kinase II-mediated phosphorylation decreases DNA-binding activity of Sp1 (2).
Phosphorylation of Sp1 upon virus infection has been reported in several studies (9, 11, 28, 31). For example, simian virus 40 infection induces both increased levels and phosphorylation of Sp1 (28). In a study of human immunodeficiency virus type 1 (HIV-1) infection, it was shown that the HIV-1-encoded Tat protein promotes DNA-PK-dependent Sp1 phosphorylation in vitro, which is associated with increased transcription activity (11).
Herpes simplex virus type 1 (HSV-1) is a large, enveloped virus with 152-kbp double-stranded DNA encoding approximately 84 proteins (39, 49). During productive replication, cascade regulation of gene expression occurs, based on stepwise activation of immediate-early, early, early late, and late promoters (24). The promoters of different expression kinetics classes are equipped for binding of not only viral transcription factors but also various cellular transcription factors, including Sp1. Sp1-binding sites are frequently found in promoters of immediate-early and early genes (48, 57), suggesting a pivotal function of Sp1 in gene expression of HSV-1. HSV-1 infection induces hyperphosphorylation of Sp1 at early stages of infection without any significant change in abundance (31). While the DNA-binding activity of Sp1 was found to be unchanged until 8 h postinfection (hpi), purified Sp1 from HSV-1-infected cells at 12 hpi had reduced transcription activity in vitro (31). However, the kinase(s) responsible for Sp1 phosphorylation induced by HSV-1 infection have remained unclear.
We and others previously reported that HSV-1 infection induces a cellular DNA damage response, with activation of the ATM signal transduction pathway (37, 53, 60). The activated form of ATM phosphorylated at Ser-1981 and the DNA damage sensor Mre11-Rad50-Nbs1 complex are recruited and retained in viral replication compartments, where transcription and replication of viral genes take place. Activation of the ATM-Rad3-related (ATR) replication checkpoint pathway, in contrast, is minimal. ATM is a member of the related phosphatidylinositol 3 (PI-3)-like kinase family, as well as ATR and DNA-PK, and displays kinase activity against serine and threonine, followed by glutamine, generally responding to a genotoxic stress such as ionizing radiation (IR) (1, 6, 12, 13, 52).
In the present report, we provide evidence for considerable ATM involvement in the hyperphosphorylation of Sp1 during HSV-1 infection rather than individual HSV-1-encoded PKs. In ATM-deficient cells or cells silenced for ATM expression by short hairpin RNA (shRNA) targeting, the levels of the hyperphosphorylated form of Sp1 did not increase even as HSV-1 infection progressed. Using mutational analysis and immunoblotting with phosphopeptide-specific antibodies, we have identified that at least Ser-56 and Ser-101 on Sp1 became phosphorylated in response to HSV-1 infection. Although ATM phosphorylated Ser-101 but not Ser-56 on Sp1 in vitro, phosphorylation of Sp1 at both sites was not detected at all upon infection in ATM-deficient cells, suggesting that other cellular kinase(s) activated by ATM could be involved in phosphorylation at Ser-56. Upon viral infection Sp1-dependent transcription in ATM expression-silenced cells was almost the same as that in ATM-intact cells, suggesting that ATM-dependent phosphorylation of Sp1 might hardly affect its transcriptional activity during viral infection. Since it is known that HSV-1 infection decreases transcriptional activity of Sp1 (31), modification(s) of Sp1 besides ATM-dependent phosphorylation might affect its transcriptional activity.
MATERIALS AND METHODS
Cells.
HeLa, HFF2 (human foreskin fibroblasts immortalized by introduction of the human telomerase reverse-transcriptase [hTERT] gene [56]), and 293T cells were grown and maintained at 37°C in Dulbecco modified Eagle medium (DMEM; Sigma) supplemented with 10% fetal calf serum (FCS). Human glioma cell lines M059J and MJ-M6 (34) were maintained in DMEM-Ham F-12 nutrient mixture (Sigma) supplemented with 10% FCS and puromycin (0.5 μg/ml). Skin fibroblasts from ataxia telangiectasia patients immortalized by introduction of hTERT gene (AT10S/T-n cells) were maintained at 37°C in DMEM supplemented with 10% FCS and G418 (200 μg/ml) (43). The 293T cells were infected with retroviruses expressing ATM shRNA or control retroviruses and selected (56). The resultant 293T cells stably expressing ATM shRNA (293T-ATM shRNA) or the control vector cells (293T-Control vector) were maintained at 37°C in DMEM supplemented with 10% FCS and hygromycin B (100 μg/ml) (56). Sf9 and SF21 cells were maintained at 27°C in Sf-900 II (Gibco) supplemented with 10% FCS.
Viruses.
HSV-1 strain 17+ was used throughout the experiments. A Us3-deficient virus, R7041 (46), and a UL13-deficient virus, R7356 (47), derived from HSV-1 strain F were kindly provided by B. Roizman. The UL39-deficient virus, ICP6Δ, derived from HSV-1 strain KOS was from a collaborator, S. K. Weller (20). Infection was performed on monolayers of cultured cells at the indicated multiplicities of infection (MOIs). After 1 h adsorption at 37°C, inoculum was removed, and monolayers were overlaid with fresh medium.
Plasmids.
For subcloning of Sp1 gene into pFastBac1 (Invitrogen) containing a sequence (RGS-6xHis-Flag) supplied by W. Nakai, two primers (5′-GGAATTCCATATGGATGAAATGACAGCTGTGG-3′ and 5′-GCTCTAGATCAGAAGCCATTGC-3′) were designed. Plasmid CMV-hSp1 (22) supplied from G. Suske was used as a template for PCR. The PCR product was double digested by NdeI and XbaI and subcloned into pFastBac1 containing a sequence (RGS-6xHis-Flag), and the resultant plasmid was designated pFB-RHF/Sp1. For construction of a mammalian expression vector for Sp1, two primers (5′-TTATAATAGGGGTACCCCACCATGCGCGG-3′ and 5′-GCTCTAGAGCTCAGAAGCCA-3′) were designed. pFB-RHF/Sp1 was used as a template for PCR. The PCR product was double digested by KpnI and XbaI and subcloned into pcDNA3.1(+) (Invitrogen), and the resultant plasmid was designated pcDNA-RHF/Sp1. Serine/threonine-to-alanine mutations were constructed with a QuikChange site-directed mutagenesis kit (Stratagene) using the primer sets given in Table 1. Multiple mutations were introduced by repetition. The plasmids were designated pcDNA-RHF/Sp1-S36A, pcDNA-RHF/Sp1-S56A, pcDNA-RHF/Sp1-S81A, pcDNA-RHF/Sp1-S85A, pcDNA-RHF/Sp1-T98A, pcDNA-RHF/Sp1-S101A, pcDNA-RHF/Sp1-T250A/S281A, pcDNA-RHF/Sp1-S291/296A, pcDNA-RHF/Sp1-S313A, pcDNA-RHF/Sp1-S351A, pcDNA-RHF/Sp1-T394A, pcDNA-RHF/Sp1-T427A/S431A, pcDNA-RHF/Sp1-S56/81/85A, pcDNA-RHF/Sp1-S56/101A, pcDNA-RHF/Sp1-S56A/T250A, and pcDNA-RHF/Sp1-S56/281A, and all mutations were confirmed by sequencing.
TABLE 1.
Primer sets for construction of serine/threonine-to-alanine mutations by site-directed mutagenesis
| Mutation site | Sequences of paired primers (5′-3′) |
|---|---|
| S36A | GGTGGTGCCTTTGCACAGGCTCGAA |
| TTCGAGCCTGTGCAAAGGCACCACC | |
| S56A | GGAGGGCAGGAGGCCCAGCCATCCC |
| GGGATGGCTGGGCCTCCTGCCCTCC | |
| S81A | AGAACAGCAACAACGCCCAGGGCCCGAGTC |
| GACTCGGGCCCTGGGCGTTGTTGCTGTTCT | |
| S85A | CTCCCAGGGCCCGGCTCAGTCAGGGGGAAC |
| GTTCCCCCTGACTGAGCCGGGCCCTGGGAG | |
| T98A | GACCTCACAGCCGCACAACTTTCAC |
| GTGAAAGTTGTGCGGCTGTGAGGTC | |
| S101A | GCCACACAACTTGCACAGGGTGCCA |
| TGGCACCCTGTGCAAGTTGTGTGGC | |
| T250A | CTCTCAGGACAGGCTCAGTATGTGA |
| TCACATACTGAGCCTGTCCTGAGAG | |
| S281A | CCTTGACTCCCAGCGCTCAGGCAGTCACGA |
| TCGTGACTGCCTGAGCGCTGGGAGTCAAGG | |
| S291A/S296A | AGCAGCTCTGGGGCCCAGGAGAGTGGCGCACAGCCTGTCA |
| TGACAGGCTGTGCGCCACTCTCCTGGGCCCCAGAGCTGCT | |
| S313A | AGCTTGGTATCAGCACAAGCCAGTT |
| AACTGGCTTGTGCTGATACCAAGCT | |
| S351A | TCAGGGACCAACGCTCAAGGCCAGA |
| TCTGGCCTTGAGCGTTGGTCCCTGA | |
| T394A | CAAAACCAGCAGGCACAGCA |
| TGCTGTGCCTGCTGGTTTTG | |
| T427A/S431A | GGGCAGACCTTTGCAGCTCAAGCCATCGCCCAGGAAACCC |
| GGGTTTCCTGGGCGATGGCTTGAGCTGCAAAGGTCTGCCC |
For subcloning of a glutathione S-transferase (GST) fusion protein with truncated Sp1 (8 to 167 amino acids), two primers (5′-CGGGATCCCGTATGGATGAAATGAC-3′ and 5′-ACTCTCGAGCACTCCAGGTAGT-3′) were designed, and pcDNA-RHF/Sp1, pcDNA-RHF/Sp1-S56A, pcDNA-RHF/Sp1-S101A, and pcDNA-RHF/Sp1-S56/101A were used as templates for PCR. The PCR products were double digested with BamHI and XhoI and subcloned into pGEX-6P-3 (Amersham Biosciences). The plasmids were designated pGEX-6P-Sp18-167, pGEX-6P-Sp18-167-S56A, pGEX-6P-Sp18-167-S101A, and pGEX-6P-Sp18-167-S56/101A.
Purification of recombinant Sp1 from insect cells.
Purification of recombinant His6 flag-tagged Sp1 from insect cells was carried out with a Bac-to-Bac system (Invitrogen). DH10Bac Escherichia coli cells were transformed with pFB-RHF/Sp1, and the resultant bacmid was transfected into Sf9 cells using Lipofectin reagent (Invitrogen). The obtained virus was designated AcRHF/Sp1. SF21 cells (5 × 107) were infected with AcRHF/Sp1 at an MOI of 5. At 60 hpi, the cells were harvested, suspended in a buffer (50 mM Tris-HCl [pH 8], 1% Nonidet P-40 [NP-40], 250 mM NaCl, 10 mM 2-mercaptoethanol [2-ME], 1 mM phenylmethylsulfonyl fluoride [PMSF]), sonicated, and centrifuged. The clarified lysate was combined with Ni-NTA (QIAGEN). After rotation for 2 h, the beads were washed twice with buffer A (20 mM Tris-HCl [pH 8], 10% glycerol, 10 mM 2-ME) containing 500 mM KCl and 20 mM imidazole, once with buffer A containing 1 M KCl, once with buffer A containing 500 mM KCl and 20 mM imidazole, and twice with buffer A containing 100 mM KCl and 20 mM imidazole and then eluted with buffer A containing 100 mM KCl and 100 mM imidazole. The eluted protein was dialyzed against dialysis buffer (20 mM Tris-HCl [pH 8], 100 mM NaCl, 20% glycerol, and 1 mM PMSF) and combined with anti-Flag M2 affinity gel (Sigma). After rotation for 3 h, the beads were washed three times with TBS+ buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol, 1 mM PMSF), and the protein was eluted with 0.1 M glycine (pH 3.5), neutralized immediately, dialyzed against dialysis buffer, and stored at −80°C.
Purification of recombinant Sp1 from bacteria.
E. coli (BL21) transformed with pGEX-6P-Sp18-167, pGEX-6P-Sp18-167-S56A, pGEX-6P-Sp18-167-S101A, and pGEX-6P-Sp18-167-S56/101A were cultured at 30°C until the optical density at 600 nm reached 0.6, and IPTG (isopropyl-β-d-thiogalactopyranoside) was added at a final concentration of 1 mM. After 5 h of incubation, the collected cells were suspended in GLB buffer (50 mM Tris-HCl [pH 7.4], 50 mM glucose, 1 mM EDTA, 10 mM 2-ME, 1 mM PMSF, 0.2% NP-40). After sonication, the lysate was clarified by centrifugation, combined with glutathione-Sepharose 4B (Amersham Biosciences), and rotated at 4°C for 90 min. After three washes with GLB containing 0.2% NP-40, the proteins were eluted with elution buffer (10 mM reduced glutathione, 50 mM Tris-HCl [pH 8], 1 mM PMSF) and dialyzed against dialysis buffer. The purified proteins were designated GST-Sp18-167, GST-Sp18-167-S56A, GST-Sp18-167-S101A, and GST-Sp18-167-S56/101A.
IP-kinase assays.
Immunoprecipitation (IP)-kinase assays were performed with some modification as described previously (6, 10, 32). 293T cells (5 × 106) were transfected with either pcDNA-Flag-ATMwt or pcDNA-Flag-ATMkd (5 μg of each; kind gifts from M. B. Kastan [6]) using Lipofectamine 2000 (Invitrogen). At 48 h posttransfection, cells were harvested, rinsed with ice-cold phosphate-buffered saline, lysed with lysis buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100 [TX-100], 5% glycerol, 1 mM PMSF, 100 mM NaF, 2 mM Na3VO4, and complete protease inhibitor [Roche]), incubated for 15 min on ice, sonicated, and clarified by centrifugation. Cell lysates (2 mg) were incubated with anti-Flag M2 affinity resin (20 μl of suspension) and rotated for 3 h at 4°C. The immunocomplex was washed three times with lysis buffer containing 0.65% TX-100, twice with Tris-LiCl buffer (100 mM Tris-HCl [pH 7.5], 0.5 M LiCl, 1 mM NaF, 1 mM Na3VO4) and once with kinase buffer (10 mM HEPES [pH 7.9], 50 mM glycerophosphate, 50 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM dithiothreitol, 1 mM NaF, and 1 mM Na3VO4) containing 5 μM ATP. For activation of ATM, the immunocomplex was incubated with kinase buffer containing 1 mM ATP for 30 min at 30°C. After incubation, the immunocomplex was washed three times with kinase buffer containing 5 μM ATP and divided into two portions for kinase reactions with either purified Sp1 or p53 as substrate. Kinase reactions were carried out by resuspending the immunocomplex in kinase buffer containing 10 μCi [γ-32P]ATP and either 500 ng of Sp1 purified from insect cells or 250 ng of p53 (Active Motif), followed by incubation for 30 min at 30°C. The reactions were terminated by addition of sodium dodecyl sulfate (SDS) gel loading buffer, and the samples were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE), followed by autoradiography. In the case of GST-Sp18-167, GST-Sp18-167-S56A, GST-Sp18-167-S101A, and GST-Sp18-167-S56/101A, 1 μg of each protein was used as a substrate.
Antibodies.
Primary antibodies were purchased from Santa Cruz (Sp1 PEP2), Abcam (HSV-1 ICP4, UL42), GeneTex (ATM-2C1), Cell Signaling Technology (ATM-S1981), Ambion (GAPDH [glyceraldehyde-3-phosphate dehydrogenase]), Sigma (Flag M2), and Biosource (DNA-dependent PK catalytic subunit). Phosphopeptide-specific rabbit antibodies were raised against serine-56 phosphopeptide, CGGGQEpSQPSPL, for anti-Sp1 (pS56) and serine-101 phosphopeptide, CTATQLpSQGANG, for anti-Sp1 (pS101), conjugated with KLH. Antibodies were purified through the specific phosphopeptide-conjugated columns and passed through the corresponding unphosphorylated peptide-conjugated columns.
Transient-transfection and infection.
HeLa cells (6 × 105) were transfected with 0.8 μg of pcDNA-RHF/Sp1 or expression vectors of mutated Sp1 using Lipofectamine 2000. At 24 h posttransfection, the transfected cells were infected with HSV-1 at an MOI of 10. At 24 hpi, the cells were harvested and subjected to immunoblot analysis.
IP.
HeLa cells transfected with pcDNA-RHF/Sp1, pcDNA-RHF/Sp1-S56A, or pcDNA-RHF/Sp1-S101A were infected with HSV-1 at an MOI of 10. At 24 hpi, the cells were harvested, suspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% TX-100, 100 mM NaF, 2 mM Na3VO4, and protease inhibitor cocktail [Sigma]), and centrifuged. The clarified lysate was combined with anti-Flag M2 affinity resin and rotated for 3 h at 4°C. The immune complex was washed three times with TBS buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl), and eluted with SDS gel loading buffer.
Immunoblot analysis.
HeLa, HFF2, M059J, MJ-M6, and AT10S/T-n cells were suspended in lysis buffer (20 mM Tris-HCl [pH 7.4], 0.5% TX-100, 300 mM NaCl, 1 mM EDTA, 0.1% SDS, 100 mM NaF, 2 mM Na3VO4, protease inhibitor cocktail [Sigma]) and incubated on ice for 40 min, followed by centrifugation to obtain clarified supernatants. 293T-ATM shRNA and 293T-Control vector cells were suspended in urea buffer (8 M urea, 0.1 M NaH2PO4, 10 mM Tris [pH 8]), sonicated, and centrifuged. For experiments with alkaline phosphatase treatment, HFF2 and HeLa cells were infected with HSV-1 and harvested at 10 and 12 hpi, respectively. Cells were suspended in AP buffer (50 mM Tris-HCl [pH 8], 0.5 M NaCl, 2% NP-40, protease inhibitor cocktail [Sigma]), stored on ice for 30 min, and then centrifuged. Whole-cell lysates (20 μg) were incubated in a reaction mixture containing 10 U of calf intestinal alkaline phosphatase (CIAP; New England Biolabs) and 10 mM MgCl2 for 30 min at 37°C. Equal amounts of proteins (2.5 to 30 μg) were separated by 7.5% (acrylamide [A]:bisacrylamide [B] = 72:1) or 10% (A:B = 30:0.8) SDS-PAGE and transferred onto Immobilon transfer membranes (Millipore). Immunoreactivity was detected by Western Lightning (Perkin-Elmer). Images were processed by LumiVision PRO 400EX (Aisin/Taitec, Inc.). Signal intensity was quantified with LumiVision Analyzer 400. The system used in the present study mounts the cooled charge-coupled device camera that has 16 bit = 65,535 grayscale wide dynamic range. It enhances the accuracy of the quantitative analysis up to 100 times compared to the ordinary quantitative analysis scanning an X-ray film into the personal computer after exposing the signal to the film.
IR.
HFF2 cells were exposed to gamma irradiation with 10 Gy and harvested at 15 min after radiation. 293T-ATM shRNA and 293T-Control vector cells were exposed to gamma irradiation with 20 Gy and harvested at 15 min after radiation.
CAT assays.
Chloramphenicol acetyltransferase (CAT) assays were carried out as described previously (27, 62). All transfections were in triplicate on 35-mm-diameter plates of 293T-ATM shRNA or 293T-Control vector cells (1.2 × 106) with either p65F1CAT (3.5 μg; a gift from A. D. Yurochko) (62) or pCAT TATA+Sp1(−55)+Sp1 (−75) (1 μg) (27) using Lipofectamine 2000 according to the manufacturer's instructions. At 24 h posttransfection, cells were infected with HSV-1 at an MOI of 5 and harvested at 12 hpi. Cell lysates were then prepared and subjected to CAT assays as described previously (27, 62). Equal amounts of proteins from each sample [0.4 μg for p65F1CAT-transfected cells, 0.2 μg for pCAT TATA+Sp1(−55)+Sp1(−75)-transfected cells] were assayed for CAT activity. Acetylated and unacetylated [14C]chloramphenicol (Amersham Biosciences) were separated by thin-layer chromatography in a chloroform-methanol (95:5) solvent. Images were obtained using a BAS2500 Image Reader (Fujifilm), the signal intensities were quantified with an Image Gauge, and the levels of activity were analyzed by calculating the percentage of the conversion of unacetylated [14C]chloramphenicol to the acetylated form.
RESULTS
Hyperphosphorylation of Sp1 is induced upon HSV-1 infection.
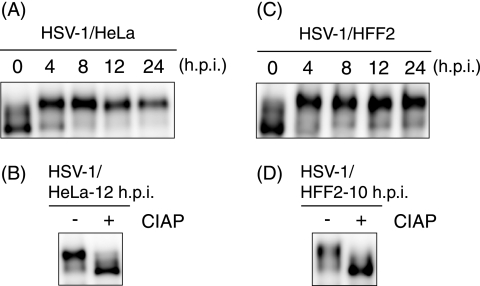
In order to confirm whether transcription factor Sp1 is hyperphosphorylated upon HSV-1 infection as reported previously (31), HeLa and HFF2 cells were infected with HSV-1. As shown in Fig. 1A, most of the Sp1 proteins were converted to slower-migrating forms in SDS-PAGE by 4 hpi in HeLa cells. Similarly, the slower-migrating forms of Sp1 in HFF2 cells became the major form by 4 hpi (Fig. 1C). CIAP treatment of the lysates from HSV-1-infected HeLa cells (Fig. 1B) or HFF2 cells (Fig. 1D) changed the slower-migrating forms of Sp1 to the faster-migrating form. Thus, Sp1 became hyperphosphorylated after HSV-1 infection without a significant change in abundance, confirming the previous finding (31).
FIG. 1.
Hyperphosphorylation of Sp1 is induced upon HSV-1 infection. HeLa (A) and HFF2 cells (C) were infected with HSV-1 at MOIs of 10 and 5, respectively, and were harvested at the indicated times postinfection. Whole-cell lysates were prepared, and equal amounts of proteins from each sample (2.5 or 30 μg) were subjected to immunoblot analysis with anti-Sp1 antibody. (B and D) Whole-cell lysates obtained from HSV-1-infected HeLa and HFF2 cells at the indicated times postinfection were treated with (+) or without (−) CIAP for 30 min at 37°C. The samples were subjected to immunoblot analysis with anti-Sp1 antibody.
Hyperphosphorylation of Sp1 is induced even upon infection of HSV-1 mutants defective for viral PK, Us3, UL13, or UL39 gene product.
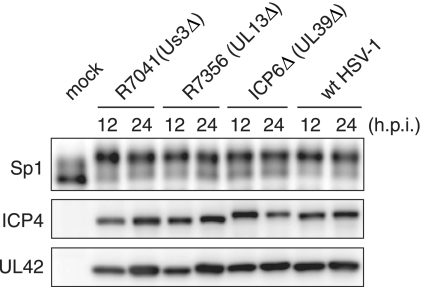
HSV-1 expresses at least three viral PKs encoded by the Us3, UL13, and UL39 genes (20, 46, 47). To examine whether Us3, UL13, or UL39 viral PK is involved in the Sp1 hyperphosphorylation, we investigated the phosphorylation state of Sp1 in HFF2 cells infected with wild type, Us3-deficient (R7041), UL13-deficient (R7356), or UL39 (viral ribonucleotide reductase large subunit)-deficient (ICP6Δ) virus (Fig. 2). The expression levels of the virus-encoded immediate-early protein (ICP4) and early protein (UL42) were almost the same among wild-type- and the mutant virus-infected cells. Since R7041 and R7356 are derived from HSV-1 strain F (46, 47) and ICP6Δ is from HSV-1 strain KOS (20), it is possible that the different gel mobilities of ICP4 proteins among wild-type HSV-1 (strain 17+) and the mutant viruses could be due to interstrain variabilities. Sp1 proteins in HFF2 cells infected with Us3-deficient (R7041), UL13-deficient (R7356), or UL39-deficient (ICP6Δ) viruses were mainly detected as hyperphosphorylated forms (the slower-migrating forms) by 12 hpi, as in the case of wild-type HSV-1 (Fig. 2). These results indicate that HSV-1 encoded PKs are not individually involved to any large extent in Sp1 hyperphosphorylation induced upon HSV-1 infection. However, the possibility of redundancy among some combination of these three PKs in Sp1 phosphorylation could not be excluded.
FIG. 2.
Hyperphosphorylation of Sp1 is induced even upon infection of HSV-1 mutants defective for viral PK, Us3, UL13 or UL39 gene product. HFF2 cells were infected with wild-type HSV-1 (wt HSV-1), Us3-defective virus (R7041), UL13-defective virus (R7356), or UL39-defective virus (ICP6Δ) at an MOI of 5. At 12 and 24 hpi, cells were harvested and whole-cell lysates were prepared. Equal amounts of proteins from each sample (15 μg) were subjected to immunoblot analysis with anti-Sp1, ICP4, and UL42 proteins specific antibodies. Mock, mock infection.
Activated ATM is involved in hyperphosphorylation of Sp1 upon HSV-1 infection.
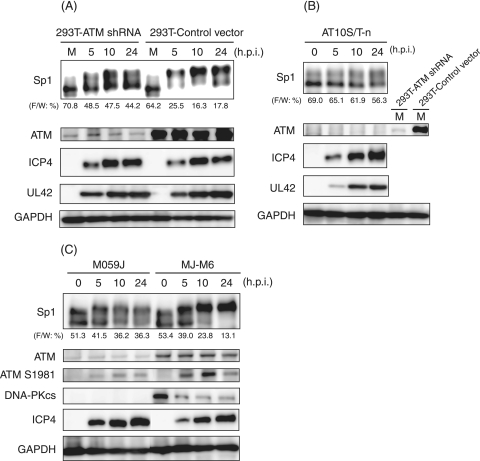
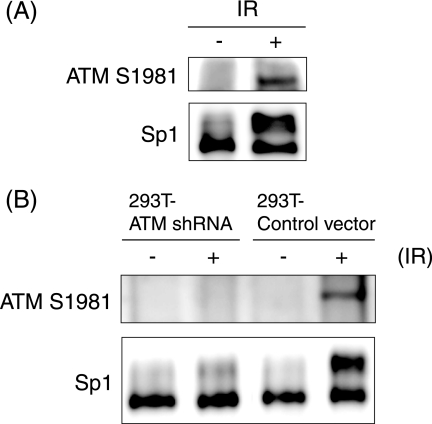
The HSV-1 infection activates ATM and elicits an ATM-dependent DNA damage signal transduction in infected cells (37, 53, 60). Since Sp1 possesses 15 putative phosphorylation sites targeted by ATM as estimated from the motifs (see Fig. 5A), we therefore examined phosphorylation states of Sp1 in ATM expression-silenced 293T cells infected with HSV-1 (Fig. 3A). The 293T cells stably expressing the ATM gene-targeted shRNA (293T-ATM shRNA) or the control vector cells (293T-Control vector) (53, 56) were infected with HSV-1. The expression level of ATM was very low in 293T-ATM shRNA cells compared to that in 293T-Control vector cells. In 293T-Control vector cells, most of the Sp1 proteins were hyperphosphorylated to slower-migrating forms by 5 hpi (Fig. 3A). In contrast, in 293T-ATM shRNA cells part of the Sp1 protein was converted to hyperphosphorylated forms, but 44% of Sp1 still remained as the faster-migrating form even at 24 hpi. It should be noted that the expression profiles of an immediate-early protein, ICP4, and an early protein, UL42, were almost the same between both cell lines, although the phosphorylation state of Sp1 was considerably different. Furthermore, as shown in Fig. 3B, in ATM-deficient AT10S/T-n cells from ataxia telangiectasia patients (43), Sp1 was originally present mostly as the faster-migrating form and only partially as the slower-migrating forms. The conversion efficiency from the faster-migrating form to slower-migrating forms was low in HSV-1-infected AT10S/T-n cells, as well as the case with ATM expression-silenced 293T cells. These observations strongly suggest that ATM is involved directly or indirectly in Sp1 hyperphosphorylation upon HSV-1 infection.
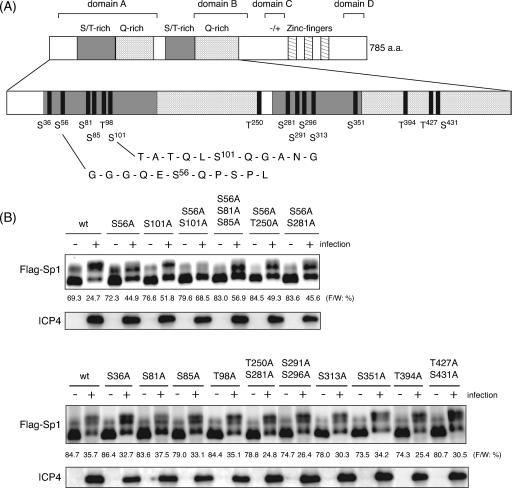
FIG. 5.
Mapping of phosphorylation sites of Sp1 upon HSV-1 infection. (A) Schematic diagram of functional domains and putative ATM phosphorylation sites of Sp1. The 785 amino acids of Sp1 have two S/T-rich regions, two Q-rich regions, and three zinc fingers. The domains A, B, C, and D correspond to multiple transcriptional activation domains. The sign “−/+” represents a region of high charge density (14). Motifs of serine (S) or threonine (T), followed by glutamine (Q) (SQ and TQ) in Sp1, are putative phosphorylation sites for ATM. Sp1 contains 15 SQ and TQ sites. The amino acid sequences around Ser-56 and Ser-101 are shown in detail. (B) HeLa cells were transfected with pcDNA-RHF/Sp1 expressing wild-type (wt) Sp1 or a variety of expression vectors for mutated Sp1 containing the indicated serine or threonine residues replaced with alanine. Cells were infected with HSV-1 at an MOI of 10 at 24 h posttransfection and harvested at 24 hpi. Whole-cell lysates were prepared, and 25-μg portions of proteins from each sample were subjected to immunoblot analysis with anti-ICP4 antibody or anti-Flag antibody to detect exogenously expressed Sp1 proteins (Flag-Sp1). Each value at the bottom of the panel of Flag-Sp1 (“F/W: %”) represents the percentage of level of the faster-migrating form to whole amounts of Flag-Sp1 as described in Materials and Methods.
FIG. 3.
ATM is involved in hyperphosphorylation of Sp1 induced by HSV-1 infection. (A and B) 293T-ATM shRNA and 293T-Control vector cells (A) and ATM defective AT10S/T-n cells (B) were infected with HSV-1 at an MOI of 10 and harvested at the indicated times postinfection. Whole-cell lysates were prepared, and equal amounts of proteins from each sample (7 to 15 μg) were subjected to immunoblot analysis with the indicated antibodies. Each value at the bottom of the panel of Sp1 (“F/W: %”) represents the percentage of the level of the faster-migrating form to whole amounts of Sp1, calculated as described in Materials and Methods. M, mock infection. (C) Human glioma M059J (DNA-PKcs null and the lower level of ATM) and MJ-M6 (M059J cells expressing DNA-PKcs) cells were infected with HSV-1 at an MOI of 2.5 and harvested at the indicated times postinfection. Whole-cell lysates (15 μg) were subjected to immunoblot analysis for expression profiles of Sp1, ATM, ATM phosphorylated at Ser-1981, DNA-PKcs, ICP4, and GAPDH. Anti-GAPDH antibody was used to confirm equal protein loading.
To further confirm the relationship between Sp1 hyperphosphorylation and ATM activation, the phosphorylation states of Sp1 were also compared in the human glioma cell lines M059J, which is DNA-dependent PK catalytic subunit (DNA-PKcs) null, and MJ-M6, which is a DNA-PKcs revertant of M059J cells transfected with the full-length DNA-PKcs cDNA expression vector (34) (Fig. 3C). It was previously shown that DNA-PKcs-deficient cells express very low levels of ATM and that recovery of DNA-PKcs partially restores ATM expression levels (25, 45). In MJ-M6 cells, the slower-migrating forms of Sp1 began to increase from 5 hpi, and almost all Sp1 proteins were converted to hyperphosphorylated forms by 10 hpi, appearing to correlate with expression levels of the activated ATM phosphorylated at Ser-1981. No change in the amount of ATM protein occurred throughout HSV-1 infection, whereas the levels of DNA-PKcs protein significantly decreased by 5 hpi. As previously reported, the degradation is dependent on the expression of the virus-encoded immediate-early protein ICP0, viral ubiquitin ligase (35, 44). In contrast, in DNA-PKcs-defective M059J cells, the slower-migrating form of Sp1 appeared from 5 hpi and subsequently did not increase. Although the ATM phosphorylated at Ser-1981 increased slowly with progression of HSV-1 infection, the expression level was very low, reflecting the phosphorylation status of Sp1 in M059J cells. The expression profile of ICP4 was almost the same in both M059J and MJ-M6 cells. Overall, the results strongly suggest that activated ATM is involved directly or indirectly in the hyperphosphorylation of Sp1 in response to HSV-1 infection.
After IR, Sp1 is hyperphosphorylated dependent on activated ATM.
In order to determine whether activation of ATM induces hyperphosphorylation of Sp1, HFF2 cells were exposed to 10 Gy of gamma irradiation, which generates double-strand DNA breaks, leading to activation of ATM-dependent DNA damage signal transduction. As shown in Fig. 4A, Sp1 was hyperphosphorylated immediately after IR accompanied by phosphorylation of Ser-1981 of ATM, leading to catalytic activation of the protein.
FIG. 4.
(A) Hyperphosphorylation of Sp1 induced upon exposure to IR. HFF2 cells were exposed to gamma irradiation (IR) with 10 Gy and harvested at 15 min post-IR. Whole-cell lysates were prepared, and 30-μg aliquots of proteins from each sample were subjected to immunoblot analysis with anti-Sp1 and ATM S1981 antibodies. (B) Sp1 hyperphosphorylation induced upon IR is correlated with the activation of ATM. The 293T-ATM shRNA and 293T-Control vector cells were exposed to IR with 20 Gy and harvested at 15 min post-IR. Whole-cell lysates were prepared, and equal amounts of proteins from each sample were subjected to immunoblot analysis with anti-Sp1 and ATM S1981 antibodies. −, No IR.
To determine whether IR-induced Sp1 phosphorylation depends on activated ATM, ATM-silenced 293T cells (293T-ATM shRNA) and 293T-Control vector cells were exposed to gamma irradiation (20 Gy) (Fig. 4B). In 293T-Control vector cells, IR induced the phosphorylation of ATM at Ser-1981 by 15 min post-IR, and simultaneously more than half of the Sp1 proteins were converted to hyperphosphorylated and slower-migrating forms. In contrast, in IR-treated ATM expression-silenced 293T cells, the phosphorylation status of Sp1 was almost unchanged. These observations with IR support the idea that the hyperphosphorylation of Sp1 is directly or indirectly dependent on activated ATM.
Mapping of Sp1 phosphorylation sites induced upon HSV-1 infection.
Related PI-3-like kinases family members, ATM, DNA-PK, and ATR (ATM-Rad3-related), display kinase activity against serine (S) and threonine (T), followed by glutamine (Q) (SQ or TQ), residues (1, 6, 12, 13). Sp1 possesses 11 SQ sites and four TQ sites (Fig. 5A). With the aim of determination of the phosphorylation site(s), we constructed expression vectors of Flag-tagged Sp1 (Flag-Sp1) containing alanine mutations of serine/threonine in each SQ/TQ site and investigated their phosphorylation in HSV-1-infected cells. As shown in Fig. 5B, most of the exogenously expressed Flag-tagged wild-type Sp1, like endogenous Sp1, was converted to slower-migrating forms upon HSV-1 infection, although a part of the recombinant protein (25 to 36%) remained as the faster-migrating form at 24 hpi. Phosphorylation patterns of Sp1 containing alanine mutations of Ser-36, Ser-81, Ser-85, Thr-98, Thr-250, Ser-281, Ser-291, Ser-296, Ser-313, Ser-351, Thr-394, Thr-427, and Ser-431 were almost the same as that of wild-type Sp1. The percentages of the remaining faster-migrating form to the total amounts of those mutated Sp1 were almost the same as or less than that of wild-type Sp1 (Fig. 5B, lower panel). In contrast, although Flag-Sp1 containing an alanine mutation of either Ser-56 or Ser-101 was also phosphorylated upon HSV-1 infection, the percentages of the faster-migrating form to total Sp1 were much higher (45 to 57%) compared to that seen with wild-type Sp1 (25%) (Fig. 5B, upper panel). Furthermore, Flag-Sp1 containing alanine mutations at both Ser-56 and Ser-101 resulted in considerably impaired conversion to the slower-migrating and hyperphosphorylated forms upon HSV-1 infection. Thus, mutation of Ser residues 56 and 101 individually or in tandem to alanine resulted in a reduction of the hyperphosphorylation of Sp1 upon HSV-1 infection.
Kim and DeLuca suggested that Sp1 is important for the expression of immediate-early and early genes whose promoters possess Sp1 binding sites and predicted that its hyperphosphorylation contributed to the downregulation of expression of these gene classes late in infection (31). However, the expression levels of ICP4 were almost constant among cells expressing wild-type Sp1 and a variety of mutated Sp1 proteins (Fig. 5B). Thus, even when the mutated Sp1 at Ser-56 and Ser-101 was expressed exogenously, we did not observe any reduced or overaccumulation of the immediate-early protein, ICP4, compared to exogenously expressed wild-type Sp1.
Ser-56 and Ser-101 on Sp1 become phosphorylated upon HSV-1 infection.
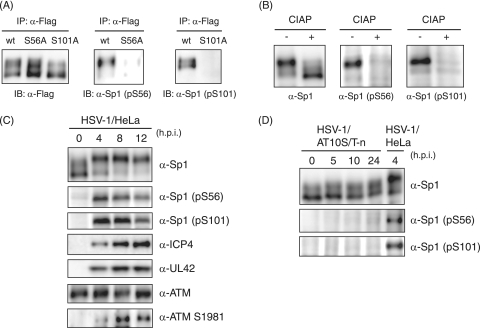
In order to confirm that Sp1 is phosphorylated at Ser-56 and Ser-101 during HSV-1 infection, we prepared phosphopeptide-specific antibodies (α-Sp1 [pS56] and α-Sp1 [pS101] antibodies) raised against phosphorylated Ser-56 and Ser-101 residues, respectively, and characterized the specificity of each antibody (Fig. 6A and B). Exogenously expressed Sp1 proteins were immunoprecipitated with anti-Flag antibody from whole-cell lysates of HeLa cells that were transfected with each expression vector—pcDNA-RHF/Sp1, pcDNA-RHF/Sp1-S56A, or pcDNA-RHF/Sp1-S101A—followed by HSV-1 infection. Immunoprecipitated wild-type and mutant Sp1 samples were subjected to immunoblot analysis with the phosphopeptide-specific antibodies (Fig. 6A). α-Sp1 (pS56) could recognize wild-type Flag-Sp1 but not Flag-Sp1-S56A. Similarly, α-Sp1 (pS101) recognized wild-type Flag-Sp1 but not Flag-Sp1-S101A. In addition, both α-Sp1 (pS56) and α-Sp1 (pS101) showed CIAP-sensitive reactivity with endogenous Sp1 in whole-cell lysates obtained from HSV-1-infected HeLa cells at 12 hpi (Fig. 6B). Thus, it was proved that these two antibodies are directed against phospho-Ser-56 and phospho-Ser-101, respectively.
FIG. 6.
Ser-56 and Ser-101 of Sp1 are phosphorylated upon HSV-1 infection. (A) HeLa cells were transfected with pcDNA-RHF/Sp1 (wt), pcDNA-RHF/Sp1-S56A, or pcDNA-RHF/Sp1-S101A and infected with HSV-1 at an MOI of 10 at 24 h posttransfection. At 24 hpi, cells were harvested and lysed. The whole-cell lysates were subjected to IP with anti-Flag M2 affinity resin, and the immunoprecipitated samples were subjected to immunoblot (IB) analysis with anti-Flag, α-Sp1 (pS56), and α-Sp1 (pS101) antibodies. (B) Whole-cell lysates obtained from HSV-1-infected HeLa cells at 12 hpi were treated with (+) or without (−) CIAP. The samples were applied for immunoblot analysis with the indicated antibodies. (C) HeLa cells were infected with HSV-1 at an MOI of 10 and harvested at the indicated times postinfection. Whole-cell lysates were prepared, and equal amounts of proteins from each sample were applied for immunoblot analysis with the indicated antibodies. (D) AT10S/T-n cells were infected with HSV-1 at an MOI of 10 and harvested at the indicated times postinfection. Whole-cell lysates were prepared and subjected to immunoblot analysis with the indicated antibodies. Whole-cell lysate from HSV-1-infected HeLa cells at 4 hpi was also applied as a positive control.
Next, we examined phosphorylation of Sp1 at Ser-56 and Ser-101 in HeLa cells and the ATM-deficient cell line, AT10S/T-n, throughout HSV-1 infection using the phosphopeptide-specific antibodies (Fig. 6C and D). In HeLa cells, phosphorylation of Sp1 at Ser-56 or Ser-101 was detected by 4 hpi and reached maximum level at 8 hpi. Similarly, the activated ATM phosphorylated at Ser-1981 was detected by 4 hpi and increased gradually. In contrast, in AT10S/T-n cells, both residues on Sp1 were not phosphorylated at all throughout infection. These observations clearly showed that both Ser-56 and Ser-101 residues on Sp1 become phosphorylated directly or indirectly by ATM in response to HSV-1 infection.
ATM phosphorylates Ser-101 but not Ser-56 on Sp1 in vitro.
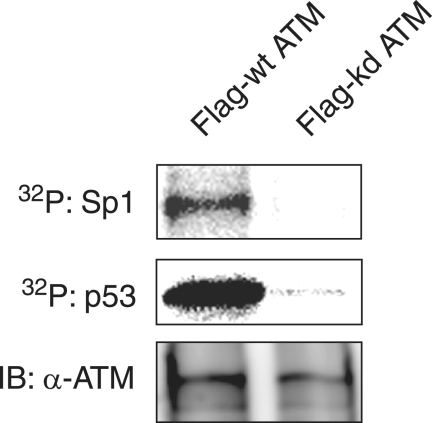
In order to investigate whether ATM can phosphorylate Sp1 directly, IP-kinase assays were performed (Fig. 7). Purified Sp1 from insect cells was subjected to an in vitro kinase assay with Flag-tagged wild-type ATM and kinase-dead ATM immunoprecipitated by anti-Flag antibody from cells transfected with each expression vector. Purified p53 known as a substrate for ATM (6) was used as a positive control. Equal amounts of Flag-tagged wild-type ATM and kinase-dead ATM were immunoprecipitated by anti-Flag antibodies. As shown in Fig. 7, Flag-tagged wild-type ATM phosphorylated Sp1 and p53 directly, whereas kinase-dead ATM did not. Thus, ATM can directly phosphorylate Sp1 in vitro.
FIG. 7.
Sp1 is phosphorylated by ATM in vitro. The lysates of 293T cells transfected with plasmid expressing Flag-tagged wild-type ATM (Flag-wt ATM) or kinase-dead ATM (Flag-kd ATM) were immunoprecipitated with anti-Flag antibody. Each of immunocomplexes containing Flag-wt ATM or Flag-kd ATM protein were incubated with purified Sp1 or p53 as substrates in the presence of [γ-32P]ATP. Sp1 and p53 were resolved by SDS-10% PAGE, followed by autoradiography. The amounts of immunoprecipitated Flag-wt ATM and Flag-kd ATM were confirmed by immunoblot analysis with anti-ATM antibody.
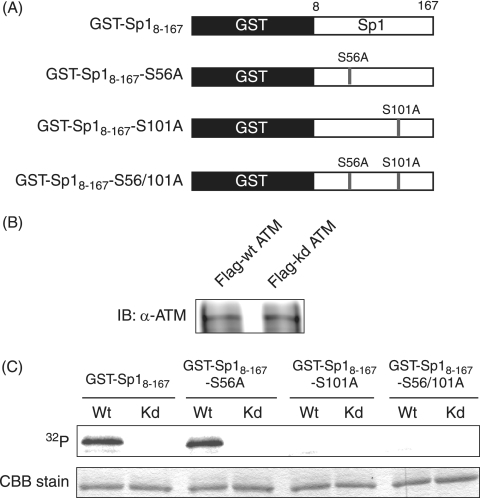
Next, to determine whether ATM can phosphorylate Ser-56 and Ser-101 on Sp1 directly, GST fusion proteins of truncated Sp1 (8 to 167 amino acids) (GST-Sp18-167,) and identical fragments with Ser-56 or Ser-101 or both mutated to alanine (GST-Sp18-167-S56A, GST-Sp18-167-S101A, and GST-Sp18-167-S56/101A) (Fig. 8A) were used as substrates in IP-kinase assays. The amounts of immunoprecipitated Flag-tagged wild-type ATM and kinase-dead ATM proved the same on immunoblot analysis (Fig. 8B). As shown in Fig. 8C, GST-Sp18-167-S56A, as well as wild-type GST-Sp18-167, was phosphorylated by wild-type ATM but not by kinase-dead ATM. In contrast, wild-type ATM could not phosphorylate GST-Sp18-167-S101A or GST-Sp18-167-S56/101A. These results clearly indicate that ATM can directly phosphorylate Sp1 at Ser-101 but not Ser-56. Considering this and the result that neither Ser-101 nor Ser-56 were phosphorylated upon HSV-1 infection in the ATM-defective cell line (Fig. 6D) together, other kinase(s) activated by ATM would phosphorylate Ser-56 on Sp1 upon HSV-1 infection.
FIG. 8.
ATM phosphorylates Sp1 at Ser-101 but not Ser-56 in vitro. (A) Schematic diagram of GST fusion proteins of truncated Sp1 and identical fragments with one or both of Ser-56 and Ser-101 mutated to alanine. (B) Lysates of 293T cells transfected with plasmid expressing Flag-tagged wild-type ATM (Flag-wt ATM) or kinase-dead ATM (Flag-kd ATM) were immunoprecipitated with anti-Flag antibody. Immunocomplexes of Flag-wt ATM or Flag-kd ATM were resolved on SDS-10% PAGE gel and subjected to immunoblot analysis with anti-ATM antibody. (C) IP-kinase assays. GST-Sp18-167, GST-Sp18-167-S56A, GST-Sp18-167-S101A, and GST-Sp18-167-S56/101A were expressed in E. coli, purified, and used as substrates for IP-kinase assays. Immunocomplexes containing Flag-wt ATM (Wt) or Flag-kd ATM (Kd) protein were each incubated with purified GST-Sp18-167, GST-Sp18-167-S56A, GST-Sp18-167-S101A, or GST-Sp18-167-S56/101A as substrates in the presence of [γ-32P]ATP. Samples were resolved by SDS-10% PAGE, followed by autoradiography. The amounts of each GST fusion protein were confirmed by Coomassie brilliant blue (CBB) staining.
ATM-dependent Sp1 phosphorylation does not affect Sp1-dependent transcriptional activity during viral infection.
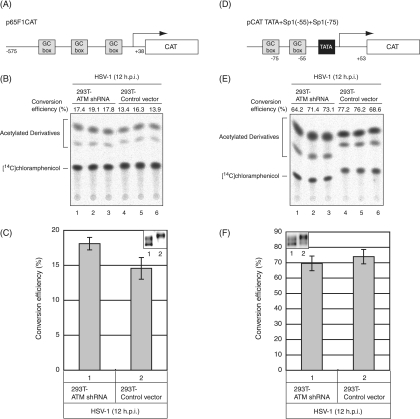
In order to examine whether ATM affects Sp1-dependent transcriptional activity upon HSV-1 infection, CAT activity from the Sp1 responsive promoter in ATM expression-silenced 293T cells (293T-ATM shRNA) was compared to those in ATM expression-intact 293T cells (293T-Control vector) (Fig. 9). Sp1 stimulates transcription from promoters containing a GC-rich recognition element, the GC-box (15, 16, 19), and is also important for the regulation of TATA-less genes that encode housekeeping proteins (23). As depicted in Fig. 9A, the CAT reporter plasmid, p65F1CAT, has a promoter sequence (−575 to +38) of p65, an NF-κB subunit, containing three GC-boxes upstream of the CAT gene without any TATA consensus sequence (62). 293T-ATM shRNA cells and 293T-Control vector cells transfected with p65F1CAT were infected with HSV-1 at 24 h posttransfection and harvested at 12 hpi. The lysates from both cells were subjected to CAT assay, and the transcriptional activities were analyzed by calculating the percentage of the conversion of unacetylated [14C]chloramphenicol to the acetylated form. In a CAT assay for p65F1CAT, the transcriptional activity from the Sp1 responsive promoter in 293T-ATM shRNA cells was almost the same (only a 1.2-fold increase) as that in 293T-Control vector cells (Fig. 9B and C). Next, another reporter plasmid, pCAT TATA+Sp1(−55)+Sp1(−75) (Fig. 9D) containing two GC-boxes and TATA consensus sequence of human cytomegalovirus (HMCV) major immediate-early gene upstream of the CAT gene (27) was used as TATA-dependent Sp1 responsive promoter. Similarly, the transcriptional activity from the promoter in 293T-ATM shRNA cells infected with HSV-1 was almost the same as that in 293T-Control vector cells (Fig. 9E and F). In 293T-Control vector cells Sp1 was detected mainly as the slower-migrating and hyperphosphorylated forms at 12 hpi. In contrast, in 293T-ATM shRNA cells, Sp1 was detected mostly as the faster-migrating form and partially as the slower-migrating form (Fig. 9C and F, inset images). Phosphorylation of Ser-56 and Ser-101 on Sp1 was observed in 293T-Control vector cells, while the phosphorylation was not in 293T-ATM shRNA cells (data not shown). As shown in Fig. 3A, the expression profiles of ICP4, whose promoter possesses several Sp1-binding sites, in 293T-ATM shRNA and 293T-Control vector cells were almost the same throughout HSV-1 infection, corresponding well with the same Sp1-dependent transcriptional activities in both cells obtained in the reporter-gene analyses of Fig. 9. Collectively, ATM-dependent Sp1 phosphorylation does not appear to affect the Sp1-dependent transcriptional activity during viral infection.
FIG. 9.
ATM-dependent Sp1 phosphorylation does not affect Sp1-dependent transcription upon infection. (A and D) Schematic illustration of reporter vectors: p65F1CAT contains three Sp1-binding sites (GC-boxes) of the TATA less p65 promoter sequence (−575 to +38) (A), and pCAT TATA+Sp1(−55)+Sp1(−75) contains two GC-boxes and TATA consensus sequence of HCMV major immediate-early gene (D). (B and E) 293T-ATM shRNA and 293T-Control vector cells were transfected with either p65F1CAT (B) or pCAT TATA+Sp1(−55)+Sp1(−75) (E), infected with HSV-1 at 24 h posttransfection at an MOI of 5, and harvested at 12 hpi. CAT assays were performed as described in Materials and Methods. All transfection experiments were in triplicate. (C and F) Data from three independent experiments in panels B and E were plotted on the graph, respectively. The levels of activity (i.e., the conversion efficiency) were determined by calculating the percentage of the conversion of unacetylated [14C]chloramphenicol to the acetylated form. In order to confirm phosphorylation states and amounts of Sp1 in 293T-ATM shRNA and 293T-Control vector cells infected with HSV-1 at 12 hpi, equal amounts of proteins from each sample were subjected to immunoblot analysis with anti-Sp1 antibody (panels C and F, inset images).
DISCUSSION
It was found here that hyperphosphorylation of the transcription factor Sp1 upon HSV-1 infection is mainly due to ATM and/or cellular kinase(s) activated by ATM rather than individual HSV-1 encoded PKs. Hyperphosphorylation of Sp1 upon HSV-1 infection was thus impaired in ATM-deficient or -expression silenced cells. Although at least two sites of Ser-56 and Ser-101 could be phosphorylated upon HSV-1 infection, ATM was here found to directly phosphorylate Sp1 at Ser-101 but not at Ser-56. To our knowledge, this is a first report describing determination of the target site on Sp1 for ATM. Involvement of ATM in Sp1 phosphorylation is also supported by the previous reports that Sp1 and ATM coimmunoprecipitate reciprocally in vivo and that Sp1 directly interacts with the kinase region of ATM in vitro (21).
Hyperphosphorylation of Sp1 is observed also in infection of other herpesviruses such as HCMV and Epstein-Barr virus (EBV). HCMV infection results in increased phosphorylated forms of Sp1, together with an increased level of Sp1 (27, 62), and the ATM DNA damage checkpoint pathway is activated in response to HCMV infection (18, 38). Also, induction of lytic replication in EBV latently infected cells results in hyperphosphorylation of Sp1 (S. Iwahori, A. Kudoh, and T. Tsurumi, unpublished result) and EBV lytic replication elicits ATM checkpoint signal transduction (33). Thus, ATM-dependent Sp1 hyperphosphorylation might be a common phenomenon during the lytic replication of herpesviruses.
Although phosphorylation of Sp1 at Ser-56 was not detected at all upon infection in ATM-deficient cells, ATM by itself could not phosphorylate Ser-56 on Sp1 in vitro, suggesting that other cellular kinase(s) activated by ATM could be involved in the Ser-56 phosphorylation. The sequence of Ser-56 followed by glutamine is known as putative phosphorylation site for related members of the PI-3-like kinase family such as ATM, ATR, and DNA-PK. Chen et al. have recently suggested that ATM is likely the kinase mediating IR-induced DNA-PKcs phosphorylation required for full activation of DNA-PKcs (7). Sp1 phosphorylation by DNA-PK has already been reported from studies of human immunodeficiency virus type 1 infection (11). However, as shown in Fig. 3C, the level of DNA-PKcs decreases significantly at early stages of HSV-1 infection, the degradation being in a virus-encoded ICP0-dependent manner as reported by others (35, 44). Also, activation of ATR is minimal during HSV-1 infection (37, 53), although the ATR PK activity is dependent upon ATM (29, 42, 58). Furthermore, it has recently been reported that HSV-1 disrupts the ATR-dependent DNA damage response through destruction of the usually tight colocalization of ATR and ATRIP (59). Thus, involvement of ATR and DNA-PK in phosphorylation of Ser-56 on Sp1 upon HSV-1 infection is unlikely. In response to IR, ATM phosphorylates and activates checkpoint kinases, Chk1 and Chk2, that play roles as signal transducers (3). We reported that the activation of Chk1 and Chk2 was induced upon HSV-1 infection (53). However, since the sequence around Ser-56 is distinct from Chk1 and Chk2 target motif (Arg-x-x-Ser) (8, 55), it is also unlikely that activated Chk1 and Chk2 are involved in the phosphorylation of Sp1 at Ser-56.
With respect to the role of Sp1 hyperphosphorylation during HSV-1 infection, Kim and DeLuca have reported purified Sp1 from HSV-1-infected cells at 12 hpi to exhibit reduced transcriptional activity in an in vitro transcription assay, although the DNA-binding activity of Sp1 was unchanged until 8 hpi (31). These researchers suggested that the reduced transcriptional activity of Sp1 may be due to the hyperphosphorylation of Sp1 and contribute to the reduced transcription of immediate-early and early genes with Sp1-binding sites in its promoters at late stages of infection (31). We also observed the reduced Sp1-dependent transcriptional activity upon HSV-1 infection in reporter gene assay (data not shown). As shown in Fig. 9, however, the transcriptional activity from the Sp1 responsive promoter in 293T-ATM shRNA cells upon HSV-1 infection was almost the same as that in 293T-Control vector cells. In addition, there was almost no difference between the expression profile of ICP4, whose promoter possesses several Sp1-binding sites, in 293T-ATM shRNA and that in 293T-Control vector cells throughout HSV-1 infection, although Sp1 hyperphosphorylation was impaired in 293T-ATM shRNA cells (Fig. 3A), a finding corresponding well with our previous report that there is no difference in the yields of HSV type 2 in 293T-ATM shRNA cells and 293T-Control vector cells (53). Thus, although at least Ser-56 and Ser-101 of Sp1 were phosphorylated dependent on ATM during HSV-1 infection, the Sp1 phosphorylation at both sites does not appear to affect Sp1-dependent transcriptional activity upon HSV-1 infection. Therefore, modification of Sp1 besides phosphorylation at Ser-56 and Ser-101 might induce the reduction of its transcriptional activity during the HSV-1 infection.
IR rapidly activates ATM, which then phosphorylates several transcription factors such as p53, ATF2, and CREB (4, 6, 51). In the present study, high doses of IR (10 and 20 Gy) induced the hyperphosphorylation of Sp1 in ATM expression-positive cells, but it did not so significantly in ATM expression-silenced cells (Fig. 4), suggesting that Sp1 is phosphorylated in an IR-induced ATM-dependent manner. A previous report demonstrated that IR at low dose (3 to 6 Gy) induces phosphorylation of Sp1 and simultaneously causes an increase in DNA-binding activity of Sp1 (40, 61). Furthermore, coexpression of Sp1 and ATM in Drosophila Schneider cells lacking endogenous Sp1 results in an ATM dose-dependent synergistic transactivation of IGF-IR promoter containing Sp1 binding sites, suggesting that Sp1 phosphorylation by ATM might increase its transactivation activity (50). In the present study, however, the transcriptional activity from the Sp1 responsive promoter in ATM-silenced cells upon HSV-1 infection was almost the same as that in ATM-intact cells. Upon HSV-1 infection, Sp1 might undergo phosphorylation besides ATM-dependent phosphorylation or other modification(s). Thus, complex modification(s) of Sp1 caused by HSV-1 infection might mask the functional change by the phosphorylation at Ser-56 and Ser-101.
Kim and DeLuca (31) first documented the hyperphosphorylation of transcription factor Sp1 after HSV-1 infection. From studies conducted with ICP4 deletion mutants, these authors speculated that ICP4 is necessary for the hyperphosphorylation of Sp1 directly or indirectly. Also, they observed partial conversion of Sp1 to the hyperphosphorylated form during infection with wild-type HSV-1 in the presence of phosphonoacetic acid, a specific inhibitor of the viral DNA polymerase (31). We and others have previously demonstrated that HSV infection elicits ATM-dependent DNA damage responses, whereas infection with a UV-inactivated virus or with a replication-defective virus does not, suggesting that viral DNA synthesis is essential for ATM activation (37, 53). In the presence of phosphonoacetic acid, the ATM DNA damage signaling upon infection is blocked at a low multiplicity of infection, although the UL42 gene product, viral early protein, is expressed (37, 53). Therefore, ICP4 may indirectly contribute to Sp1 hyperphosphorylation through expression for viral replication proteins synthesizing viral DNA. Since synthesized viral DNA structure triggers activation of ATM-dependent DNA damage responses upon HSV infection, newly syhthesized viral DNA rather than expressed viral protein(s) appears to be necessary for the hyperphosphorylation of Sp1.
Acknowledgments
We thank G. Suske, Philipps-Universität, Marburg, Germany, for CMV-hSp1; M. B. Kastan, St. Jude Children's Research Hospital, for pcDNA-Flag-ATMwt and pcDNA-Flag-ATMkd; A. D. Yurochko, Louisiana State University Health Sciences Center, for p65F1CAT; W. Nakai, Osaka University (Japan), for pFastBac1 containing a sequence (RGS-6xHis-Flag); B. Roizman, The University of Chicago, for R7041 and R7356; M. Fujita and Y. Tatsumi, National Cancer Center Research Institute (Japan), for the 293T-ATM shRNA and 293T-Control vector cell lines; A. Kurimasa, Tottori University (Japan), for the M059J and MJ-M6 cell lines; and K. Ishizaki and H. Nakamura, Aichi Cancer Center Research Institute (Japan), for the AT10S/T-n cells. We also thank H. Goto and Y. Nishikawa, Aichi Cancer Center Research Institute (Japan), for instruction for the preparation of phosphopeptide-specific antibodies and for assistance with cell culture, respectively.
This study was supported by grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, Culture, and Technology of Japan (19041078, 18012058, and 18390147 to T.T.) and partly by the Uehara Memorial Foundation. Y.S. and A.K. were supported by a Research Fellowship of the Japanese Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, S. A., D. A. Barry, R. W. Leggett, and C. R. Mueller. 1997. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J. Biol. Chem. 272:13489-13495. [DOI] [PubMed] [Google Scholar]
- 3.Bartek, J., and J. Lukas. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3:421-429. [DOI] [PubMed] [Google Scholar]
- 4.Bhoumik, A., S. Takahashi, W. Breitweiser, Y. Shiloh, N. Jones, and Z. Ronai. 2005. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol. Cell 18:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouwman, P., and S. Philipsen. 2002. Regulation of the activity of Sp1-related transcription factors. Mol. Cell Endocrinol. 195:27-38. [DOI] [PubMed] [Google Scholar]
- 6.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 7.Chen, B. P., N. Uematsu, J. Kobayashi, Y. Lerenthal, A. Krempler, H. Yajima, M. Lobrich, Y. Shiloh, and D. J. Chen. 2007. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 282:6582-6587. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P., C. Luo, Y. Deng, K. Ryan, J. Register, S. Margosiak, A. Tempczyk-Russell, B. Nguyen, P. Myers, K. Lundgren, C. C. Kan, and P. M. O'Connor. 2000. The 1.7-Å crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell 100:681-692. [DOI] [PubMed] [Google Scholar]
- 9.Chu, S., and T. J. Ferro. 2005. Sp1: regulation of gene expression by phosphorylation. Gene 348:1-11. [DOI] [PubMed] [Google Scholar]
- 10.Chun, H. H., R. B. Cary, F. Lansigan, J. Whitelegge, D. J. Rawlings, and R. A. Gatti. 2004. ATM protein purified from vaccinia virus expression system: DNA binding requirements for kinase activation. Biochem. Biophys. Res. Commun. 322:74-81. [DOI] [PubMed] [Google Scholar]
- 11.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 72:2615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortez, D., G. Glick, and S. J. Elledge. 2004. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA 101:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. [DOI] [PubMed] [Google Scholar]
- 14.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 15.Dynan, W. S., and R. Tjian. 1983. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 32:669-680. [DOI] [PubMed] [Google Scholar]
- 16.Dynan, W. S., and R. Tjian. 1983. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35:79-87. [DOI] [PubMed] [Google Scholar]
- 17.Fojas de Borja, P., N. K. Collins, P. Du, J. Azizkhan-Clifford, and M. Mudryj. 2001. Cyclin A-CDK phosphorylates Sp1 and enhances Sp1-mediated transcription. EMBO J. 20:5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaspar, M., and T. Shenk. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. USA 103:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gidoni, D., W. S. Dynan, and R. Tjian. 1984. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. Nature 312:409-413. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein, D. J., and S. K. Weller. 1988. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology 166:41-51. [DOI] [PubMed] [Google Scholar]
- 21.Gueven, N., K. Keating, T. Fukao, H. Loeffler, N. Kondo, H. P. Rodemann, and M. F. Lavin. 2003. Site-directed mutagenesis of the ATM promoter: consequences for response to proliferation and ionizing radiation. Genes Chromosomes Cancer 38:157-167. [DOI] [PubMed] [Google Scholar]
- 22.Hagen, G., S. Muller, M. Beato, and G. Suske. 1994. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 13:3843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haidweger, E., M. Novy, and H. Rotheneder. 2001. Modulation of Sp1 activity by a cyclin A/CDK complex. J. Mol. Biol. 306:201-212. [DOI] [PubMed] [Google Scholar]
- 24.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoppe, B. S., R. B. Jensen, and C. U. Kirchgessner. 2000. Complementation of the radiosensitive M059J cell line. Radiat. Res. 153:125-130. [DOI] [PubMed] [Google Scholar]
- 26.Hung, J. J., Y. T. Wang, and W. C. Chang. 2006. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol. Cell. Biol. 26:1770-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isomura, H., M. F. Stinski, A. Kudoh, T. Daikoku, N. Shirata, and T. Tsurumi. 2005. Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J. Virol. 79:9597-9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, S. P., J. J. MacDonald, S. Lees-Miller, and R. Tjian. 1990. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell 63:155-165. [DOI] [PubMed] [Google Scholar]
- 29.Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith, J. Lukas, and S. P. Jackson. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell. Biol. 8:37-45. [DOI] [PubMed] [Google Scholar]
- 30.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079-1090. [DOI] [PubMed] [Google Scholar]
- 31.Kim, D. B., and N. A. DeLuca. 2002. Phosphorylation of transcription factor Sp1 during herpes simplex virus type 1 infection. J. Virol. 76:6473-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozlov, S., N. Gueven, K. Keating, J. Ramsay, and M. F. Lavin. 2003. ATP activates ataxia-telangiectasia mutated (ATM) in vitro. Importance of autophosphorylation. J. Biol. Chem. 278:9309-9317. [DOI] [PubMed] [Google Scholar]
- 33.Kudoh, A., M. Fujita, L. Zhang, N. Shirata, T. Daikoku, Y. Sugaya, H. Isomura, Y. Nishiyama, and T. Tsurumi. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156-8163. [DOI] [PubMed] [Google Scholar]
- 34.Kurimasa, A., S. Kumano, N. V. Boubnov, M. D. Story, C. S. Tung, S. R. Peterson, and D. J. Chen. 1999. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 19:3877-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leggett, R. W., S. A. Armstrong, D. Barry, and C. R. Mueller. 1995. Sp1 is phosphorylated and its DNA binding activity down-regulated upon terminal differentiation of the liver. J. Biol. Chem. 270:25879-25884. [DOI] [PubMed] [Google Scholar]
- 37.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 102:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, M. H., K. Rosenke, K. Czornak, and E. A. Fortunato. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69(Pt. 7):1531-1574. [DOI] [PubMed] [Google Scholar]
- 40.Meighan-Mantha, R. L., A. T. Riegel, S. Suy, V. Harris, F. H. Wang, C. Lozano, T. L. Whiteside, and U. Kasid. 1999. Ionizing radiation stimulates octamer factor DNA binding activity in human carcinoma cells. Mol. Cell. Biochem. 199:209-215. [DOI] [PubMed] [Google Scholar]
- 41.Milanini-Mongiat, J., J. Pouyssegur, and G. Pages. 2002. Identification of two Sp1 phosphorylation sites for p42/p44 mitogen-activated protein kinases: their implication in vascular endothelial growth factor gene transcription. J. Biol. Chem. 277:20631-20639. [DOI] [PubMed] [Google Scholar]
- 42.Myers, J. S., and D. Cortez. 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 281:9346-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura, H., H. Fukami, Y. Hayashi, T. Kiyono, S. Nakatsugawa, M. Hamaguchi, and K. Ishizaki. 2002. Establishment of immortal normal and ataxia telangiectasia fibroblast cell lines by introduction of the hTERT gene. J. Radiat. Res. 43:167-174. [DOI] [PubMed] [Google Scholar]
- 44.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng, Y., R. G. Woods, H. Beamish, R. Ye, S. P. Lees-Miller, M. F. Lavin, and J. S. Bedford. 2005. Deficiency in the catalytic subunit of DNA-dependent protein kinase causes down-regulation of ATM. Cancer Res. 65:1670-1677. [DOI] [PubMed] [Google Scholar]
- 46.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajcani, J., V. Andrea, and R. Ingeborg. 2004. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes 28:293-310. [DOI] [PubMed] [Google Scholar]
- 49.Roizman, B. 1996. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc. Natl. Acad. Sci. USA 93:11307-11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shahrabani-Gargir, L., T. K. Pandita, and H. Werner. 2004. Ataxia-telangiectasia mutated gene controls insulin-like growth factor I receptor gene expression in a deoxyribonucleic acid damage response pathway via mechanisms involving zinc-finger transcription factors Sp1 and WT1. Endocrinology 145:5679-5687. [DOI] [PubMed] [Google Scholar]
- 51.Shi, Y., S. L. Venkataraman, G. E. Dodson, A. M. Mabb, S. LeBlanc, and R. S. Tibbetts. 2004. Direct regulation of CREB transcriptional activity by ATM in response to genotoxic stress. Proc. Natl. Acad. Sci. USA 101:5898-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 53.Shirata, N., A. Kudoh, T. Daikoku, Y. Tatsumi, M. Fujita, T. Kiyono, Y. Sugaya, H. Isomura, K. Ishizaki, and T. Tsurumi. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336-30341. [DOI] [PubMed] [Google Scholar]
- 54.Spengler, M. L., and M. G. Brattain. 2006. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J. Biol. Chem. 281:5567-5574. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, C., L. Smith, and N. B. La Thangue. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell. Biol. 5:401-409. [DOI] [PubMed] [Google Scholar]
- 56.Tatsumi, Y., N. Sugimoto, T. Yugawa, M. Narisawa-Saito, T. Kiyono, and M. Fujita. 2006. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J. Cell Sci. 119:3128-3140. [DOI] [PubMed] [Google Scholar]
- 57.Wagner, E. K., J. F. Guzowski, and J. Singh. 1995. Transcription of the herpes simplex virus genome during productive and latent infection. Prog. Nucleic Acids Res. Mol. Biol. 51:123-165. [DOI] [PubMed] [Google Scholar]
- 58.Westphal, C. H. 1997. Cell-cycle signaling: Atm displays its many talents. Curr. Biol. 7:R789-R792. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson, D. E., and S. K. Weller. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, C. R., C. Wilson-Van Patten, S. M. Planchon, S. M. Wuerzberger-Davis, T. W. Davis, S. Cuthill, S. Miyamoto, and D. A. Boothman. 2000. Coordinate modulation of Sp1, NF-κB, and p53 in confluent human malignant melanoma cells after ionizing radiation. FASEB J. 14:379-390. [DOI] [PubMed] [Google Scholar]
- 62.Yurochko, A. D., M. W. Mayo, E. E. Poma, A. S. Baldwin, Jr., and E. S. Huang. 1997. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J. Virol. 71:4638-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]