Abstract
The Epstein-Barr virus (EBV) latent membrane protein 1 (LMP1) gene is considered the EBV oncogene as it is necessary for EBV-mediated transformation of B lymphocytes and itself transforms rodent fibroblasts. LMP1 activates the NF-κB, phosphatidylinositol 3-kinase (PI3K)-Akt, mitogen-activated protein kinase, and Jun N-terminal protein kinase signaling pathways through its two signaling domains, carboxyl-terminal activating regions 1 and 2 (CTAR1 and CTAR2). CTAR1 and CTAR2 induce signal transduction pathways through their direct (CTAR1) or indirect (CTAR2) recruitment of tumor necrosis factor receptor-associated factors (TRAFs). CTAR1 is necessary for LMP1-mediated transformation as well as activation of PI3K signaling and induction of cell cycle markers associated with G1/S transition. In this study, activation of PI3K-Akt signaling and deregulation of cell cycle markers were mapped to the TRAF-binding domain within CTAR1 and to the residues between CTAR1 and CTAR2. LMP1 CTAR1 also activated the MEK1/2-extracellular signal-regulated kinase 1/2 signaling pathway, and this activation was necessary for LMP1-induced transformation of Rat-1 fibroblasts. Dominant-negative forms of TRAF2 and TRAF3 inhibited but did not fully block LMP1-mediated transformation. These findings identify a new signaling pathway that is uniquely activated by the TRAF-binding domain of LMP1 and is required for transformation.
Epstein-Barr virus (EBV) is a ubiquitous herpesvirus that is associated with a variety of malignancies, including nasopharyngeal carcinoma, Hodgkin's lymphoma, posttransplant lymphoma, and others (29, 42, 55, 56). The EBV-encoded latent membrane protein 1 (LMP1) gene is considered the EBV oncogene as it is essential for EBV-mediated B-lymphocyte transformation and itself can transform rodent fibroblasts (2, 28, 51). LMP1 is frequently expressed in EBV malignancies, including nasopharyngeal carcinoma and Hodgkin's lymphoma (19, 55).
LMP1 consists of 386 amino acids that form a short 23 amino-terminal cytoplasmic end, six hydrophobic transmembrane domains, and a long carboxyl-terminal tail that contains the two main signaling domains, carboxyl-terminal activating regions 1 and 2 (CTAR1 and CTAR2). LMP1 self-associates in the plasma membrane. Not requiring a ligand, LMP1 acts as a constitutively active tumor necrosis factor receptor (TNFR) by mediating signaling through the recruitment of TNFR-associated factors (TRAFs) by CTAR1 or through recruitment of adapter molecules, such as the TNFR-associated death domain or the receptor-interacting protein 1 by CTAR2 (24, 26, 38). CTAR1 recruits TRAF1/2 or TRAF3/5 heterodimers through a consensus TRAF-binding domain (PQQAT), whereas CTAR2 recruits TRAF2 and TRAF6 through adapter molecules (29).
LMP1 activates multiple signaling pathways, including NF-κB, mitogen-activated protein kinase, c-Jun N-terminal kinase, and phosphatidylinositol 3-kinase (PI3K)-Akt pathways (10, 17, 26, 33, 37, 41, 43, 49). LMP1 also induces the deregulation of various cellular markers associated with G1/S cell cycle transition, including the upregulation of the inhibitor of differentiation 1 (Id1) and Id3, downregulation of cyclin-dependent kinase inhibitors p27Kip1 and p16INK4a, upregulation of cyclin-dependent kinase 2 (CDK2), and upregulation of retinoblastoma (Rb) (18, 30, 40).
Although both CTAR1 and CTAR2 recruit TRAFs, they yield different effects on the signal transduction pathways they activate. CTAR1, but not CTAR2, is necessary for rodent fibroblast transformation and EBV-mediated B-lymphocyte transformation (26, 33). Although CTAR2 more strongly activates NF-κB in reporter assays, CTAR1 induces a more complex signal, inducing both canonical and noncanonical forms of NF-κB, including p50/p50 and p50/p52 dimers (15, 27, 35, 50). While CTAR2 activates the Jun N-terminal protein kinase (JNK) signaling cascade, CTAR1 activates PI3K-Akt signaling, resulting in the phosphorylation of the Akt target, glycogen synthase kinase 3β (GSK3β). CTAR1 also has unique effects on cellular gene expression, including the induction of epithelial growth factor receptor (EGFR), TRAF1, Id1, and Id3 while repressing p27Kip1 (11, 12, 14, 18, 33, 36). The activation of PI3K-Akt, but not NF-κB, is required for Rat-1 fibroblast transformation (33).
The presence or absence of a specific TRAF can determine the outcome of the signal being transduced by LMP1. Although CTAR2, but not CTAR1, activates JNK signaling in epithelial cells, overexpression of TRAF1 allows CTAR1 to activate JNK signaling (16). In TRAF2 and TRAF5 knockout mouse embryo fibroblasts (MEFs), LMP1-mediated NF-κB activation is similar to wild-type MEFs, and this activation is highly dependent on TRAF6 (32). In contrast, in mouse lymphoma cells, NF-κB and immunoglobulin M secretion are almost completely TRAF3 dependent (53, 54).
In this study, the roles of the TRAF-binding domain, TRAF2, and TRAF3 in rodent fibroblast transformation and signal transduction were evaluated. LMP1 mutants with mutations in the carboxyl-terminal tail and the TRAF-binding domain revealed that CTAR1 mediates the majority of PI3K-Akt signaling and deregulation of cellular markers associated with G1/S transition and induces a decrease in the protein levels of the desmosome-associated protein plakoglobin. The activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) was uniquely activated by CTAR1, and this activation was necessary for LMP1-mediated transformation of rodent fibroblasts. Inhibition of TRAF2 or TRAF3 signaling using dominant-negative TRAFs (dnTRAFs) impaired LMP1-induced transformation of Rat-1 cells and decreased activation of ERK1/2. These data identify a signaling pathway that is uniquely activated by CTAR1 and is necessary for LMP1-mediated transformation.
MATERIALS AND METHODS
Plasmids.
LMP1 and LMP1 mutants were cloned into the pBabe-puromycin vector with a hemagglutinin (HA) tag at the amino terminus. LMP1-A5 (204-208AAAAA), LMP1Δ204-208, LMP1-204/6AA, and LMP1-208A were subcloned from plasmids described in reference 35 into pBabe-puromycin with the same primers used to clone full-length LMP1 in reference 18. Truncation mutants with unaltered CTAR1 were constructed using the full-length LMP1 construct, and truncation mutants with a mutated CTAR1 (A5) were constructed using the LMP1-A5 construct. PCR amplification of the LMP1 mutants used the same 5′ primer (LMP1-HA5′ used to clone full-length LMP1), but different 3′ primers were used to produce the various constructs. The 1-187 construct was produced with 3′ primer 3TMSTOP (ATCACGAGGAATTCCTATTAATGGTAATACATCCAGATTAAAATCGCC), 1-195 with the 3′ primer 3TM195 (ATCACGAGGAATTCCTATTAGTGTTCATCACTGTGTCGTTGTC), 1-203 with the 3′ primer 3TM203 (ATCACGAGGAATTCCTATTAGTGCGGGAGGGAGTCATC), 1-208 with the 3′ primer TM-208 (ATCACGAGGAATTCCTATTAGGTAGCTTGTTGAGGGTGCG), 1-208-A5 with the 3′ primer TM-2085A (ATCACGAGGAATTCCTATTAGGCAGCTGCTGCAGCGTG), 1-220 and 1-220 A5 with the 3′ primer 3TM220 (ATCACGAGGAATTCCTATTAGTTGGAGTTAGAGTCAGATTCATGG), LMP1-378Stop and A5-378Stop with the 3′ primer 378STOP (ATCACGAGGAATTCCTATTAGCCGTGGGGGTCGTCATC), and LMP1-Y384G and A5-Y384G with the 3′ primer Y384G (ATCACGAGGAATTCCTATTATTAGTCATAGCCGCTTAGCTGAACTGG). PCR amplification was monitored by restriction enzyme digestion and insertion into the BamHI and EcoRI sites of pBabe-puromycin-M3 (with a puromycin or hygromycin cassette and containing a triple myc tag [M3]).
An expression cassette encoding three tandem myc epitope tags was moved from M3-LMP1 (18) to pBabe-puromycin for retroviral expression of myc-tagged proteins. The myc tag was amplified with M3EBH5′ (CGCCGGATCCAGATCTAAGCTTGCCGCTCGAGCCACCATGGAACAAAAG) and M3Bam3′ (CGTGTTCCATATGGATCCGAG), digested with BglII and BamHI, and cloned into the BamHI site of pBabe-puromycin. Cloning in frame with the myc tag results in triple-myc-tagged proteins.
The dnTRAF6 construct was a gift from Ilona Jaspers (8). dnTRAF2 and dnTRAF3 were subcloned in frame into pBabe-puromycin-M3 (containing a triple myc tag) from plasmids described in reference 37. dnTRAF2 was PCR amplified with the 5′ primer TRF2DN5′ (CGACGGATCCATAGGGAGGTGGAGAGCCTGCCG) and the 3′ primer TRF2DN3′ (ATCACGAGGTCGACTTAGAGCCCTGTCAGGTCCACAATGG). dnTRAF3 was PCR amplified with the 5′ primer TRF3DN5′ (CGACGGATCCTAGAGGAAGCAGACAGCATGAAGAGCAG) and the 3′ primer TRF3DN3′ (ATCACGAGGTCGACTCAGGGATCGGGCAGATCCGAAG). PCR amplification was followed by restriction enzyme digestion and insertion into the BamHI and SalI sites of pBabe-puromycin-M3 (puromycin and hygromycin). The 1-231 construct was described previously (18).
LMP1 was cloned into retroviral packaging vector pHSCG (kindly provided by Lishan Su, University of North Carolina at Chapel Hill [9]) coexpressing green fluorescent protein under the control of the cytomegalovirus promoter. HA-tagged LMP1 was amplified using HA-EcoRI (GCCGAATTCATGGCTTACCCATACGATGTTCCAG) and LMP1 SalI (GCGGTCCAGAATGTGGCTTTTCAGCCTAGAC) primers, digested with EcoRI and SalI, and ligated into the EcoRI and XhoI sites of pHSCG by compatible ends.
Retrovirus production and transduction.
Retroviruses were produced as described previously (18). Briefly, subconfluent HEK-293T cells were triple transfected with 5 μg of pBabe-puromycin or pHSCG or each of the pBabe-puromycin-HA-tagged LMP1 constructs or pHSCG-LMP1, 5 μg of pVSVG, and 5 μg of pGag/Pol using Fugene 6 (Roche) transfection reagent according to the manufacturer's directions. Following overnight incubation at 37°C, the medium was replaced with fresh medium (Dulbecco modified Eagle medium [DMEM] [GIBCO] with 10% fetal bovine serum [FBS] [Sigma] and antibiotic/antimycotic [GIBCO]), and the cells were incubated at 33°C overnight. Cell supernatant containing the retrovirus was clarified by centrifugation at 1,000 × g for 5 min. Rat-1 cells were then transduced with the clarified supernatant and 4 μg/ml Polybrene overnight.
Cell culture and stable cell lines.
Rat-1 fibroblasts and HEK-293T cells were maintained in DMEM supplemented with 10% FBS (Sigma) and antibiotic/antimycotic (GIBCO). Rat-1 cells were prepared fresh and maintained for no more than five passages for each experiment. Stable cell lines with pBabe-puromycin, the LMP1 constructs, or the dnTRAFs were established by transduction with recombinant retrovirus followed by selection with 5 μg/ml of puromycin (Sigma). Alternatively, cells stably expressing dnTRAF were kept under hygromycin (Cellgro) at a concentration of 200 μg/ml.
Focus formation and soft agars.
Subconfluent Rat-1 cells were transduced with recombinant retrovirus in six-well plates overnight. Cells were then maintained by changing the medium every other day for 10 to 14 days. To assay for focus formation in the presence of U0126 (Calbiochem), stable cell lines were established as described previously and seeded in duplicate six-well plates. Upon reaching confluence and every other day thereafter for 7 to 10 days, fresh medium containing 10 μM U0126 or vehicle control (dimethyl sulfoxide from Sigma) was added to the cells. Cells were then stained with 1% crystal violet in 50% ethanol and imaged with a stereomicroscope.
Soft agar assays were performed as described previously (33, 46). Briefly, Bacto agar medium (5% Bacto agar in DMEM with 10% FBS and antibiotic/antimycotic to a final concentration of 0.5%) was poured into a 12-well plate and allowed to solidify. A total of 1.0 × 105 to 2.0 × 105 cells/well were resuspended in Bacto agar medium, overlaid on the initial layer of solidified Bacto agar medium, and allowed to solidify. Medium was added on top of the solidified Bacto agar medium and changed every 2 or 3 days for 14 to 21 days. For the dnTRAF soft agars, medium was supplemented with 5 μg/ml of puromycin to maintain stable expression of the dnTRAFs. Cells were imaged with phase-contrast microscopy and fluorescence microscopy to verify green fluorescent protein expression.
Cell harvesting and Western blotting.
Cell lines were grown to confluence and harvested as described previously (18). Briefly, after the cells were harvested, they were washed with ice-cold phosphate-buffered saline (GIBCO), centrifuged at 1,000 × g, and lysed with radioimmunoprecipitation assay buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 0.1% deoxycholate, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and protease and phosphatase inhibitor cocktails from Sigma). Lysates were incubated on ice for 10 min and clarified by centrifugation, and protein concentration was determined using the Bio-Rad DC protein assay system. Lysates were boiled in SDS sample buffer for 5 min, 12.5 or 25.0 μg of protein was separated by SDS-polyacrylamide gel electrophoresis and transferred to an Optitran membrane (Whatman) for Western blot analysis, and efficient transfer was confirmed with Ponceau S staining (Fluka). Primary antibodies used included HA probe (Covance), CS1-4 (Dako), c-myc tag, β-actin, TRAF6, phospho-ERK1/2 (Y473), total ERK1/2, poly(ADP-ribose) polymerase (PARP) (Santa Cruz), plakoglobin (Abcam), CDK2, Rb (BD Biosciences), total GSK3 (Upstate Biotechnology), phospho-Rb (Oncogene), p27Kip1 (Calbiochem and Cell Signaling), phospho-GSK3β (S9), phospho-CDK2 (T160), phospho-Akt (S473), phosphorylated protein kinase c zeta isoform (PKC-ζ) (T410/403), and total Akt (Cell Signaling). Densitometry analysis was done with Image J software.
RESULTS
CTAR1 is necessary for rodent fibroblast transformation.
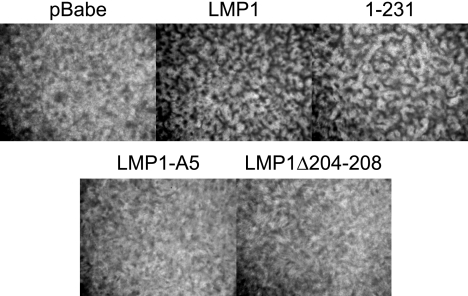
The LMP1 gene is considered the EBV oncogene as it is able to block contact inhibition and induce anchorage-independent growth in Rat-1 fibroblasts (18, 33, 51). Deletion mutants of LMP1 indicated that CTAR1 but not CTAR2 was required for rodent fibroblast transformation (33). To assess the role of the TRAF-binding site in rodent fibroblast transformation, two TRAF-binding domain mutants were developed. LMP1-A5 has the TRAF-binding domain mutated from PQQAT to AAAAA, and LMP1Δ204-208 has a deletion of the TRAF-binding domain encompassing amino acids 204 to 208. To determine whether these constructs were able to block contact inhibition, Rat-1 fibroblasts were transduced with pBabe-puromycin (vector), LMP1, 1-231 (LMP1 deleted for amino acids 232 to 386), LMP1-A5, and LMP1Δ204-208. Ten days posttransduction, cells were analyzed for block of contact inhibition as indicated by focus formation (Fig. 1). As previously described, LMP1 and 1-231, both of which contain CTAR1, induced focus formation; however, foci were not produced by pBabe-puromycin and the two TRAF-binding domain mutants, LMP1-A5 and LMP1Δ204-208. Western blot analysis on cells transduced in parallel confirmed expression of all four constructs, indicating that the failure to block contact inhibition was not due to failed transduction or expression (data not shown). These data reveal that the TRAF-binding domain, amino acids 204 to 208, is necessary for rodent fibroblast transformation.
FIG. 1.
The TRAF-binding domain is necessary for LMP1-mediated block of contact-inhibited growth. Rat-1 fibroblasts were transduced with pBabe-puromycin (pBabe), LMP1, 1-231, LMP1-A5, or LMP1Δ204-208, maintained for 10 days, stained with crystal violet, and observed for focus formation.
Role of CTAR1 in LMP1 signal transduction.
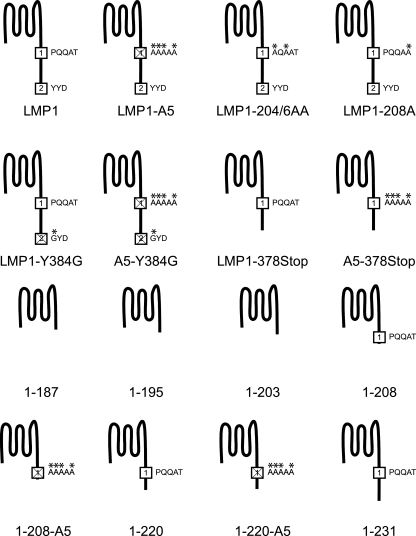
To further evaluate the role of CTAR1 in the induction of several signal transduction pathways, additional LMP1 mutants were constructed (Fig. 2). Two full-length LMP1 mutants of CTAR1, LMP1-204/6AA (two alanine substitutions at amino acids 204 and 206) and LMP1-208A (alanine substitution at amino acid 208), were subcloned to evaluate the role of the TRAF-binding site within CTAR1. The serial truncation LMP1 mutants ranged from the start of the carboxyl-terminal tail at amino acid 188 to amino acid 231. Finally, a CTAR2 deletion mutant (LMP1-378Stop) and a point mutant that has been described to abrogate most CTAR2 activity (LMP1-Y384G) were constructed with a functional CTAR1 and a mutant CTAR1 (A5-378Stop and A5-Y384G) (4, 20). All LMP1 constructs were tagged hemagglutinin at the amino terminus.
FIG. 2.
Graphical representations of LMP1 mutants. All constructs are tagged at the amino terminus with HA. CTAR1 and CTAR2 are represented by squares with the number 1 or 2 in the square, respectively. Substitutions to CTAR1 or CTAR2 are highlighted with an asterisk above the mutated residue. The mutants in the bottom two rows are truncation mutants.
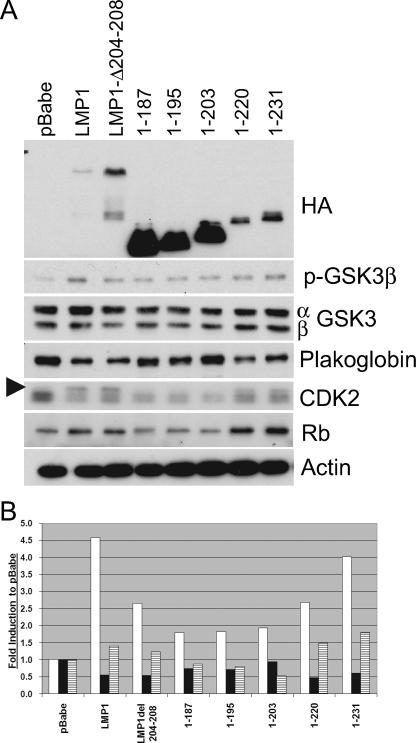
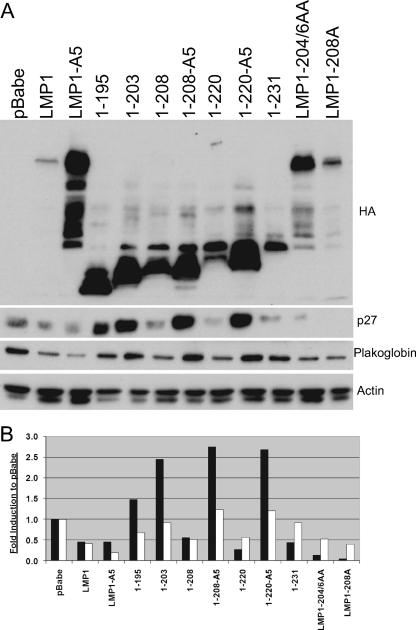
Rat-1 fibroblasts stably expressing pBabe-puromycin, LMP1, LMP1Δ204-208, 1-187, 1-195, 1-203, 1-220, and 1-231 were assayed for their ability to modulate three aspects of transformation that have been linked to LMP1: PI3K-Akt signaling, cell cycle deregulation, and cellular adhesion (Fig. 3A). The levels of expression of these targets were normalized to actin and are presented as changes in induction from that of vector control (Fig. 3B). Expression of LMP1 was confirmed using antibodies against the amino-terminal HA tag. The variation in the levels of LMP1 expression has been shown to result from ubiquitination and proteasomal degradation mediated by CTAR1-TRAF binding (44). As reported previously, full-length LMP1 induced activation of the PI3K-Akt signaling pathway as represented by phosphorylation and inactivation of the Akt target, GSK3β. The LMP1 mutants 1-220 and 1-231, which are deleted for CTAR2, also slightly increased levels of phosphorylated GSK3β, although 1-231 had the strongest induction. The deletion mutants (1-187, 1-195, and 1-203) were impaired in their effects on GSK3β. Interestingly, deletion of the TRAF-binding domain in full-length LMP1 (LMP1Δ204-208) retained approximately 60% of the activity of full-length LMP1 (Fig. 3B).
FIG. 3.
CTAR1 induces activation of PI3K-Akt signaling and deregulates markers associated with cell cycle progression. (A) Rat-1 cells stably expressing pBabe-puromycin (pBabe), LMP1, or LMP1 mutants were assayed by Western blot analysis for expression of LMP1 constructs with antibodies against HA, phosphorylated GSK3β (p-GSK3β) (S9), total GSK3 (α and β forms), plakoglobin, CDK2 (hyperphosphorylated CDK2 is marked by the arrowhead), total Rb, and actin as a loading control. (B) Quantitative analysis of protein levels of pGSK3b (white bars), plakoglobin (black bars), and Rb (stippled bars). Values are changes in induction compared to the level induced by pBabe-puromycin (pBabe).
Increased levels of hyperphosphorylated CDK2 and increased levels of total Rb, cellular markers involved in G1/S cell cycle progression, were observed in LMP1, LMP1Δ204-208, 1-220, and 1-231. Total Rb levels have been demonstrated to be increased concomitant with the hyperphosphorylated form of Rb in the presence of LMP1, although the effect of the increase in Rb levels remains to be determined (34). Plakoglobin, a cellular protein associated with cell-cell adhesion, was also downregulated by LMP1, LMP1Δ204-208, 1-220, and 1-231. The ability of the nontransforming LMP1Δ204-208 to modulate GSK3β, plakoglobin, CDK2, and Rb to levels similar to those by the transforming 1-220 is surprising. Comparison of the truncation mutants (1-187, 1-195, and 1-203) to those that contain CTAR1 (1-220, and 1-231) indicated that the TRAF-binding domain is required to affect these cellular pathways. However, comparison of full-length LMP1 and LMP1Δ204-208 revealed that in addition to the TRAF-binding domain, another region in the carboxy terminus can also contribute to these effects on cell expression and regulation.
Mutations in CTAR1 ameliorate LMP1 transforming potential.
Two residues within the TRAF-binding domain, the proline at amino acid 204 and the glutamine at position 206, that constitute CTAR1 have been found to have an important role in TRAF binding (35). To further evaluate the roles of the residues within CTAR1 in rodent fibroblast transformation, Rat-1 cells were transduced with pBabe-puromycin, LMP1, LMP1-A5, 1-195, 1-203, 1-208, 1-208-A5, 1-220, 1-220-A5, 1-231, LMP1-204/6AA, and LMP1-208A and assayed for the ability to block contact inhibition and induce foci (Fig. 4). Cells transduced with LMP1, 1-220, 1-231, LMP1-208A, and LMP1-204/6AA formed foci, while cells transduced with truncation mutants 1-195, 1-203, and 1-208 did not form foci. Cells transduced with LMP1-204/6AA were impaired in this ability as indicated by smaller foci. Cells transduced with LMP1-208A formed foci to levels similar to those of cells transduced with LMP1, indicating that the threonine residue at position 208 in the TRAF-binding site is not necessary for transformation. Although cells transduced with 1-208 did not form foci even though the TRAF-binding site is intact, previous studies have shown that the aspartic acid residue at position 210 is necessary for TRAF3 binding, suggesting that 1-208 could be nontransforming due to this TRAF-binding defect (52). Furthermore, substitution of CTAR1 to all alanine residues (LMP1-A5 or 1-220-A5) or deletion of CTAR1 (1-195 or 1-203) inhibited transformation. These data indicate that the TRAF-binding site is required for transformation and that additional residues between amino acids 208 and 220 contribute to its function.
FIG. 4.
Transformation by LMP1 CTAR1 mutants. Rat-1 cells were transduced with pBabe-puromycin (pBabe), LMP1, or LMP1 mutants, maintained for 14 days, stained with crystal violet, and observed for focus formation.
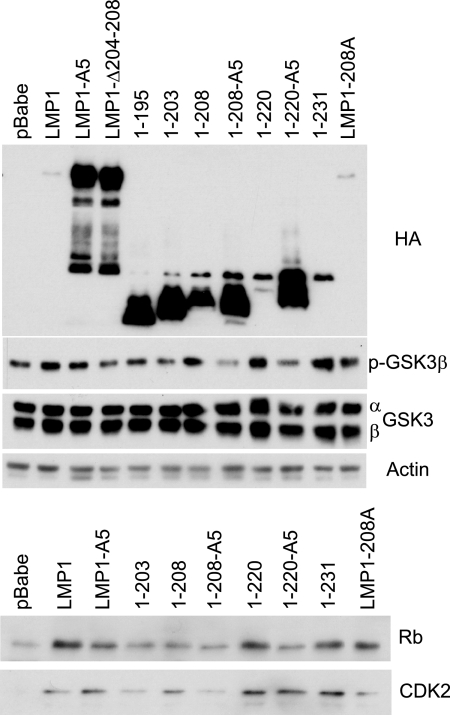
Effects of mutation of the TRAF-binding domain on LMP1 signaling.
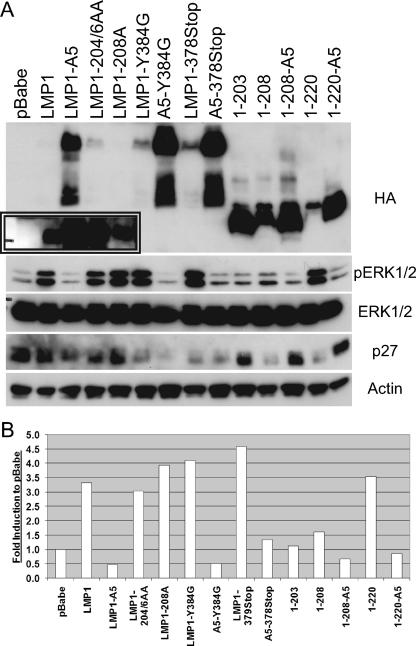
To evaluate the roles of CTAR1 and the amino acids surrounding CTAR1 in LMP1-mediated signal transduction, Rat-1 cells stably expressing LMP1 and several LMP1 mutants were established and their ability to induce various signal transduction pathways was assessed through Western blot analysis (Fig. 5). LMP1, 1-208, 1-220, 1-231, and LMP1-208A had increased phospho-GSK3β. Mutation (LMP1-A5) or deletion (LMP1Δ204-208) of the TRAF-binding site within full-length LMP1 slightly decreased this induction (Fig. 5). In contrast, mutation of the TRAF-binding site within truncated LMP1 (1-208-A5 or 1-220-A5) abolished the effect of LMP1 on the level of phospho-GSK3β (Fig. 5). Similarly, total Rb levels were upregulated by LMP1, LMP1-A5, 1-220, 1-231, and LMP1-208A (Fig. 5). Although full-length LMP1Δ204-208, LMP1-A5, and 1-208 are nontransforming, they all affected the levels of phosphorylated GSK3β and total Rb. Within the truncated forms of LMP1, 1-208, or 1-220, mutation of the TRAF-binding site inhibited the ability of LMP1 to modulate GSK3β and Rb. Similar effects were observed with total levels of CDK2 (Fig. 5). With some of the mutants that had reduced but detectable activity, these markers were not uniformly affected in all experiments. For example, in the experiment presented, 1-208 clearly affected pGSK3β and CDK2 but had minimal effects on Rb. The overall assessment of the effects of LMP1 and the various mutants on these cellular proteins is presented in Table 1. These data confirm that sequences between CTAR1 and CTAR2 can compensate for the loss of the TRAF-binding domain and modulate the levels of pGSK3β, Rb, and CDK2.
FIG. 5.
Effects on LMP1-induced inactivation of GSK3β and cell cycle markers by LMP1 CTAR mutants. Rat-1 cells stably expressing pBabe-puromycin (pBabe), LMP1, or LMP1 mutants were assayed for LMP1 expression with antibodies against HA, phosphorylated GSKβ (p-GSKβ), total GSK3 (α and β subunits), actin as a loading control, total Rb, and total CDK2.
TABLE 1.
Summary of activities of LMP1 and LMP1 mutantsa
| Construct | Transforming ability | Inhibition by U0126 | Level of proteinb
|
|||
|---|---|---|---|---|---|---|
| p-GSK3β | p27Kip1 | Plakoglobin | p-ERK1/2 | |||
| LMP1 | Yes | Yes | ++++ | −−−− | −−− | ++++ |
| LMP1-A5 | No | Yes | +++ | −−−− | −−−− | NC |
| LMP1-Δ204-208 | No | Yes | ++ | −−−− | −−−− | NC |
| LMP1-204/6AA | Yesc | Yes | ++ | −−− | −−− | +++ |
| LMP1-208A | Yes | Yes | +++ | −− | −−− | ++++ |
| LMP1-Y384G | Yes | Yes | ++ | −−− | −−− | ++++ |
| A5-Y384G | No | Yes | +++ | −−−− | −−−− | NC |
| LMP1-378Stop | Yes | Yes | ++ | −−− | −−− | ++++ |
| A5-378Stop | No | Yes | +++ | −−− | −−−− | NC |
| 1-187 | No | Yes | + | ++ | − | NC |
| 1-195 | No | Yes | + | ++ | − | NC |
| 1-203 | No | Yes | + | +++ | NC | NC |
| 1-208 | No | Yes | +++ | −−−− | −−−− | NC |
| 1-208-A5 | No | Yes | + | +++ | + | NC |
| 1-220 | Yes | Yes | ++++ | −−−− | −−−− | ++++ |
| 1-220-A5 | No | Yes | + | +++ | + | NC |
| 1-231 | Yes | Yes | ++++ | −−−− | −− | ++++ |
LMP1 and LMP1 mutants that are transforming are shown in boldface type.
The level of protein (phosphorylated GSK3β [p-GSK3β] and phosphorylated ERK1/2 [pERK1/2]) compared to the level of the control is shown. Plus signs indicate a level higher than that of the control, while minus signs indicate a level lower than that of the control. The number of symbols indicates levels of change from vector control, with more symbols indicating a greater level of change. NC, no change compared to that of the control.
Decreased transformation potential.
Rat-1 cells stably expressing LMP1 and several LMP1 mutants were also assayed for their ability to deregulate the cell cycle marker p27Kip1 as well as the cell adhesion protein plakoglobin (Fig. 6A). LMP1, LMP1-A5, 1-208, 1-220, 1-231, LMP1-204/6AA, and LMP1-208A decreased the protein levels of p27Kip1 and plakoglobin. Similar to the effects on pGSK3β, Rb, and CDK2, mutation of the TRAF-binding site in full-length LMP1 (LMP1-A5) did not affect the LMP1-induced downregulation of p27Kip1 and plakoglobin. However, the TRAF-binding site in 1-208 or 1-220 was absolutely required for the LMP1-mediated decrease of p27Kip1 and plakoglobin (Fig. 6). In addition, the mutations within the TRAF domain in LMP1-204/6AA and LMP1-208A did not affect the ability of LMP1 to decrease p27Kip1 and plakoglobin. These findings reveal that the effect of LMP1 on these cellular markers does not correlate with the ability to transform Rat-1 cells, as LMP1-A5 and 1-208 are nontransforming but still modulate these cellular markers. The data suggest that the TRAF-binding site is the main effector in the induction of PI3K-Akt signaling, as measured by GSK3β phosphorylation, the deregulation of cell cycle markers Rb and p27Kip1, and the cell adhesion marker plakoglobin. However, the data also confirm that in the absence of the TRAF-binding site, sequences between CTAR1 and CTAR2 can also modulate these cellular markers. Interestingly, all of the truncated forms of LMP1 (1-195, 1-203, 1-208-A5, and 1-220-A5) that lack the TRAF domain and the sequences between CTAR1 and CTAR2 actually increased the levels of p27Kip1 and plakoglobin compared to the vector control.
FIG. 6.
Effects on deregulation of p27Kip1 and plakoglobin levels by LMP1 CTAR mutants. (A) Rat-1 cells stably expressing pBabe-puromycin (pBabe), LMP1, or LMP1 mutants were assayed for LMP1 expression with antibodies against HA, p27Kip1, plakoglobin, or actin as a loading control by Western blot analysis. (B) Quantitative analysis of p27Kip1 (black bars) and plakoglobin (white bars) protein levels. Values shown are changes in induction compared to the level induced by pBabe-puromycin (pBabe).
ERK1/2 activation is necessary for LMP1-mediated transformation.
LMP1 has been shown to induce activation of mitogen-activated protein kinase and ERK1/2 in Rat-1 fibroblasts and in T cells (7, 43). To determine the role of CTAR1 in ERK1/2 activation, Rat-1 cells stably expressing pBabe-puromycin, LMP1, or various LMP1 constructs were assayed for ERK1/2 activation by Western blot analysis (Fig. 7A). Densitometry analysis was performed to quantitate the levels of phosphorylated ERK1/2 (Fig. 7B). All of the LMP1 mutants that contained the TRAF-binding domain (LMP1-204/6AA, LMP1-208A, LMP1-Y384G, LMP1-378Stop, and 1-220) had significantly increased levels of phosphorylated ERK1/2. Mutation or deletion of CTAR2 (LMP1-Y384G, A5-Y384G, LMP1-378Stop, or A5-378Stop), whether in the presence or absence of a wild-type TRAF-binding site, did not affect the levels of phosphorylated ERK1/2 or p27Kip1. In contrast to that observed with phosphorylated GSK3β, Rb, p27Kip1, and plakoglobin, activation of ERK1/2 was detected only in cells expressing LMP1 constructs that were transforming. LMP1-A5 and 1-220-A5, which are nontransforming, had significantly reduced levels of phospho-ERK1/2 compared to LMP1 and 1-220, which are transforming. These data indicate that ERK1/2 activation is mediated strictly by the TRAF-binding site and that the CTAR1-mediated activation of ERK1/2 is a major factor in LMP1-mediated transformation of rodent fibroblasts.
FIG. 7.
CTAR1 is necessary for LMP1-induced activation of ERK1/2. (A) pBabe-puromycin (pBabe), LMP1, or LMP1 mutants stably expressed in Rat-1 cells were assayed by Western blot analysis for LMP1 expression (longer exposure shown in inset) with antibodies against HA, phosphorylated ERK1/2 (pERK1/2) (Y204), total ERK1/2, p27Kip1, or actin as a loading control. (B) Quantitative analysis of phosphorylated ERK1/2 protein levels. Values shown are changes in induction compared to the level induced by pBabe-puromycin (pBabe).
To evaluate the requirement of MEK1/2 and ERK1/2 signaling to LMP1-mediated transformation, parallel sets of stable Rat-1 cells were treated for 7 days with dimethyl sulfoxide or 10 μM U0126, a MEK1/2 chemical inhibitor. Cells were then observed for focus formation or assessed for ERK1/2 activation by Western blot analysis. U0126 treatment abolished the LMP1-mediated block of contact inhibition as measured by focus formation. U0126 treatment equally inhibited focus formation induced by the various LMP1 mutants (Table 1). U0126 also blocked LMP1-induced ERK1/2 activation, and LMP1 expression levels in the presence and absence of U0126 were confirmed by Western blot analysis to ensure that the effects on contact inhibition were not due to lack of LMP1 expression (data not shown). These data indicate that ERK1/2 activation is necessary for LMP1-mediated transformation of Rat-1 fibroblasts. The relationship of activation of these pathways to transformation is presented in Table 1.
LMP1-mediated transformation is impaired by dnTRAF2 and dnTRAF3.
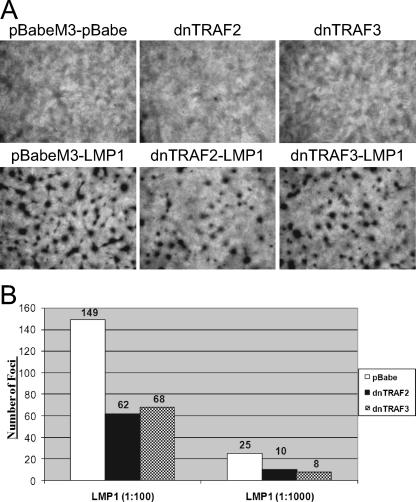
The interaction of CTAR1 with several TRAFs, including TRAF1, -2, -3, and -5, mediates the activation of signal transduction pathways, such as NF-κB and induction of key progrowth cellular molecules like the EGFR (5, 12, 13, 35, 37, 53). To determine whether TRAF2 and/or TRAF3 mediates the ability of LMP1 to block contact-inhibited growth, Rat-1 cell lines stably expressing pBabe-puromycin-M3, dnTRAF2, or dnTRAF3 were established and then transduced with serial dilutions of either pBabe-puromycin or LMP1. Cells on duplicate plates were harvested, and expression of the dnTRAFs and LMP1 was confirmed by Western blot analysis indicating reduced levels of nuclear NF-κB isoforms p50 and p65 (data not shown). dnTRAFs contain the TRAF-binding domain but have been deleted of their RING and zinc finger domains, thus allowing interaction with their target molecule while impairing their ability to induce signaling upon their recruitment. Fourteen days posttransduction, cells were observed for focus formation (Fig. 8A). Transduction of the pBabe-puromycin-M3 (vector) stable cells with pBabe-puromycin (pBabe-puromycin-M3-pBabe-puromycin) or the stable cells expressing only dnTRAF2 or dnTRAF3 did not show signs of focus formation. In contrast, transduction of the pBabe-puromycin-M3, dnTRAF2, or dnTRAF3 stable cells with LMP1 resulted in focus formation. Although dnTRAF2 or dnTRAF3 did not completely ablate focus formation, they markedly reduced the number of foci.
FIG. 8.
LMP1-mediated transformation of Rat-1 fibroblasts is inhibited by dnTRAF2 and dnTRAF3 but not dnTRAF6. (A) Rat-1 fibroblasts stably expressing pBabe-puromycin-M3 (pBabe-M3), dnTRAF2, or dnTRAF3 were transduced with pBabe-puromycin (pBabe) or LMP1, maintained for 14 days, stained with crystal violet, and observed for focus formation. (B) Quantitative analysis of focus formation assay of stable Rat-1 cells expressing pBabe-puromycin (pBabe), dnTRAF2, or dnTRAF3 transduced with pBabe-puromycin or LMP1 at two serial dilutions. Experiment was performed in triplicate with representative values of a single experiment shown.
Quantitative analysis from a representative experiment of the foci indicate that dnTRAF2 and dnTRAF3 repressed focus formation by approximately 2.5-fold (Fig. 8B). Furthermore, anchorage-independent growth was also impaired, but not fully blocked, by either dnTRAF2 or dnTRAF3 (data not shown). To confirm the specificity of the inhibition on LMP1-mediated transformation by dnTRAF2 and dnTRAF3, Rat-1 cells stably expressing dnTRAF6 were transduced with LMP1 and assayed for focus formation. TRAF6 is recruited to LMP1 and signals specifically through CTAR2, not CTAR1 (47), and as such LMP1-mediated focus formation was not impaired by the stable expression of dnTRAF6 (data not shown). These data suggest that TRAF2 and TRAF3, but not TRAF6, contribute to LMP1-mediated transformation of rodent fibroblasts. The inability of dnTRAF2 and dnTRAF3 to fully block LMP1-mediated transformation suggests a redundancy in TRAF function. Indeed, in TRAF2 and TRAF5 knockout MEFs, LMP1 can still mediate nuclear translocation of the NF-κB subunit p65 (31).
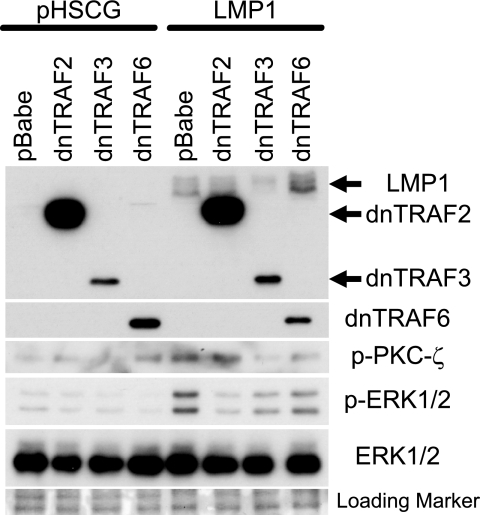
To determine the roles of TRAF2, TRAF3, and TRAF6 in the modulation of LMP1-mediated activation of ERK1/2, Rat-1 cells stably expressing pBabe-puromycin, dnTRAF2, dnTRAF3, or dnTRAF6 were transduced with vector (pHSCG) or LMP1, followed by Western blot analysis (Fig. 9). Expression of LMP1 was confirmed with a pool of monoclonal antibodies to LMP1, while expression of myc-tagged dnTRAF2 and dnTRAF3 and of dnTRAF6 was confirmed with anti-myc or anti-TRAF6. Interestingly, expression of dnTRAF2 was considerably higher than expression of dnTRAF3. Expression of phosphorylated ERK1/2 was significantly inhibited by dnTRAF2 and dnTRAF3 and partially affected by dnTRAF6. This finding suggests that the effects of dnTRAF2 and dnTRAF3 on Rat1 transformation are likely mediated in part by their effects on ERK activation.
FIG. 9.
LMP1-induced activation of ERK1/2 is inhibited by dnTRAF2 and dnTRAF3. Rat-1 cells stably expressing pBabe-puromycin (pBabe), dnTRAF2, dnTRAF3, or dnTRAF6 were transduced with pHSCG or LMP1 and assayed for expression with antibodies against LMP1, dnTRAF2, dnTRAF3, dnTRAF6, phosphorylated PKCζ (p-PKCζ), phosphorylated ERK1/2 (p-ERK1/2), and total ERK1/2. Ponceau S-stained membrane was used to confirm equal loading.
ERK1/2 activation can be mediated through the PI3K-dependent activation of PKC-ζ, which then activates MEK1/2 (3, 39). To determine the role of PKC-ζ in LMP1-induced activation of ERK1/2, Rat-1 cells stably expressing LMP1 and dnTRAF2, dnTRAF3, or dnTRAF6 were analyzed by Western blotting. LMP1 expression increased the levels of phosphorylated PKC-ζ in the control Rat-1 cells containing the vector (pBabe-puromycin). Rat-1 cells stably expressing high levels of dnTRAF2 did not impair PKC-ζ phosphorylation, while dnTRAF3 completely inhibited PKC-ζ phosphorylation despite its lower levels of expression. Expression of dnTRAF6 expression moderately impaired this activity; however, dnTRAF6 expression activated PKC-ζ in the absence of LMP1. Although dnTRAF2 almost entirely inhibited the LMP1-induced ERK1/2 activation, it did not affect PKC-ζ phosphorylation. In contrast, dnTRAF3 inhibited the activation of ERK1/2 to a lesser degree than dnTRAF2 but completely blocked activation of PKC-ζ. Total levels of PKC-ζ were not detected due to poor reactivity of the PKC-ζ antiserum; therefore, equal protein loading was confirmed by Ponceau S staining.
These data suggest that LMP1 activation of ERK1/2 is mediated through TRAF2 and TRAF3 while the phosphorylation of PKC-ζ requires TRAF3 but not TRAF2. Due to the differences in expression of both dnTRAF2 and dnTRAF3, it is difficult to determine the exact contribution of TRAF2 and TRAF3 to the activation of PKC-ζ and/or ERK1/2. However, the ability of LMP1 to induce ERK phosphorylation in the absence of phosphorylated, and activated, PKC-ζ suggests that PKC-ζ phosphorylation is not necessary for LMP1-mediated ERK activation.
DISCUSSION
The data presented in this study evaluated the roles of the TRAF-binding site in LMP1-mediated transformation and LMP1-mediated signal transduction. Several studies have characterized the ability of LMP1 to recruit TRAFs (TRAF1, -2, -3, and -5) to CTAR1 through a consensus TRAF-binding site at positions 204 to 208 (5, 25, 35, 37, 38, 52, 53). Substitution of the proline and glutamine residues to alanines at positions 204 and 206 in the TRAF-binding site has been demonstrated to disrupt all TRAF1 and TRAF2 binding and most, but not all, TRAF3 binding, respectively (35). In agreement with these data, substitution of the proline and glutamine residues to alanines in the TRAF-binding site (LMP1-204/6AA), which has decreased TRAF3 binding (35), impaired LMP1-induced focus formation. Accordingly, LMP1 with the substitution of threonine at position 208 to alanine, which does not affect TRAF binding, transformed rodent fibroblasts to levels similar to those transformed by wild-type LMP1 (36).
TRAF binding to LMP1 is also aided by the residues surrounding the TRAF-binding site. While the aspartic acid residue at position 209 is not necessary for TRAF binding, the aspartic acid residue at position 210 is required for TRAF binding (5, 52). The aspartic acid at position 210 is also part of a destruction box motif that binds the E3-ubiquitin ligase component homologue of Slimb (HOS), which contributes to regulating LMP1-mediated IκBα-dependent NF-κB signaling (34, 48). These data are corroborated by the finding in this study that 1-208 is nontransforming even though an intact TRAF-binding site is present. Interestingly, although 1-208 is nontransforming, it is still able to modulate effects on GSK3β, Rb, p27Kip1, and plakoglobin, indicating that some signal transduction properties remain intact. Furthermore, these data indicated that activation of these pathways is not sufficient for Rat-1 fibroblast transformation. Recently, the aspartic acid at position 211 and the serine at position 215 have been demonstrated to be phosphorylation sites within LMP1 (6). While these residues have not been shown to affect TRAF binding, their close proximity to the TRAF-binding site could regulate TRAF-LMP1 through their phosphorylation status.
CTAR1 has been demonstrated to recruit TRAF1-TRAF2 or TRAF3-TRAF5 heterodimers, with the binding of the heterodimers being mutually exclusive (13). Signal transduction mediated by the LMP1-TRAF interaction is more complex than simply recruiting the TRAFs to LMP1. In knockout TRAF2 and TRAF5 MEFs, TRAF6 is essential for NF-κB signaling, but in mouse lymphoma cells, TRAF3 is central for such activation (31, 53, 54). TRAF-LMP1 interactions are further complicated by the role of adapter molecules in mediating binding, especially in CTAR2-TRAF binding. The finding that coexpression of either dnTRAF2 or dnTRAF3 inhibited, but did not fully block, LMP1-mediated transformation indicates that TRAF2 and TRAF3 are important factors in transformation but that other TRAFs (e.g., TRAF1 and TRAF5) can also contribute to transformation.
The data in this study extend the important and unique properties of CTAR1, which is necessary for B-lymphocyte transformation by EBV as well as for LMP1-mediated transformation of rodent fibroblasts (25, 33). CTAR1 has the unique ability to induce expression of EGFR and TRAF1 and can deregulate molecules involved in G1/S cell cycle progression, such as Id1, the CDK inhibitor p27Kip1, CDK2, and Rb (13, 18, 27, 30, 35-37). Although CTAR1 is not as effective as CTAR2 in activation of NF-κB reporter constructs, CTAR1 induces specific forms of NF-κB through both canonical and noncanonical pathways and is primarily responsible for the activation of the PI3K-Akt signaling pathway (1, 11, 32, 33, 37, 45, 50).
In this study, the activation of the PI3K-Akt-GSK3β signaling pathway, the deregulation of several cellular markers involved in G1/S cell cycle progression and the cell adhesion protein plakoglobin were mapped to the TRAF-binding site within CTAR1, which was required in the context of a truncated molecule. However, deletion or mutation of CTAR1 in full-length LMP1 revealed that another signaling motif in LMP1 can augment LMP1-mediated activation of the PI3K-Akt-GSK3β pathway, deregulate cell cycle markers, such as p27Kip1, Rb, and CDK2, and decrease levels of plakoglobin. The motif associated with this redundancy in LMP1-mediated signaling does not appear to be CTAR2, as mutation of both CTAR1 and CTAR2 (A5-Y384G or A5-378Stop) could still decrease levels of p27 Kip1.
The absolute requirement for CTAR1 in a truncated form of LMP1 but not in full-length LMP1 suggests that the motif that provides signaling redundancy is located between amino acids 220 and 378. Importantly, this motif is not sufficient for transformation, as ablation of CTAR1 in any context yields a nontransforming phenotype. Putative Janus kinase 3-binding sites have been identified in the region between CTAR1 and CTAR2; however, the ability of LMP1 to bind and activate Janus kinase 3 has been controversial (21, 22). Another motif of interest is a putative TRAF-binding site between amino acids 320 and 324, although deletion of amino acids 232 to 351 did not affect TRAF binding to CTAR1 (23).
In this study, ERK1/2 activation was mapped to CTAR1, and this activation was shown to be necessary for LMP-1 mediated transformation of rodent fibroblasts. LMP1-mediated transformation and ERK1/2 activation are mediated by MEK1/2, as inhibition of MEK1/2 by the chemical inhibitor U0126 blocked both transformation and ERK1/2 activation. ERK1/2 responds to various mitogens and growth factors and can initiate changes in cellular proliferation and differentiation. The ability of LMP1 to affect RECK, a metastasis suppressor gene, is dependent on ERK activation, and LMP1-induced ERK1/2 activation has been demonstrated to be mediated by TRAF2 and TRAF5 in T cells (7, 31).
The requirement for ERK1/2 activation in transformation was supported by the inhibition of ERK1/2 activation by dnTRAF2 and the partial inhibition by dnTRAF3, both of which decreased transformation. These data suggest that TRAF2 and TRAF3 are important factors in LMP1-mediated activation of ERK1/2 in the context of rodent fibroblast transformation. A major difference between the transformation of rodent fibroblasts by LMP1 and B-lymphocyte immortalization by EBV is that activation of NF-κB is not required for rodent fibroblast transformation but is absolutely essential for immortalization of B lymphocytes. Activation of PI3 kinase is required for both rodent fibroblast and B-lymphocyte transformation. It will be interesting to determine the requirement for ERK1/2 activation in the transformation of B lymphocytes by EBV. Different cellular environments are likely to require the activation and inactivation of different cellular pathways. Indeed, LMP1 signaling has been demonstrated to be regulated in part by the ratio of TRAFs bound to its signaling domains (53), and different cell types likely have different endogenous levels of the different TRAFs.
We have previously shown that CTAR1-induced activation of PI3K-Akt signaling, but not IκBα-dependent NF-κB-signaling, is necessary for Rat-1 fibroblast transformation (33). The data presented in this study indicate that while the activation of PI3K-Akt signaling and deregulation of cell cycle markers associated in G1/S transition are necessary, these properties are not sufficient for rodent fibroblast transformation. The activation of ERK1/2 signaling by only those LMP1 constructs that have a transforming phenotype underscores the importance of ERK1/2 activation in LMP1-mediated transformation. It is becoming clear that LMP1 induces a variety of stimuli to affect cellular expression and induce transformation and that at least several of these signal transduction pathways are needed to work in unison for transformation to take place.
Acknowledgments
We thank Kathy Shair for the initial identification of LMP1 effects on plakoglobin, Ilona Jaspers for the dnTRAF6 construct, Lishan Su for the pHSCG construct, Rachael Liesman for help with subcloning LMP1-204/6AA and LMP1-208A, and Stephanie Mazzuca for assistance with Western blots. We also thank Betsy Edwards, Aron Marquitz, and Natalie Thornburg for critical reviews of the manuscript.
This work was supported by National Institutes of Health grants CA 19014 and CA 32979 to N.R.-T.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Atkinson, P. G., H. J. Coope, M. Rowe, and S. C. Ley. 2003. Latent membrane protein 1 of Epstein-Barr virus stimulates processing of NF-kappa B2 p100 to p52. J. Biol. Chem. 278:51134-51142. [DOI] [PubMed] [Google Scholar]
- 2.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 3.Berra, E., M. T. Diaz-Meco, I. Dominguez, M. M. Municio, L. Sanz, J. Lozano, R. S. Chapkin, and J. Moscat. 1993. Protein kinase C zeta isoform is critical for mitogenic signal transduction. Cell 74:555-563. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, P., J. E. Floettmann, A. Mehl, M. Jones, and M. Rowe. 2001. Mechanism of action of a novel latent membrane protein-1 dominant negative. J. Biol. Chem. 276:1195-1203. [DOI] [PubMed] [Google Scholar]
- 5.Busch, L. K., and G. A. Bishop. 2001. Multiple carboxyl-terminal regions of the EBV oncoprotein, latent membrane protein 1, cooperatively regulate signaling to B lymphocytes via TNF receptor-associated factor (TRAF)-dependent and TRAF-independent mechanisms. J. Immunol. 167:5805-5813. [DOI] [PubMed] [Google Scholar]
- 6.Chien, K. Y., Y. S. Chang, J. S. Yu, L. W. Fan, C. W. Lee, and L. M. Chi. 2006. Identification of a new in vivo phosphorylation site in the cytoplasmic carboxyl terminus of EBV-LMP1 by tandem mass spectrometry. Biochem. Biophys. Res. Commun. 348:47-55. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, H. C., J. D. Lay, W. C. Hsieh, H. C. Wang, Y. Chang, S. E. Chuang, and I. J. Su. 2005. Epstein-Barr virus LMP1 inhibits the expression of SAP gene and upregulates Th1 cytokines in the pathogenesis of hemophagocytic syndrome. Blood 106:3090-3096. [DOI] [PubMed] [Google Scholar]
- 8.Ciencewicki, J., L. Brighton, W. D. Wu, M. Madden, and I. Jaspers. 2006. Diesel exhaust enhances virus- and poly(I:C)-induced Toll-like receptor 3 expression and signaling in respiratory epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 290:L1154-L1163. [DOI] [PubMed] [Google Scholar]
- 9.Coffield, V. M., W. S. Helms, Q. Jiang, and L. Su. 2004. Galpha13 mediates a signal that is essential for proliferation and survival of thymocyte progenitors. J. Exp. Med. 200:1315-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson, C. W., A. G. Eliopoulos, S. M. Blake, R. Barker, and L. S. Young. 2000. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology 272:204-217. [DOI] [PubMed] [Google Scholar]
- 11.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694-3704. [DOI] [PubMed] [Google Scholar]
- 12.Devergne, O., E. D. Cahir McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devergne, O., E. Hatzivassiliou, K. M. Izumi, K. M. Kaye, M. F. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eliopoulos, A. G., S. M. Blake, J. E. Floettmann, M. Rowe, and L. S. Young. 1999. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J. Virol. 73:1023-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eliopoulos, A. G., and A. B. Rickinson. 1998. Epstein-Barr virus: LMP1 masquerades as an active receptor. Curr. Biol. 8:R196-R198. [DOI] [PubMed] [Google Scholar]
- 16.Eliopoulos, A. G., E. R. Waites, S. M. Blake, C. Davies, P. Murray, and L. S. Young. 2003. TRAF1 is a critical regulator of JNK signaling by the TRAF-binding domain of the Epstein-Barr virus-encoded latent infection membrane protein 1 but not CD40. J. Virol. 77:1316-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731-1742. [DOI] [PubMed] [Google Scholar]
- 18.Everly, D. N., Jr., B. A. Mainou, and N. Raab-Traub. 2004. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein-Barr virus and regulation of p27/Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J. Virol. 78:13470-13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fahraeus, R., H. L. Fu, I. Ernberg, J. Finke, M. Rowe, G. Klein, K. Falk, E. Nilsson, M. Yadav, P. Busson, T. Tursz, and B. Kallin. 1988. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int. J. Cancer 42:329-338. [DOI] [PubMed] [Google Scholar]
- 20.Floettmann, J. E., and M. Rowe. 1997. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C-terminus and requires oligomerisation for NF-kappaB activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 21.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higuchi, M., E. Kieff, and K. M. Izumi. 2002. The Epstein-Barr virus latent membrane protein 1 putative Janus kinase 3 (JAK3) binding domain does not mediate JAK3 association or activation in B-lymphoma or lymphoblastoid cell lines. J. Virol. 76:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi, K. M., E. D. Cahir McFarland, E. A. Riley, D. Rizzo, Y. Chen, and E. Kieff. 1999. The residues between the two transformation effector sites of Epstein-Barr virus latent membrane protein 1 are not critical for B-lymphocyte growth transformation. J. Virol. 73:9908-9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izumi, K. M., E. D. Cahir McFarland, A. T. Ting, E. A. Riley, B. Seed, and E. D. Kieff. 1999. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-κB activation. Mol. Cell. Biol. 19:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izumi, K. M., K. M. Kaye, and E. D. Kieff. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 94:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-kappaB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaye, K. M., O. Devergne, J. N. Harada, K. M. Izumi, R. Yalamanchili, E. Kieff, and G. Mosialos. 1996. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-kappa B activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc. Natl. Acad. Sci. USA 93:11085-11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In B. N. Fields, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, S. E. Straus, and D. M. Knipe (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins Publishers, Philadelphia, PA. [Google Scholar]
- 30.Li, H. M., Z. H. Zhuang, Q. Wang, J. C. Pang, X. H. Wang, H. L. Wong, H. C. Feng, D. Y. Jin, M. T. Ling, Y. C. Wong, A. G. Eliopoulos, L. S. Young, D. P. Huang, and S. W. Tsao. 2004. Epstein-Barr virus latent membrane protein 1 (LMP1) upregulates Id1 expression in nasopharyngeal epithelial cells. Oncogene 23:4488-4494. [DOI] [PubMed] [Google Scholar]
- 31.Luftig, M., E. Prinarakis, T. Yasui, T. Tsichritzis, E. Cahir-McFarland, J. Inoue, H. Nakano, T. W. Mak, W. C. Yeh, X. Li, S. Akira, N. Suzuki, S. Suzuki, G. Mosialos, and E. Kieff. 2003. Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc. Natl. Acad. Sci. USA 100:15595-15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luftig, M., T. Yasui, V. Soni, M. S. Kang, N. Jacobson, E. Cahir-McFarland, B. Seed, and E. Kieff. 2004. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc. Natl. Acad. Sci. USA 101:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mainou, B. A., D. N. Everly, Jr., and N. Raab-Traub. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917-6924. [DOI] [PubMed] [Google Scholar]
- 34.Mainou, B. A., and N. Raab-Traub. 2006. LMP1 strain variants: biological and molecular properties. J. Virol. 80:6458-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, W. E., J. L. Cheshire, and N. Raab-Traub. 1998. Interaction of tumor necrosis factor receptor-associated factor signaling proteins with the latent membrane protein 1 PXQXT motif is essential for induction of epidermal growth factor receptor expression. Mol. Cell. Biol. 18:2835-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, W. E., H. S. Earp, and N. Raab-Traub. 1995. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J. Virol. 69:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, W. E., G. Mosialos, E. Kieff, and N. Raab-Traub. 1997. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J. Virol. 71:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389-399. [DOI] [PubMed] [Google Scholar]
- 39.Naranatt, P. P., S. M. Akula, C. A. Zien, H. H. Krishnan, and B. Chandran. 2003. Kaposi's sarcoma-associated herpesvirus induces the phosphatidylinositol 3-kinase-PKC-zeta-MEK-ERK signaling pathway in target cells early during infection: implications for infectivity J. Virol. 77:1524-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtani, N., P. Brennan, S. Gaubatz, E. Sanij, P. Hertzog, E. Wolvetang, J. Ghysdael, M. Rowe, and E. Hara. 2003. Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J. Cell Biol. 162:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paine, E., R. I. Scheinman, A. S. Baldwin, Jr., and N. Raab-Traub. 1995. Expression of LMP1 in epithelial cells leads to the activation of a select subset of NF-κB/Rel family proteins. J. Virol. 69:4572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raab-Traub, N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431-441. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, M. L., and N. R. Cooper. 1998. Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology 240:93-99. [DOI] [PubMed] [Google Scholar]
- 44.Rothenberger, S., K. Burns, M. Rousseaux, J. Tschopp, and C. Bron. 2003. Ubiquitination of the Epstein-Barr virus-encoded latent membrane protein 1 depends on the integrity of the TRAF binding site. Oncogene 22:5614-5618. [DOI] [PubMed] [Google Scholar]
- 45.Saito, N., G. Courtois, A. Chiba, N. Yamamoto, T. Nitta, N. Hironaka, M. Rowe, and S. Yamaoka. 2003. Two carboxyl-terminal activation regions of Epstein-Barr virus latent membrane protein 1 activate NF-kappaB through distinct signaling pathways in fibroblast cell lines. J. Biol. Chem. 278:46565-46575. [DOI] [PubMed] [Google Scholar]
- 46.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultheiss, U., S. Puschner, E. Kremmer, T. W. Mak, H. Engelmann, W. Hammerschmidt, and A. Kieser. 2001. TRAF6 is a critical mediator of signal transduction by the viral oncogene latent membrane protein 1. EMBO J. 20:5678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang, W., O. A. Pavlish, V. S. Spiegelman, A. A. Parkhitko, and S. Y. Fuchs. 2003. Interaction of Epstein-Barr virus latent membrane protein 1 with SCFHOS/beta-TrCP E3 ubiquitin ligase regulates extent of NF-kappaB activation. J. Biol. Chem. 278:48942-48949. [DOI] [PubMed] [Google Scholar]
- 49.Thornburg, N. J., W. Kulwichit, R. H. Edwards, K. H. Shair, K. M. Bendt, and N. Raab-Traub. 2006. LMP1 signaling and activation of NF-kappaB in LMP1 transgenic mice. Oncogene 25:288-297. [DOI] [PubMed] [Google Scholar]
- 50.Thornburg, N. J., R. Pathmanathan, and N. Raab-Traub. 2003. Activation of nuclear factor-kappaB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 63:8293-8301. [PubMed] [Google Scholar]
- 51.Wang, D., D. Liebowitz, and E. Kieff. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831-840. [DOI] [PubMed] [Google Scholar]
- 52.Wu, S., P. Xie, K. Welsh, C. Li, C. Z. Ni, X. Zhu, J. C. Reed, A. C. Satterthwait, G. A. Bishop, and K. R. Ely. 2005. LMP1 protein from the Epstein-Barr virus is a structural CD40 decoy in B lymphocytes for binding to TRAF3. J. Biol. Chem. 280:33620-33626. [DOI] [PubMed] [Google Scholar]
- 53.Xie, P., and G. A. Bishop. 2004. Roles of TNF receptor-associated factor 3 in signaling to B lymphocytes by carboxyl-terminal activating regions 1 and 2 of the EBV-encoded oncoprotein latent membrane protein 1. J. Immunol. 173:5546-5555. [DOI] [PubMed] [Google Scholar]
- 54.Xie, P., B. S. Hostager, and G. A. Bishop. 2004. Requirement for TRAF3 in signaling by LMP1 but not CD40 in B lymphocytes. J Exp. Med. 199:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, and J. Cohen. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 56.Young, L. S., and A. B. Rickinson. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757-768. [DOI] [PubMed] [Google Scholar]