Abstract
Noroviruses are the causative agents of the majority of viral gastroenteritis outbreaks in humans. During the past 15 years, noroviruses of genotype GGII.4 have caused four epidemic seasons of viral gastroenteritis, during which four novel variants (termed epidemic variants) emerged and displaced the resident viruses. In order to understand the mechanisms and biological advantages of these epidemic variants, we studied the genetic changes in the capsid proteins of GGII.4 strains over this period. A representative sample was drawn from 574 GGII.4 outbreak strains collected over 15 years of systematic surveillance in The Netherlands, and capsid genes were sequenced for a total of 26 strains. The three-dimensional structure was predicted by homology modeling, using the Norwalk virus (Hu/NoV/GGI.1/Norwalk/1968/US) capsid as a reference. The highly significant preferential accumulation and fixation of mutations (nucleotide and amino acid) in the protruding part of the capsid protein provided strong evidence for the occurrence of genetic drift and selection. Although subsequent new epidemic variants differed by up to 25 amino acid mutations, consistent changes were observed in only five positions. Phylogenetic analyses showed that each variant descended from its chronologic predecessor, with the exception of the 2006b variant, which is more closely related to the 2002 variant than to the 2004 variant. The consistent association between the observed genetic findings and changes in epidemiology leads to the conclusion that population immunity plays a role in the epochal evolution of GGII.4 norovirus strains.
Since the beginning of viral gastroenteritis outbreak surveillance in the early 1990s, noroviruses have become recognized as the major cause of reported outbreaks of acute viral gastroenteritis worldwide. Noroviruses form a genus within the family Caliciviridae and are genetically and antigenically highly variable. Currently, five distinct genogroups (GGs) are recognized. Strains belonging to GGI, GGII, and GGIV are known to cause infections in humans. The GGs have been subdivided further into genotypes, defined by a minimum amino acid sequence identity over the complete capsid sequence of 80% (1).
The strains most commonly identified as the cause of outbreaks belong to genotype GGII.4. In The Netherlands, this was the case for 68% of all norovirus outbreaks that were characterized during 12 years of surveillance and for up to 81% of all health care-related outbreaks. Since their first detection in The Netherlands in January 1995, the GGII.4 strains have consistently been present in the Dutch population (46). These observations are in agreement with those of other surveillance studies worldwide (3, 4, 15, 17, 29, 36, 55).
During the past 15 years, four epidemic norovirus seasons have occurred, in the winters of 1995-1996, 2002-2003, 2004-2005, and 2006-2007. These worldwide epidemics were invariantly caused by the predominant genotype, GGII.4, and were attributed to the emergence of new variant lineages of this genotype (4, 31, 35, 52, 53). These genetic variants, which have been identified previously by partial sequencing of either the RNA-dependent RNA polymerase (RdRp) or the capsid gene, have been given several names across the world. Here they are referred to by using the first year of their detection, supplemented where necessary with an extra suffix. The following variants have been identified: <1996, 1996, 2002, 2004, 2006a, and 2006b.
The pattern of emergence of new lineages followed by large-scale epidemics suggests that new variants obtained one or more decisive advantages over the previously circulating predominant variant. It is unknown what the nature of this advantage is, but its basis is likely to be found in VP1, since this protein is needed for essential properties and functions in the viral life cycle, such as antigenicity, host specificity, host cell binding and virus entry properties, and assembly of new particles.
Noroviruses have a positive-strand RNA genome of ∼7.6 kb, which is subdivided into three open reading frames (ORFs). ORF1 encodes a polyprotein which is posttranslationally processed into the nonstructural proteins, including the RdRp. Conserved regions within the RdRp are commonly used as targets for diagnostic PCR assays. At the National Institute for Public Health and the Environment in The Netherlands (RIVM), region A (nucleotides 4279 to 4604; Lordsdale genome numbering [GenBank accession no. X86557]) is commonly used for genotyping outbreak strains. The second ORF (ORF2) encodes the major structural protein VP1. Ninety dimers of this capsid protein form a T=3 icosahedral shell (41). In the virion, a small number of copies of the protein encoded by ORF3 are present. The precise role of this protein is not clear, although it has been suggested that it functions both in upregulation of VP1 expression and as a histone-like protein in stabilizing the capsid-RNA complex (2, 19, 22).
The understanding of immunity against noroviruses remains limited. Between the different GGs and genotypes, antigenic differences as well as cross-reactivities have been demonstrated using virus-like particles and polyclonal antisera (20). Short-term immunity was reported, but preexisting antibodies were not protective against reinfection with the same genotype (25, 39, 56). Studies looking at neutralizing antibodies have not been possible due to the lack of cell culture or small-animal model systems (13). The high level of genetic diversity between different GGs and even between genotypes within the same GG resulting from the high mutation rate and from recombination events contributes to a large degree of antigenic diversity.
Host genetic factors determining the presence or absence of virus receptors also play an important role in susceptibility (21, 23). These receptors, the histo-blood group antigens, show virus strain-specific binding patterns, determining the ability of virus to infect potential host cells. Because noroviruses belonging to GGII.4 have the broadest range of binding to the histo-blood group antigens of all genotypes assayed to date, this may explain part of the relative success of these viruses (24). Other success factors may include a higher stability of the viral particles outside the host, a higher replication rate, or other factors that need to be investigated more thoroughly.
To obtain more insight into the genetic and structural bases of the selective advantage of new GGII.4 variants over the old GGII.4 variants, we determined the complete capsid sequences of a systematic sample of GGII.4 norovirus outbreak strains found in The Netherlands during 13 years of surveillance of viral gastroenteritis and studied their genetic diversity and predicted structure (46). Because a high-resolution three-dimensional (3D) model of GGII noroviruses was lacking at the time this study was initiated, a homology model of the capsid protein was made in silico based on the known 3D structure of the Norwalk virus (NV; GGI.1) capsid protein.
MATERIALS AND METHODS
Strain selection.
Norovirus outbreak strains for which the capsid sequence was determined were selected from the norovirus surveillance database used at RIVM. In this Bionumerics database (Applied Maths BVBA, Sint-Martens-Latem, Belgium), epidemiological and virological data for all norovirus strains found in The Netherlands since January 1994 are collected as previously described (46). The norovirus surveillance system is a laboratory-based passive reporting system to which municipal health services can submit patient samples from suspected viral gastroenteritis outbreaks. Because there is mandatory reporting for outbreaks of illness in institutions and RIVM has been the only laboratory providing diagnostic services for noroviruses in The Netherlands, the collection represents a national sample of reported outbreaks.
As the first step, all GGII.4 strains detected between January 1994 and December 2004 were selected from the database. A phylogenetic tree was made based on partial polymerase sequences of 145 nucleotides for the older sequences to 250 nucleotides for the strains isolated after 2001 (amplified with primer pair JV12 and JV13 or modifications thereof [region A]) (51, 53). The branching of the tree was used to guide the selection of outbreak strains for this study, with at least two strains per branch selected when sufficient material was available. Following reports of unusual outbreaks in the spring of 2006 (27, 28), six strains from this period were included in the study. A minimum spanning tree (MST) was made on the basis of 145 nucleotides of the polymerase gene, using the default settings in Bionumerics, to give an overview of the distribution of strains available in the database. An MST is a tree that connects all samples from a database in such a manner that the summed distance between all samples or branches is minimized. An MST is particularly useful for representing large (genomic) data sets with relatively high similarity levels and, as such, has been shown to enable representation of microevolution or population modeling (44, 45). Another condition is that the data set should represent the biodiversity for the organism under study and therefore should have been gathered over a time period that is short relative to the expected rate of change for the organism. During tree formation, the sample with the largest number of related samples is chosen as the root node, and subsequent branches are added in order of relatedness.
Viral RNA isolation, cDNA preparation, and sequencing.
Stool specimens taken in selected outbreaks were collected from the biobank. Specimens were stored as undiluted stools at 4°C, as 10% fecal suspensions at 4°C, and as RNA extracts frozen at −80°C. Where available, extracted RNAs were used as the template. When this failed to yield a PCR product, a new RNA extraction was done from diluted stool or fecal samples. Sequencing of these samples was done as described previously (11). Briefly, RNA was reverse transcribed in overlapping fragments, using avian myeloblastosis virus reverse transcriptase (Invitrogen), and subsequently, the obtained cDNA was amplified and sequenced using an ABI Prism BigDye Terminator v3.0 ready reaction cycle sequencing kit. The primers that were used were the following: TCTCAGATCTGAGCACGTGG (GR19A), AACAGTTAAGATTGGGACG (GR19B), GTCTCTTGTCGAGTTCTCACG (GR20), GGTGAATTGAACACTACCCAGC (GR21), CTCGACCCGTGCCCACAAAGC (GR22), CATTATAATGCACGCCTGCGCC (GR23), GGGTCAACCAGTTCTACACAC (GR24), CCAGCTGAAGAACCTAGTCTCG (GR25), ACGTGCCCAGGCAAGAGCCAAT (GR-JS1), TAACATCTACTATTATATGGG (GR-JS2), TCATATTTGCAGCAGTCCCA (GR20A), CTCTGAAGGTGCAGATGTTG (GR21A), TGTGAATCCAGACACAGGTAG (GR24A), and ACGGGCCGCATCTGCTGTGGAA (GR25A).
Data processing.
DNA sequences were processed using SeqMan and EditSeq (DNAStar Inc., Konstanz, Germany) and aligned and analyzed using the BioEdit sequence alignment editor (Isis Pharmaceuticals Inc.). Alignments were done manually or using ClustalW alignment algorithms in BioEdit. Informative sites were determined by ProSeq 2.91. Sites were considered informative when at least two strains had an identical amino acid mutation in the alignment. Informative sites discriminating subsequent epidemic variants were also determined. Epidemic variants were defined as GGII.4 strains that were dominant for at least one outbreak season following initial detection. Silent mutations (nucleotide mutations which caused no amino acid mutation) or replacement mutations (nucleotide mutations which caused amino acid mutations) were determined using ProSeq 2.91, with an insertion considered a single mutation.
Phylogenetic analyses were done in Bionumerics, using neighbor joining and the unweighted-pair group method using average linkages, with 1,000 bootstrap resamplings and no correction and with the gap cost set at 5%. Trees were plotted using the program Treeview (version 1.6.6) (37) or Treecon (version 1.3b) (50), with the exception of the MST, which was calculated as well as plotted in Bionumerics. For phylogenetic analysis of the partial polymerase and capsid sequences, the following sequences from GenBank were included: Grimsby strain (Hu/NoV/GGII.4/Grimsby/1995/UK; GenBank accession no. AJ004864), Farmington Hills strain (Hu/NoV/GGII.4/Farmington Hills/2002/USA; accession no. AY502023), Hunter strain (Hu/NoV/GII.4/Hunter 284E/2004/AU [accession no. DQ078794] for the capsid and Hu/NoV/GII.4/Hunter 532D/2004/AU [accession no. DQ078801] for the partial polymerase sequence), and Camberwell strain (Hu/NoV/GGII.4/1994/AU [accession no. U46500] and others for the capsid analysis).
Sequences were checked for possible recombination events by using Simplot (version 3.2), where the window size was varied from 80 to 150, with steps of 20 nucleotides, and a distance model with Jukes-Cantor correction was used. The capsid and RdRp sequences were analyzed independently, as well as after concatemerization of region A and the capsid sequences, to look for possible crossover in the joining region.
Homology modeling.
The three-dimensional structure of the NV capsid protein (PDB code 1IHM) (40) was used as a template for homology modeling of the GGII.4 capsid protein. Sequence alignments were made using the program MUSCLE (14). Compared to the NV capsid protein, the GGII.4 capsid protein has four insertions of three to seven amino acids which cannot be modeled. Generally, such insertions are located in surface-exposed loops of proteins. Based on the alignment of the two sequences and on the 3D structure of the NV capsid protein, the most likely places for insertion were predicted. The GGII.4 capsid protein also has one deletion of two amino acids compared to the NV capsid protein. The place of this deletion can be modeled and was chosen in the same way as that for the insertions. Homology modeling was performed with WhatIf/Yasara Twinset software (Yasara) (54).
Nucleotide sequence accession numbers.
The complete capsid nucleotide sequences determined in this study are accessible in the DNA DataBank of Japan under accession numbers AB303922 through AB303941 and EF126961 through EF126966.
RESULTS
Comparative phylogenies of the polymerase and capsid genes and sequence analysis.
A phylogenetic tree (unweighted-pair group method using average linkages) was made to enable us to make a representative selection of strains for capsid sequencing. All GGII.4 strains found in The Netherlands from the start of viral gastroenteritis surveillance in January 1994 up to August 2006 (n = 574) for which partial polymerase sequences (region A) were available were included (tree not shown). The minimum identity was 89.44%. The strains segregated into three major branches, with multiple outbreak strains per branch (n = 166, 161, and 180); some smaller clusters; and outlying strains that did not fit into any of the major groups. The major groups were each subdivided into smaller clusters. Strains were selected using this tree, aiming to obtain two sequences from the largest clusters from each major group and from five outlying strains. The total number of strains analyzed was 26.
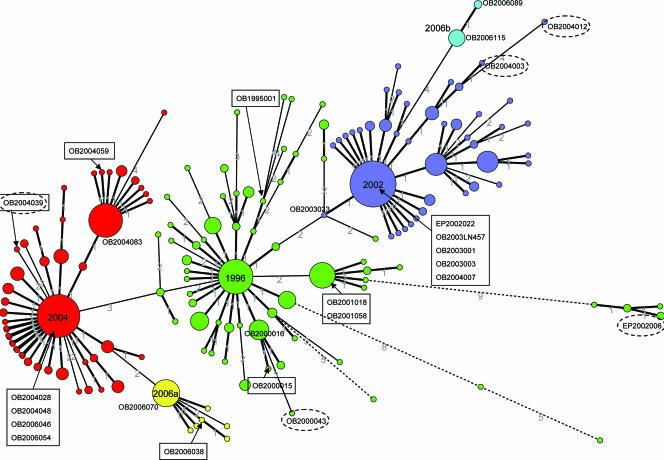
The MST for the partial polymerase sequences is shown in Fig. 1. The total distance of the tree is 230 nucleotides. This tree was not used to make the selection of the strains. However, it illustrates the grouping of the different variants and the positioning of the selected strains because it takes into account the localization of nucleotide changes. Strains OB2000043, EP2002006, OB2004003, OB2004012, and OB2004039 are considered outliers based on their positions in the neighbor-joining tree for all polymerase sequences (not shown).
FIG. 1.
MST, based on alignment of 145 nucleotides of the polymerase gene sequences (region A) of all GGII.4 strains found in The Netherlands between January 1995 and August 2006 (n = 574). Colors represent different variants, as indicated in the figure. The sizes of the circles are drawn to scale with their member counts. The smallest circles represent 1 strain, and the largest circle (the center of the 2002 cluster) represents 70 strains. Genetic distances between the circles, in numbers of nucleotides, are given on connecting lines. The total distance is 230 nucleotides. Strains included in this study are indicated. The strains shown as circles with dotted lines are considered outliers.
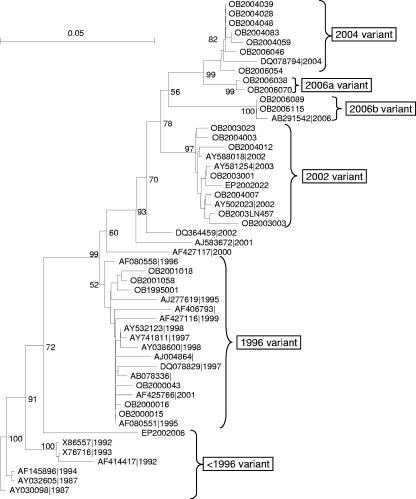
RNA sequences of the complete capsid genes were determined and aligned (see Fig. S1 in the supplemental material). All capsid sequences found belonged to GGII.4. The neighbor-joining tree for the capsid sequences is shown in Fig. 2. Strains from GenBank were included for reference. Polymerase-based and capsid-based groupings were congruent for all strains, and thus no intragenotypic recombination was observed. SimPlot analysis of complete capsids as well as of complete capsids combined with the sequences of region A revealed no potential intergenotypic recombination sites (data not shown). The relatively high level of homology between all strains may obscure possible recombination events, and therefore recombination between strains belonging to the same variant of GGII.4 cannot be ruled out.
FIG. 2.
Neighbor-joining tree for complete capsid amino acid sequences. Type strains from GenBank were used in order to emphasize and confirm the groupings. Branch lengths are drawn to scale. Bootstrap values are percentages of 1,000 iterations.
It should be noted that three strains showed different groupings upon analysis of region A in the RdRp from those obtained in the analysis of the capsids. Three of the five strains that were outliers when comparing partial polymerase nucleotide sequences (OB2004039, OB2000043, and OB2004003) (Fig. 1) did fit into the capsid amino acid tree and fell into their respective variant groups (Fig. 2). Although OB2004012 clustered with the 2002 variant, it was still an outlying strain. EP2002006 was phylogenetically more similar to the eldest strains from GenBank and was therefore used as an ancestral strain in further analyses.
Analysis of the capsid gene and changes over time.
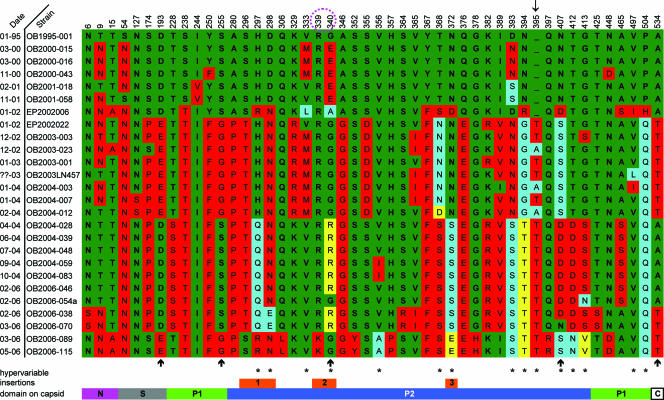
Informative sites in the capsid sequences were then determined. An alignment of all informative sites in the capsid is represented in Fig. 3. Sites were considered informative when at least two strains had an identical amino acid or nucleotide mutation in the alignment.
FIG. 3.
Fixed amino acid changes (informative sites) in capsid sequences of GGII.4 outbreak strains collected between 1995 and 2006. The informative sites throughout the protein are listed from left to right. Amino acid numbering is indicated at the top, and outbreak dates (month-year of isolation, e.g., 01-95 is January 1995) and names are given on the left. From top to bottom, the same color indicates identical amino acids, and different colors are distinct amino acids. Colors were assigned by frequency; amino acids that occurred most are shown in green, followed by red, blue, and yellow (diminishing frequencies). The amino acids circled in magenta are part of the additional RGD motif present in the 2002 variant and the earliest strain. The arrow at the top indicates where an amino acid insertion occurred. The orange bars at the bottom indicate the locations of insertions in GGII.4 compared to NV and correspond to insertions 1 to 3 in Fig. 5A. Asterisks indicate hypervariable sites (with more than one mutation), and the arrows below the sequences indicate the sites where an amino acid mutation occurs at each variant change (not including 2006a). Domains are indicated in the bar below the figure.
At the amino acid level, 48 sites (9% of 541 amino acids) were informative (Table 1). Thirty of these sites were located in the P2 domain (24% of the amino acids in this domain). In all other domains, the numbers of informative sites were significantly lower (chi-square test; P < 0.001), as follows: 3 of 40 amino acids (8%) in the N-terminal domain, 4 of 182 amino acids (2%) in the shell domain, 10 of 184 amino acids (5%) in the P1 domain, and 1 of 10 amino acids (10%) in the C-terminal domain. Hypervariable sites (sites at which three or more different amino acids were found over the 12-year period) were found only in both protruding domains, with 13 in P2 and 2 in P1 (Fig. 3). One amino acid insertion was observed, at position 395, and was first detected in strains from The Netherlands in January 2002. This insertion was located in a highly variable loop region on top of the P2 domain.
TABLE 1.
Informative sites in GGII.4 capsid sequencesa
| Domain | aa level
|
nt level
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aa positions | Total length (aa) | No. (%) of informative aa | nt positions | Total length (nt) | No. (%) of informative nt | No. (%) of mutations at codon position
|
No. (%) of silent mutations | No. (%) of replacement mutations | |||
| First | Second | Third | |||||||||
| N-terminal domain | 1-40 | 40 | 3 (8) | 1-120 | 120 | 10 (8) | 2 (20) | 2 (20) | 6 (60) | 7 (70) | 3 (30) |
| S | 41-222 | 182 | 4 (2) | 121-666 | 546 | 70 (13) | 8 (11) | 2 (3) | 60 (86) | 66 (94) | 4 (6) |
| P1 | 223-276, 402-531 | 184 | 10 (5) | 667-828, 1204-1593 | 552 | 91 (16) | 17 (19) | 5 (5) | 69 (76) | 75 (82) | 16 (18) |
| P2 | 227-401 | 125 | 30 (24) | 829-1203 | 375 | 90 (24) | 18 (20) | 20 (22) | 52 (58) | 50 (56) | 40 (44) |
| C-terminal domain | 531-541 | 10 | 1 (10) | 1593-1623 | 30 | 6 (20) | 1 (17) | 0 (0) | 5 (83) | 5 (83) | 1 (17) |
| Total | 541 | 48 (9) | 1623 | 267 (16) | 46 (3) | 29 (2) | 192 (12) | 203 | 64 | ||
Percentages for informative sites are given as fractions of the total domain length, in numbers of amino acids or nucleotides. Position numbers are given for GGII.4 strains. aa, amino acid(s); nt, nucleotide(s).
At the nucleotide level, 267 sites were found to be informative (Table 1; see Fig. S1 in the supplemental material). The P2 domain had a higher percentage of informative mutations than did the other domains: 24% of the nucleotide mutations in P2 were informative, 8% of those in the N-terminal domain were informative, 13% of those in the shell domain were informative, 16% of those in the P1 domain were informative, and 20% of those in the C-terminal region were informative. The differences with P2 were significant for the N-terminal and shell domains (chi-square test; P < 0.01), not for the P1 and C-terminal domains. Relatively high percentages of first- and second-position nucleotide mutations (20% and 22%, respectively) were seen for the P2 region compared to both the percentages found for the N-terminal and shell domains taken together (13% and 5%, respectively) and those found for the P1 domain (19% and 5%, respectively). Of these mutations, 44% were replacement mutations, versus 30% (3 of 10 mutations) in the N-terminal domain, 6% (4 of 70 mutations) in the shell domain, 18% (16 of 91 mutations) in the P1 domain, and 17% (1 of 6 mutations) in the C-terminal region. These differences with P2 are significant for comparisons of the P2 domain with the shell and P1 domains or the whole capsid sequence (chi-square test; P < 0.001).
As shown in Fig. S1 in the supplemental material and in Fig. 3, changes in informative sites occurred stepwise rather than gradually, with the steps coinciding in time with the emergence of each respective new epidemic variant (2002, 2004, 2006a, and 2006b). When consecutive variants were compared and EP2002006 was considered the precursor (<1996) of the 1996 variant, the numbers of stable amino acid mutations per emerging new variant were 14 (<1996 variant versus 1996 variant), 25 (1996 variant versus 2002 variant), 21 (2002 variant versus 2004 variant), 8 (2004 variant versus 2006a variant), 25 (2004 variant versus 2006b variant), and 23 (2002 variant versus 2006b variant). Both 2006a and 2006b were compared to the 2004 variant, since this was their temporal precursor. The 2006b variant was also compared to the 2002 variant, since these variants are genetically more closely related based on phylogenetic clustering (neighbor joining) (Fig. 2).
Prevalence of GGII.4 variants in The Netherlands from January 1995 to February 2007.
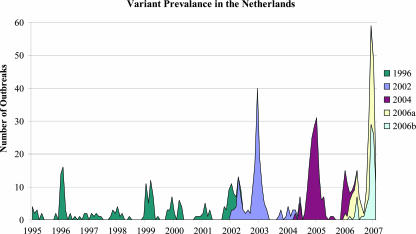
Since the capsid changes showed clustering in time of GGII.4 variants and the capsid-based variant assignment was consistent with that based on the partial RdRp sequences used for routine surveillance, we plotted the presence of the different GGII.4 variant types in The Netherlands over time (Fig. 4). This figure shows that new variants invariantly replaced their predecessors within 5 months of cocirculation.
FIG. 4.
Graph showing the prevalence of GGII.4 variant types in The Netherlands between January 1995 and February 2007. Genotype and variant type assignments were done based on partial polymerase sequence data (region A) available from the Dutch norovirus surveillance database.
Analysis of structural polymorphism of the capsid protein.
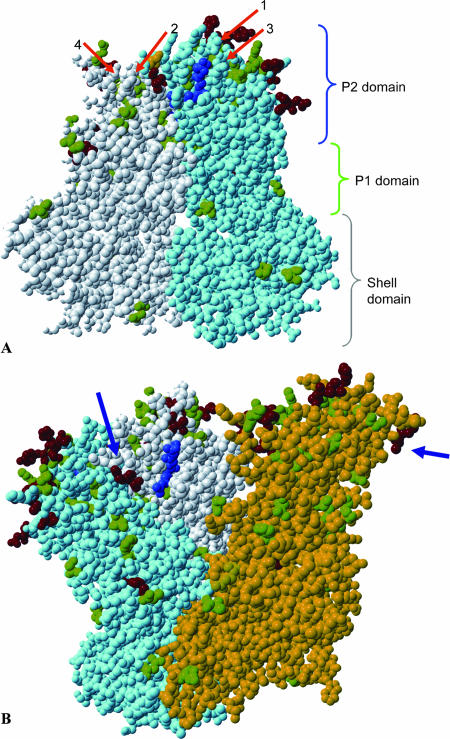
The sequence of OB2004039, a 2004 variant, was used as a reference for modeling of the basic 3D structure of capsid proteins used in this study. Amino acid differences in all other strains were plotted in this 3D model. Compared to the NV capsid protein, the GGII.4 capsid protein has four insertions, of six, three, seven, and three amino acids, with the first three occurring in the P2 region and the fourth occurring in the second coding region of P1. These insertions were not modeled because of the poor reliability of such predictions. However, we predict that these insertions are located close together three-dimensionally, both intra- and interdimerically. Three of the four insertions, all located in the P2 domain, had one or more informative sites, in contrast to the fourth insertion, located in the P1 domain, which had none. The locations of the insertions are shown as orange bars in Fig. 3 (note that the fourth insertion is not shown in this figure) and as orange arrows in Fig. 5A. In Fig. 5B, the inter- and intradimerical interactions of one dimer pair and one-half of the neighboring dimer are shown. In this figure, an extra RGD motif that is present in the earliest strain as well as in the 2002 variant strains is indicated by blue arrows. This motif is located at amino acids 339 to 341.
FIG. 5.
Informative sites mapped on 3D model of GGII.4 capsid proteins. Sites with two distinct amino acid changes over the 12-year period are depicted in green, and sites with three or more amino acid changes are shown in red. The conserved RGD motif is shown in blue. (A) Dimeric subunit of two capsids, with one in gray and one in light blue. The extra RGD motif is indicated in yellow. The locations of the insertions compared to the NV capsid, which have not been modeled, are indicated by orange arrows 1 to 4. The brackets on the right indicate the different domains. The shell domain is indicated in grey, the P1 domain is shown in green, and the P2 domain is shown in blue. (B) Three capsid proteins, including a dimer with one-half of a neighboring dimer. The gray and light blue areas form one dimer, and the yellow capsid belongs to another dimer. The inserted RGD motif is indicated by the blue arrows.
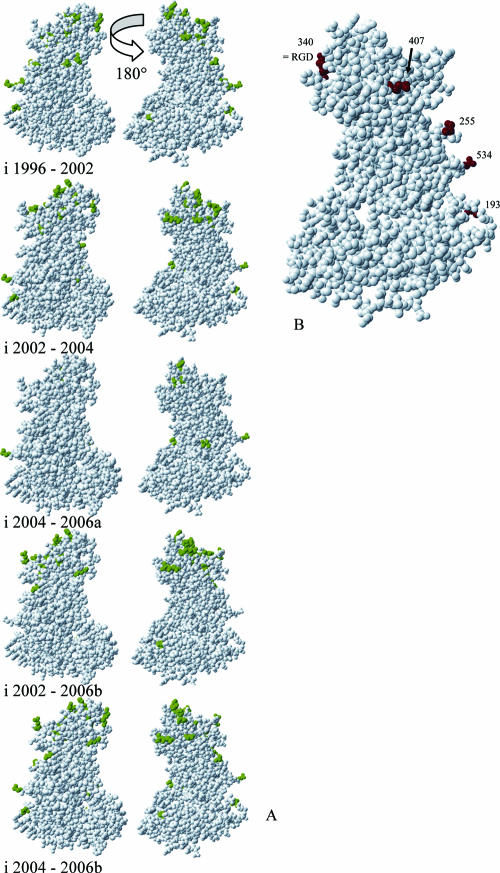
Most of the informative sites mapped to the surface of the P2 domain. Subsequent variants were compared pairwise to identify possible informative sites that consistently changed with every new epidemic variant. When informative sites of ensuing variant pairs (Fig. 6A, panels i to v) were listed, five amino acids changed between every variant pair when 2006a was not included in the analysis (Fig. 3 and 6B). These were amino acids 193 (D1996→E2002→D2004→E2006b), 255 (S1996→G2002→S2004→G2006b), 340 (E1996→G2002→R2004→G2006b), 407 (N1996→S2002→D2004→S2006b), and 534 (A1996→T2002→A2004→T2006b).
FIG. 6.
(A) (i to v) Changes in informative sites (green) derived from amino acid comparisons between subsequent epidemic variants. For each comparison, two views of the capsid protein are given, with one frontal view and one from the rear. For the 2006b variant, two comparisons were made, with the phylogenetic precursor (2002) and the chronologic precursor (2004). (B) Amino acids that change between every subsequent variant group, with 2006a not included.
DISCUSSION
During the past 15 years, four worldwide epidemics of acute gastroenteritis caused by emerging variants of GGII.4 noroviruses have been described. Emerging new variant lineages replaced the previously circulating dominant types rapidly and completely (Fig. 4) (28, 31). The mechanisms underlying the emergence of these new lineages as well as the biological advantages they possessed over other circulating strains are not yet well understood. Most biological properties that are relevant for variables such as stability, assembly, antigenicity, host cell binding, and host specificity are incorporated into the major capsid protein of the virus. We studied the genetic variation in capsids of successive variant lineages to find possible clues about the improved fitness of the successive emerging variants.
In the analysis of the informative sites, for both the nucleotide sequences and the amino acid sequences, mutations were fixed at a number of sites. Every successive variant had a number of distinct, lineage-defining mutations, which were found throughout the capsid sequence. The highest densities of informative sites were located on the surfaces of the protruding regions of the capsid (Fig. 3 and 5). The P2 domain had significantly more mutations than the rest of the capsid protein and, more specifically, many more replacement mutations (0.11 per nucleotide, versus 0.01 to 0.03 per nucleotide for the other domains of the capsid) (Table 1). This is clear evidence of selective force providing new variant viruses having certain mutations with an advantage over previously circulating variants.
The subsequent variants of GGII.4 accumulated mutations in chronological order, and each descended from its predecessor in time, with the exception of the 2006b variant. At the amino acid level, this variant seemed more related to the 2002 variant (Fig. 2). This newly emerging variant is likely a descendant of a virus strain older than the 2004 variant that has accumulated quite a few mutations while not causing many outbreaks in the population.
The situation that is currently unfolding is highly intriguing. In the spring of 2006, two distinct new variants emerged, named 2006a and 2006b. These two new variants have been detected and reported worldwide, often in cruise ship-related outbreaks (27, 28). It has not been reported before that two norovirus variants can cause epidemic-scale outbreaks simultaneously. The 2006a variant shows 8 amino acid mutations compared to its predecessor, the 2004 variant, whereas the 2006b variant shows 25 amino acid mutations compared to the 2004 variant, its temporal and therefore immunologic predecessor. Both 2006 variants emerged almost simultaneously (42). It will be interesting to see if both variants continue to cause outbreaks simultaneously in the population or if one proves to be more successful than the other, perhaps with differing patterns across the world.
The viral strains used for this analysis all originated from our outbreak surveillance database. Strains that are intermediate between the epidemic variants are likely to have reduced viral fitness and are therefore less likely to be detected on the basis of sampling from outbreaks. Although we did look for intermediate strains bridging the different variants by choosing to sequence a number of outliers from the polymerase alignment, no capsid sequences that could be considered intermediates between the different variants were found. EP2006006 does not fit with any variant of the strains included in this study. It does, however, show resemblance to the older strains from GenBank that were included in the neighbor-joining tree. Since no real intermediate strains were found, the origin of emerging variants or the reservoir in which they accumulate their defining mutations thus remains a subject for speculation. The most logical place is the general population. While not causing (many) outbreaks, strains may circulate in the population and not come to the attention of surveillance, slowly accumulating mutations until the built-up genetic variety results in enough antigenic variety to be able to successfully cause (more) outbreaks and become a dominant variant. Alternatively, animal reservoirs, a limited number of which have been recognized (7, 8, 16), or chronically infected patients (18, 34) may be places where the virus can accumulate mutations.
Neutralizing epitopes were previously reported for the surface-exposed P2 domain, and a role in antigenicity was indicated for this domain in several studies with human as well as animal caliciviruses (6, 9, 30, 32, 33, 38, 43, 48, 49).
Tan and coworkers reported the conserved RGD motif to be involved in host cell binding (12, 47). Highly variable regions were found in close spatial proximity to the conserved RGD motif (Fig. 5). Tan and coworkers also reported three amino acids, neighboring the RGD motif, which were suggested to have a role in ligand (histo-blood group antigen) binding specificity (47). One of these surrounding amino acids, designated IV in their paper, is an informative site in our study. Before the 2002 variant, this amino acid was Q376, and it mutated into E376 from the 2002 variant onward. Studies are needed to determine if these mutations lead to changes in host binding specificities.
A second RGD motif (amino acids 339 to 341) was present in the earliest strain sequenced as well as in the 2002 variant. Since it was absent from variants after 2002, it does not seem to confer a great binding advantage. The location of this motif, in spatial proximity to the reported conserved RGD motif on the surface of the molecule and as an insertion compared to the NV genome, does suggest a possible role in ligand binding.
The five amino acids that were informative when comparing all chronologic sets of variants were spread over the capsid. One that stands out is amino acid 340 (E1996→G2002→R2004→G2006b), which in the 2002 variant was also part of an additional RGD motif. The functional implications of these mutations remain to be determined. For our structural analyses of the polymorphisms in the GGII.4 variants, we used a computer-derived model of the VP1 protein. After submission of the present study, Cao et al. published a paper on the cocrystallization of the P protein of a GGII.4 strain norovirus with its receptor (5). No differences between our computer model and this high-resolution structure were found to be of influence to the data presented here.
One could speculate that the location and positioning of the P2 domain of the capsid might explain part of the great prevalence of mutations in this area. The protruding region is connected to the shell domain with a hinge region, and an additional point of flexibility between P1 and P2 was reported (10, 40). This provides flexibility to slightly adjust to the position of the protruding region on top of the shell domain if needed, thus allowing for more conformational changes and thus for more mutations in this region than in the rest of the protein (10). This does not explain the epidemiological observations, however, and therefore we do not think it is the complete story.
Similarly, a possible advantage that new lineages of GGII.4 might have obtained by the accumulation of mutations is increased stability. However, even though increased stability of the viral particles outside the host would increase the number of infectious particles of the more stable variant available for infection, it does not explain the rapid and complete replacement of previous variants that circulated in the population (31). Improved binding or a broadened host range also does not provide a tight explanation for the replacement of previously circulating strains.
The most likely advantage for new variants over older ones is that of immune evasion. Noroviruses, particularly strains of GGII.4, are highly prevalent in the population. During epidemic seasons, up to 86% of norovirus outbreaks were caused by the predominant genotype, GGII.4, followed by a sharp drop in the prevalence of this genotype in the subsequent season (46). Then, only after the emergence of a genetically distinct new lineage of this genotype, the prevalence of GGII.4 strains rose again to cause a new epidemic.
A similar pattern of so-called epochal evolution has been described very elegantly for influenza A virus (H3N2) (26), where periods of phenotypic stasis are separated by the stepwise emergence of phenotypically distinct new variants, as we also see here for noroviruses. During the periods of phenotypic (and antigenetic) stasis, neutral or almost neutral mutations do occur and accumulate if they are beneficial or at least not disadvantageous. For influenza virus, this pattern of evolution and emergence of genetically novel variants is attributed to host population immunity and subsequent antigenic escape by the virus. The striking parallel observed here for norovirus suggests that this pattern of epidemics is driven by (population) immunity as well.
No long-term immunity to norovirus infection has been reported so far. Short-term protective antibodies have been reported, however, and repeated exposure, which is likely to occur with the high prevalence of norovirus, will lengthen the duration of specific protection. Studies with NV in volunteers suggested that immune protection wanes after 6 months without reexposure (25, 39).
In agreement with the hypothesis that immunity to the predominant GGII.4 variant built up in the population, Nilsson and coworkers reported on the in vivo evolution of a GGII.3 strain infecting a chronically ill immunocompromised patient (34). They observed the accumulation of amino acid mutations in the capsid protein and suggested that these changes gave rise to a new phenotype, through immune response-driven evolution. Similar to our findings, they found most amino acid mutations in the P2 domain of the capsid. This observation supports the idea that new variants may possibly emerge from chronically infected patients.
The data presented in this paper underpin observations that the elevated numbers of norovirus outbreaks in the winter seasons of 1995-1996, 2002-2003, 2004-2005, and 2006-2007 (4, 31, 46, 53) were mainly, if not solely, due to the emergence of new variants of the GGII.4 genotype. The gradual increase in nucleotide mutations in the sequences of norovirus GGII.4 strains confirms that genetic drift occurs in the virus. Additionally, the stepwise fixation of numbers of amino acid mutations in the capsid of this predominant genotype, mainly in the surface-exposed P2 domain, is likely to be caused by selective pressure due to population immunity, which resulted in emerging variants which have caused worldwide epidemic rises in outbreak numbers.
Further immunological studies of this variation in the capsid protein are urgently needed to shed light on the mechanisms of immune evasion utilized by the most prevalent genotype of norovirus.
Supplementary Material
Acknowledgments
This work was supported by the European Commission, DG Research Quality of Life Program, under the 6th Framework (EVENT; SP22-CT-2004-502571).
Footnotes
Published ahead of print on 3 July 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Bertolotti-Ciarlet, A., S. E. Crawford, A. M. Hutson, and M. K. Estes. 2003. The 3′ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein. J. Virol. 77:11603-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193:413-421. [DOI] [PubMed] [Google Scholar]
- 4.Bull, R. A., E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarty, S., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2005. Evolutionary trace residues in noroviruses: importance in receptor binding, antigenicity, virion assembly, and strain diversity. J. Virol. 79:554-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheetham, S., M. Souza, R. McGregor, T. Meulia, Q. Wang, and L. J. Saif. 2007. Binding patterns of human norovirus-like particles to buccal and intestinal tissues of gnotobiotic pigs in relation to A/H histo-blood group antigen expression. J. Virol. 81:3535-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheetham, S., M. Souza, T. Meulia, S. Grimes, M. G. Han, and L. J. Saif. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 80:10372-10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R., J. D. Neill, M. K. Estes, and B. V. Prasad. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. USA 103:8048-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, R., J. D. Neill, J. S. Noel, A. M. Hutson, R. I. Glass, M. K. Estes, and B. V. Prasad. 2004. Inter- and intragenus structural variations in caliciviruses and their functional implications. J. Virol. 78:6469-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bruin, E., E. Duizer, H. Vennema, and M. P. Koopmans. 2006. Diagnosis of norovirus outbreaks by commercial ELISA or RT-PCR. J. Virol. Methods 137:259-264. [DOI] [PubMed] [Google Scholar]
- 12.D'Souza, S. E., M. H. Ginsberg, and E. F. Plow. 1991. Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem. Sci. 16:246-250. [DOI] [PubMed] [Google Scholar]
- 13.Duizer, E., K. J. Schwab, F. H. Neill, R. L. Atmar, M. P. Koopmans, and M. K. Estes. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79-87. [DOI] [PubMed] [Google Scholar]
- 14.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Farkas, T., S. Nakajima, M. Sugieda, X. Deng, W. Zhong, and X. Jiang. 2005. Seroprevalence of noroviruses in swine. J. Clin. Microbiol. 43:657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallimore, C. I., J. Green, D. Lewis, A. F. Richards, B. A. Lopman, A. D. Hale, R. Eglin, J. J. Gray, and D. W. Brown. 2004. Diversity of noroviruses cocirculating in the north of England from 1998 to 2001. J. Clin. Microbiol. 42:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallimore, C. I., D. Lewis, C. Taylor, A. Cant, A. Gennery, and J. J. Gray. 2004. Chronic excretion of a norovirus in a child with cartilage hair hypoplasia (CHH). J. Clin. Virol. 30:196-204. [DOI] [PubMed] [Google Scholar]
- 19.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansman, G. S., K. Natori, H. Shirato-Horikoshi, S. Ogawa, T. Oka, K. Katayama, T. Tanaka, T. Miyoshi, K. Sakae, S. Kobayashi, M. Shinohara, K. Uchida, N. Sakurai, K. Shinozaki, M. Okada, Y. Seto, K. Kamata, N. Nagata, K. Tanaka, T. Miyamura, and N. Takeda. 2006. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 87:909-919. [DOI] [PubMed] [Google Scholar]
- 21.Harrington, P. R., J. Vinje, C. L. Moe, and R. S. Baric. 2004. Norovirus capture with histo-blood group antigens reveals novel virus-ligand interactions. J. Virol. 78:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbert, T. P., I. Brierley, and T. D. Brown. 1996. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 77:123-127. [DOI] [PubMed] [Google Scholar]
- 23.Huang, P., T. Farkas, S. Marionneau, W. Zhong, N. Ruvoen-Clouet, A. L. Morrow, M. Altaye, L. K. Pickering, D. S. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19-31. [DOI] [PubMed] [Google Scholar]
- 24.Huang, P., T. Farkas, W. Zhong, M. Tan, S. Thornton, A. L. Morrow, and X. Jiang. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 79:6714-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, P. C., J. J. Mathewson, H. L. DuPont, and H. B. Greenberg. 1990. Multiple-challenge study of host susceptibility to Norwalk gastroenteritis in US adults. J. Infect. Dis. 161:18-21. [DOI] [PubMed] [Google Scholar]
- 26.Koelle, K., S. Cobey, B. Grenfell, and M. Pascual. 2006. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science 314:1898-1903. [DOI] [PubMed] [Google Scholar]
- 27.Koopmans, M., J. Harris, L. Verhoef, E. Depoortere, J. Takkinen, and D. Coulombier. 2006. European investigation into recent norovirus outbreaks on cruise ships: update. Euro. Surveill. 11:E060706.5. [DOI] [PubMed] [Google Scholar]
- 28.Kroneman, A., H. Vennema, J. Harris, G. Reuter, C. H. von Bonsdorff, K. O. Hedlund, K. Vainio, V. Jackson, P. Pothier, J. Koch, E. Schreier, B. E. Bottiger, and M. Koopmans. 2006. Increase in norovirus activity reported in Europe. Euro. Surveill. 11:E061214.1. [DOI] [PubMed] [Google Scholar]
- 29.Kroneman, A., H. Vennema, Y. Van Duijnhoven, E. Duizer, and M. Koopmans. 2004. High number of norovirus outbreaks associated with a GGII.4 variant in The Netherlands and elsewhere: does this herald a worldwide increase? Eurosurveill. Wkly. 8:041223. [Google Scholar]
- 30.Lochridge, V. P., K. L. Jutila, J. W. Graff, and M. E. Hardy. 2005. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 86:2799-2806. [DOI] [PubMed] [Google Scholar]
- 31.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torven, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szucs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363:682-688. [DOI] [PubMed] [Google Scholar]
- 32.Matsuura, Y., Y. Tohya, M. Mochizuki, K. Takase, and T. Sugimura. 2001. Identification of conformational neutralizing epitopes on the capsid protein of canine calicivirus. J. Gen. Virol. 82:1695-1702. [DOI] [PubMed] [Google Scholar]
- 33.Neill, J. D., S. V. Sosnovtsev, and K. Y. Green. 2000. Recovery and altered neutralization specificities of chimeric viruses containing capsid protein domain exchanges from antigenically distinct strains of feline calicivirus. J. Virol. 74:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson, M., K. O. Hedlund, M. Thorhagen, G. Larson, K. Johansen, A. Ekspong, and L. Svensson. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77:13117-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 36.Okada, M., T. Ogawa, I. Kaiho, and K. Shinozaki. 2005. Genetic analysis of noroviruses in Chiba Prefecture, Japan, between 1999 and 2004. J. Clin. Microbiol. 43:4391-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 38.Parker, T. D., N. Kitamoto, T. Tanaka, A. M. Hutson, and M. K. Estes. 2005. Identification of genogroup I and genogroup II broadly reactive epitopes on the norovirus capsid. J. Virol. 79:7402-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parrino, T. A., D. S. Schreiber, J. S. Trier, A. Z. Kapikian, and N. R. Blacklow. 1977. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N. Engl. J. Med. 297:86-89. [DOI] [PubMed] [Google Scholar]
- 40.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 41.Prasad, B. V., M. E. Hardy, X. Jiang, and M. K. Estes. 1996. Structure of Norwalk virus. Arch. Virol. 12(Suppl.):237-242. [DOI] [PubMed] [Google Scholar]
- 42.ProMED-mail. 26 June 2006, posting date. Viral gastroenteritis update 2006 (02). Promed-mail 20060626.1776. http://www.promedmail.org.
- 43.Radford, A. D., K. Willoughby, S. Dawson, C. McCracken, and R. M. Gaskell. 1999. The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. J. Virol. 73:8496-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schouls, L. M., A. van der Ende, I. van de Pol, C. Schot, L. Spanjaard, P. Vauterin, D. Wilderbeek, and S. Witteveen. 2005. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 43:2741-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schouls, L. M., H. G. van der Heide, L. Vauterin, P. Vauterin, and F. R. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siebenga, J. J., H. Vennema, E. Duizer, and M. P. Koopmans. 2007. Gastroenteritis caused by norovirus GGII.4, The Netherlands, 1994-2005. Emerg. Infect. Dis. 13:144-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, M., W. Zhong, D. Song, S. Thornton, and X. Jiang. 2004. E. coli-expressed recombinant norovirus capsid proteins maintain authentic antigenicity and receptor binding capability. J. Med. Virol. 74:641-649. [DOI] [PubMed] [Google Scholar]
- 49.Tohya, Y., N. Yokoyama, K. Maeda, Y. Kawaguchi, and T. Mikami. 1997. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J. Gen. Virol. 78:303-305. [DOI] [PubMed] [Google Scholar]
- 50.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 51.Vennema, H., E. de Bruin, and M. Koopmans. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233-235. [DOI] [PubMed] [Google Scholar]
- 52.Vinje, J., S. A. Altena, and M. P. Koopmans. 1997. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 176:1374-1378. [DOI] [PubMed] [Google Scholar]
- 53.Vinje, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 54.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 55.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 190:27-36. [DOI] [PubMed] [Google Scholar]
- 56.Wyatt, R. G., R. Dolin, N. R. Blacklow, H. L. DuPont, R. F. Buscho, T. S. Thornhill, A. Z. Kapikian, and R. M. Chanock. 1974. Comparison of three agents of acute infectious nonbacterial gastroenteritis by cross-challenge in volunteers. J. Infect. Dis. 129:709-714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.