Abstract
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) exon0 (orf141) has been shown to be required for the efficient production of budded virus (BV). The deletion of exon0 reduces the level of BV production by up to 99% (X. Dai, T. M. Stewart, J. A. Pathakamuri, Q. Li, and D. A. Theilmann, J. Virol. 78:9633-9644, 2004); however, the function or mechanism by which EXON0 affects BV production is unknown. In this study, we further elucidated the function of EXON0 by investigating the localization of EXON0 in infected Sf9 cells and in virions and by identifying interactions between EXON0 and other viral proteins. In addition, electron microscopy was used to study the cellular localization of nucleocapsids in cells transfected with an exon0 knockout (KO) virus. The results showed that EXON0 was localized to both the cytoplasm and the nuclei of infected Sf9 cells throughout the infection. Western blotting results also showed that EXON0 was purified along with BV and occlusion-derived virus (ODV). The fractionation of BV into the nucleocapsid and envelope components showed that EXON0 localized to the BV nucleocapsid. Yeast two-hybrid screening, coimmunoprecipitation, and confocal microscopy revealed that it interacted with nucleocapsid proteins FP25 and BV/ODV-C42. Cells transfected with the exon0 KO virus exhibited normally appearing nucleocapsids in the nuclei in numbers equal to those in the nuclei of cells transfected with the EXON0 repaired virus. In contrast, the numbers of nucleocapsids in the cytoplasm of cells transfected with the exon0 KO virus were significantly lower than those in the cytoplasm of cells transfected with the repaired virus. These results support the conclusion that EXON0 is required in the BV pathway for the efficient egress of nucleocapsids from the nucleus to the cytoplasm.
Autographa californica multiple nucleopolyhedrovirus (AcMNPV), an archetype of the Baculoviridae, has a large double-stranded DNA genome of approximately 134 kbp and comprises 154 predicted genes. AcMNPV has a nuclear site of replication, many virus-encoded enzymes for DNA transcription and replication, and a complex morphogenic pathway that produces two distinct forms of infectious virions in infected cells, budded virus (BV) and occlusion-derived virus (ODV) (52). exon0 (orf141) is a highly conserved gene found in all lepidopteran baculoviruses of the genus Nucleopolyhedrovirus. The deletion of exon0 reduces the level of BV production by up to 99%, and the infection of Sf9 cells with the exon0 knockout (KO) virus is restricted to a single cell or a few neighboring cells. However, viral replication and polyhedron production are unaffected (8), suggesting that exon0 plays a key role in the pathway that is specific for the synthesis of BV.
Baculovirus BVs are thought to enter insect and mammalian cells via adsorptive endocytosis (16, 55), including clathrin-mediated endocytosis and low-pH-dependent membrane fusion (1, 18, 21, 23). After the nucleocapsids are released from the endosomes, they are transported into the nuclei, where viral transcription and DNA replication occur, resulting in the production of BV and ODV (10). These two virion forms have different functions in the viral life cycle. ODV is required for interhost transmission, whereas BV is required for the dissemination of a viral infection throughout the tissues of an infected host. The two virion forms are genetically identical but differ in their envelope compositions and tissue tropisms and are produced at different times during infection (6, 13). At late times postinfection (pi), nucleocapsids are synthesized in the nucleus and are initially shuttled out of the nucleus and transported to the cytoplasmic membrane, from which they bud, forming BVs. At very late times pi, nucleocapsids are retained in the nucleus, where they become occluded in occlusion bodies to form ODVs. The molecular events that occur during baculovirus replication have been extensively studied (35), but the mechanisms by which the nucleocapsids are selected to become either ODVs or BVs and how the nucleocapsids are transported to the cell membrane are still unknown.
Several viral gene products have been shown to affect the synthesis of BV and ODV. For example, the AcMNPV VP1045 and VLF-1 proteins are nucleocapsid proteins of both BV and ODV and have been shown to be required for the assembly of progeny nucleocapsids in the nucleus (36, 54, 60). GP41 is an O-glycosylated protein that affects both BV and ODV but is only a structural component of ODV (37). The baculovirus core gene 38K (ac98), which expresses a nuclear protein, has previously been shown to be essential for the formation of normal nucleocapsids, affecting both BV and ODV production, but has not been shown to be a structural component of the nucleocapsid (58). Other than EXON0, very few proteins have been shown to be specific for the synthesis of BV, but one of the most intensively studied of such proteins is the envelope glycoprotein GP64. GP64 is required for efficient virion budding from the plasma membrane and is essential for BV attachment and membrane fusion (1, 16, 33, 38).
In this study, we present evidence to show that EXON0 is a structural component of both BV and ODV and a component of the BV nucleocapsid. Using yeast two-hybrid screening, immunoprecipitation, and confocal microscopy, we show that EXON0 interacts with the known nucleocapsid proteins FP25 and BV/ODV-C42. In the absence of EXON0, nucleocapsids are not efficiently transported from the nucleus to the cytoplasm but the nucleocapsid assembly remains unchanged.
MATERIALS AND METHODS
Cells and viruses.
Spodoptera frugiperda Sf9 cells were maintained in 10% fetal bovine serum-supplemented TC100 medium at 27°C. AcMNPV recombinant bacmids were derived from the commercially available bacmid bMON14272 (Invitrogen Life Technologies) and propagated in Escherichia coli strain DH10B.
Construction of HA-tagged EXON0.
To tag EXON0 with the influenza virus hemagglutinin (HA) epitope (CYPYDVPDYASL) at the N terminus, exon0 was amplified using primers 608 (5′-AGATCTATGTACCCCTACGACGTGCCCGACTACGCCATAAGAACCAGCAGTCACGTG-3′) and 609 (5′-GTTGCGTTGCCCGTTATC-3′) and p2ZeoKS-exon0 (8) as a template. Inverse PCR was used to amplify the linear fragments, which were then treated with T4 polynucleotide kinase and gel purified. Amplified fragments were self-ligated overnight at 16°C, and E. coli DH5α competent cells were transformed with these amplification products. Zeocin-resistant colonies were selected and identified by restriction digestion, and the relevant genotype was confirmed by sequencing. The resulting plasmid was named p2Zeo-HA-exon0.
Construction of bMON14272 exon0 KO virus and the HA-EXON0 repaired bacmid.
The first 114 bp of exon0 are shared with and spliced to ie0 mRNA. In addition, the TAA stop codon of exon0 overlaps with the putative late transcription initiation motif DTAAG of orf142, which is one of the 29 core baculovirus genes and encodes a structural protein of ODV (5). To prevent any possible intragenomic homologous recombination, we ensured that the wild-type (WT) locus and the repair constructs inserted at the polyhedrin locus carried no homologous sequences. To accomplish this, we constructed a new KO bacmid in which both the complete open reading frame (ORF) of exon0 and the promoters of exon0 and ie0 were deleted. The late gp64 promoter L2 (14) was inserted upstream of orf142 to drive this gene and so prevent any possible disruption of its expression. IE0 is not essential for viral replication, and in addition, the ie0 KO virus ie1 KO-IE1 produces WT levels of BV (25, 50); thus, the new KO virus provided the backbone genome for the analysis of exon0 presented in this study. The AcMNPV bacmid (bMON14272) was used to generate the KO virus by recombination in E. coli as previously described (17, 22, 27). A zeocin resistance gene was amplified using primers 665 (5′-GGTCACGTAGGCACTTTGCGCACGGCACTAGGGCTGTGGAGGGGACAGGCGGATCTCTGCAGCACGTGTT-3′) and 666 (5′-CTCTTTCCAGAGTCAACAAGTTGCCGCCACCACTCATTTTGCGTTGCGTGCTTATCTTTTTATCTTAGACATGATAAGATACATTGATGA-3′) and p2ZeoKS as a template. These primers contain 50 and 43 bp homologous to the upstream and downstream flanking regions of exon0, respectively. The PCR fragment of the zeocin resistance gene amplified with primers 665 and 666 was gel purified and electroporated into E. coli BW25113-pKD46 cells, which already contained the DNA of the AcMNPV bacmid bMON14272. The electroporated cells were incubated at 37°C for 4 h in 3 ml of SOC medium (2.0% tryptone, 0.5% yeast extract, 8.5 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 20 mM glucose [pH 7.0]) and placed onto agar medium containing 30 μg of zeocin per ml and 50 μg of kanamycin per ml. The plates were incubated at 37°C overnight, and colonies resistant to zeocin and kanamycin were selected for further confirmation of the relevant gentoype by PCR.
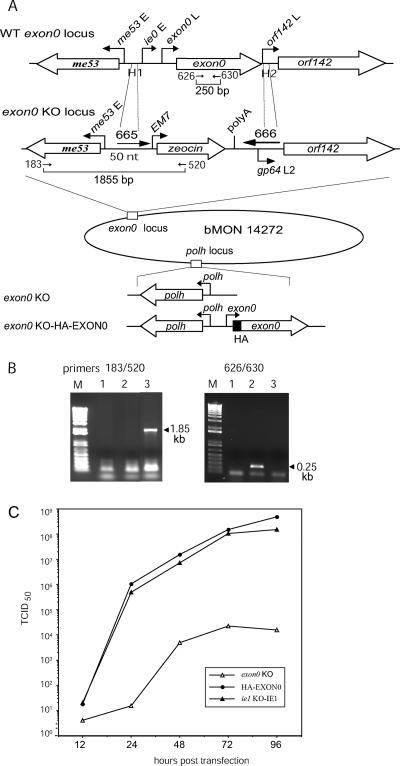
Two different pairs of primers were used to confirm that exon0 had been deleted from the exon0 locus of the AcMNPV bacmid genome (see Fig. 1A). Primers 183 (5′-CGCAACAGGATCCGAACCAGCAGTC-3′) and 520 (5′-CTTTTGGATCCACAACAGGCAATTTGAT-3′) were used to detect the correct insertion of the zeocin resistance cassette. A fragment of 1,855 bp that was amplified with primers 183 and 520 from the exon0 KO bacmid confirmed the correct insertion of the zeocin resistance gene into the exon0 locus. Primers 626 and 630 (5′-TGTATATGCGTAGGAGAGCC-3′ and 5′-CTCGCAACTGTTTCAAGTAC-3′, respectively) were used to confirm the absence of the exon0 ORF. These primers amplify a 250-bp fragment from the WT AcMNPV bacmid (see Fig. 1B). One of the recombinant bacmids confirmed by PCR was selected and named bMON14272 exon0 KO.
FIG. 1.
Construction of the exon0 KO bacmid and viral growth curves. (A) Schematic of the exon0 locus of WT AcMNPV and that of the bMON14272 exon0 KO showing the relative locations and orientations of me53, exon0, and orf142. The linear PCR fragment (middle) consisting of the EM7 promoter, the zeocin resistance gene, simian virus 40 poly(A), and gp64 late promoter L2 (14) was amplified with a 70-mer primer (665) and a 90-mer primer (666). Primer 665 contains a 50-nt region of homology (H1) complementary to the intergenic region upstream of the ie0 early promoter, and primer 666 contains a 43-nt region of homology (H2) complementary to the immediate downstream region of the exon0 ORF. A 16-nt gp64 late promoter (L2) was included in primer 666 to replace the orf142 promoter. In the bMON14272 exon0 KO bacmid, the complete exon0 ORF and the exon0 and ie0 promoters were replaced by the zeocin resistance cassette. The bMON14272 exon0 KO bacmid was repaired at the polyhedrin (polh) locus with the polyhedrin gene alone (exon0 KO virus) or with the polyhedrin gene and the gene encoding HA-tagged EXON0 (exon0 KO-HA-EXON0 virus) as described in Materials and Methods. E, early promoter; L, late promoter. (B) The deletion of exon0 was confirmed by PCR analysis. The primers used to confirm the deletion of the exon0 ORF and the correct insertion of the zeocin resistance gene cassette are shown in panel A and indicated by arrows. The presence of 1.85 kb from primers 183 and 520 and the absence of 0.25 kb from primers 626 and 630 proved the correct insertion of the zeocin resistance cassette and the loss of exon0 in bMON14272 exon0 KO. M, 1-kb-plus DNA ladder; lane 1, negative control; lane 2, WT AcMNPV; lane 3, bMON14272 exon0 KO. (C) Growth curves of exon0 KO, exon0 KO-HA-EXON0 (HA-EXON0), and ie1 KO-IE1 viruses. Sf9 cells were transfected with these bacmids, and the virus supernatants were harvested at various time points posttransfection and analyzed by using an end point dilution assay. Data points represent the averages of results from two independent titrations. BV production by the exon0 KO virus was reduced to 0.01% of that by the WT virus and the repaired virus.
To introduce HA-tagged EXON0 back into bMON14272 exon0 KO, rescue transfer vectors were constructed using the plasmid backbone pFAcT (8). pFAcT contains two Tn7 transposition excision sites that allow the genes cloned between the sites to be transposed into the mini ATT region located in the AcMNPV bacmid. p2Zeo-HA-exon0 was digested with XhoI and XbaI. The excised fragment, containing the native late promoter of exon0, the exon0 ORF, and an Orgyia pseudotsugata multiple nucleopolyhedrovirus (OpMNPV) ie1 polyadenylation signal, was cloned into the XhoI and XbaI sites of pFAcT to generate pFAcT-HA-exon0. In the pFAcT backbone, the AcMNPV polyhedrin gene is included in the transposed DNA cassette. bMON14272 exon0 KO bacmids containing the pFAcT or pFAcT-HA-exon0 cassette were generated by Tn7-mediated transposition as previously described by Luckow et al. (26), and the resulting viruses were named exon0 KO and exon0 KO-HA-EXON0.
Time course analysis of BV production.
Sf9 cells (1.0 × 106 cells/35-mm-diameter well of a six-well plate) were transfected with 1.0 μg of each bacmid construct (exon0 KO virus, exon0 KO-HA-EXON0, and the control virus ie1 KO-IE1). At various time points posttransfection, the supernatant containing the BV was harvested and cell debris was removed by centrifugation (8,000 × g for 5 min). The BV titers in Sf9 cell supernatants were determined in duplicate by end point dilution using 96-well microtiter plates.
Evaluation of cellular localization of EXON0 by nuclear and cytoplasmic fractionation.
Sf9 cells (2.0 × 106/35-mm-diameter well of a six-well plate) were infected with the exon0 KO-HA-EXON0 virus at a multiplicity of infectivity (MOI) of 10. At 6, 12, 18, 24, 36, 48, and 72 h postinfection (hpi), cells were washed with phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl [pH 7.4]), scraped with a rubber policeman, and collected by centrifugation (800 × g for 5 min). Cells were resuspended in 0.2 ml of NP-40 lysis buffer (10 mM Tris [pH 7.9], 10 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.5% NP-40 [vol/vol]), and the suspension was gently mixed and kept on ice for 5 min. Nuclei were pelleted by centrifugation at 1,000 × g for 3 min. The supernatant containing the cytosolic fraction was transferred into a new tube and mixed with an equal volume of 2× protein sample buffer (2× PSB; 0.25 M Tris-Cl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 10% 2-mercaptoethanol, 0.02% bromophenol blue). The nuclei were resuspended in the original volume of lysis buffer and mixed with an equal volume of 2× PSB. The nuclear fraction was sheared with a 25-gauge syringe and analyzed by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Immunofluorescence.
Sf9 cells were infected with exon0 KO-HA-EXON0 at an MOI of 10. At 12, 18, 24, 36, 48, and 72 hpi, the supernatant was removed and the cells were washed three times in PBS, followed by fixation in 3% paraformaldehyde in PBS for 10 min. The fixed cells were washed three times in PBS for 15 min each time, followed by permeabilization in 0.15% Triton X-100 in PBS for 15 min. The cells were then blocked for 60 min in blocking buffer (2% bovine serum albumin in PBS), followed by 1 h of incubation with either mouse monoclonal anti-HA antibody (1:200; HA11 [Covance]) alone or mouse monoclonal anti-HA antibody (1:200) coupled with either rabbit anti-FP25 antibody (1:1,000) or rabbit anti-BV/ODV-C42 antibody (1:1,000) (3, 4). The cells were washed three times in blocking buffer for 10 min each time, followed by 1 h of incubation with a fluorescence-tagged antibody. Samples incubated with HA antibody alone were further incubated with an Alexa 635-conjugated goat anti-mouse antibody (1:500; Molecular Probes). Samples incubated with rabbit anti-FP25 or anti-BV/ODV-C42 antibody were further incubated with Alexa 488-conjugated goat anti-rabbit immunoglobulin G (IgG; 1:500 [Molecular Probes]). The cells were subsequently washed three times in PBS for 10 min each time, stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) at 200 ng/ml, and examined with a Leica confocal microscope.
Purification of BV and ODV.
Sf9 cells were infected with exon0 KO-HA-EXON0 at an MOI of 0.1. Six days pi, 80 ml of medium was collected and centrifuged at 8,000 × g in a Beckman JA 17 rotor. The supernatant was then centrifuged at 100,000 × g (21,000 rpm) in a Beckman SW28 rotor at 4°C to pellet the BV. The BV pellet was resuspended in 0.4 ml of 0.1× Tris-EDTA, loaded onto a 25 to 60% sucrose gradient, and centrifuged at 100,000 × g (24,000 rpm) in a Beckman SW41 rotor for 90 min. The BV band was collected, diluted twice, and centrifuged at 100,000 × g (24,000 rpm) in a Beckman SW41 rotor for 30 min at 4°C. The virus pellet was resuspended in 200 μl of 0.1× Tris-EDTA. The protein concentration was determined by the Bradford assay (2).
In a 250-μl reaction mixture, 250 μg of BV was incubated in a solution of 1.0% NP-40 and 10 mM Tris, pH 8.5, at room temperature for 30 min with gentle agitation. The solution was then layered onto a 4-ml 30% (wt/vol) glycerol-10 mM Tris (pH 8.5) cushion and centrifuged at 150,000 × g (34,000 rpm) in a Beckman SW60 rotor for 60 min at 4°C. The envelope proteins were recovered from the top of the cushion by trichloroacetic acid precipitation. The pelleted nucleocapsids were dissolved in 10 mM Tris (pH 7.4).
Polyhedra and ODV were extracted from the cells infected with exon0 KO-HA-EXON0 at 6 days pi and purified as previously described (6).
Immunoprecipitation.
Sf9 cells (3 × 108) were infected with either WT AcMNPV E2 or exon0 KO-HA-EXON0 at an MOI of 10. At 24 hpi, the cells were collected and washed with PBS, pelleted at 800 × g for 5 min, resuspended in 2 ml of EBC (50 mM Tris-Cl [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40, 0.2 mM sodium orthovanadate, 1% sodium fluoride, 1% protease inhibitor cocktail [Sigma]), and placed on ice for 10 min. The cells were passed through a cold French pressure cell twice at 1,000 lb/in2. Fifteen microliters of the lysate was removed for Western blotting. The lysate was centrifuged at 17,949 × g (13,000 rpm) in an Eppendorf 5417 C centrifuge at 4°C for 10 min. Forty microliters of anti-HA antibody-immobilized agarose beads (Sigma) were equilibrated with EBC buffer and mixed with the 2 ml of lysate supernatant, and the mixture was incubated overnight on an orbital shaker at 4°C. The incubation mix was then transferred to a Bio-Rad column, and the beads were washed once with 0.5 ml of EBC buffer, followed by five washes with 0.5 ml of NETN buffer (20 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.5% NP-40) containing 400 mM NaCl. The beads were eluted twice using 60 μl of 100 mM glycine-HCl (pH 2.2) after 1 min of incubation. Two eluates were combined, and the pH was raised with 1.5 M Tris-HCl (pH 8.8) to a final pH of 8.0. The eluate was vacuum concentrated to 45 μl, mixed with 45 μl of 2× PSB, boiled for 10 min at 100°C, subjected to SDS-PAGE, and examined by Western blotting.
Western blot analysis.
Protein samples from the nuclear and cytoplasmic fractions, purified BV or ODV, and the protein immunoprecipitation eluates from the HA agarose beads were mixed with equal volumes of 2× PSB, and the mixtures were incubated at 100°C for 10 min. Protein samples were separated by 10% SDS-PAGE and transferred onto Millipore Immobilon-P membrane with the Bio-Rad Mini-PROTEAN II and liquid transfer apparatuses, respectively, in accordance with the manufacturers' recommended protocols. Western blots were probed with one of the following primary antibodies: (i) mouse monoclonal HA antibody (HA11 [Covance]; 1:1,000), (ii) mouse monoclonal OpMNPV VP39 antibody (1:3,000), (iii) mouse monoclonal IE1 antibody (1:5,000), (iv) rabbit polyclonal FP25 antibody (1:5,000), (v) rabbit polyclonal BV/ODV-C42 antibody (1:5,000), (vi) mouse monoclonal GP64 A. californica V5 antibody (1:5,000), or (vii) OpMNPV polyhedrin mouse monoclonal antibody (1:10,000). Horseradish peroxidase-conjugated rabbit anti-mouse secondary antibody (1:15,000) or horseradish peroxidase-conjugated goat anti-rabbit antibody (1:10,000) was used with the enhanced chemiluminescence system (Amersham).
Yeast two-hybrid screening.
The Saccharomyces cerevisiae strain YRG-2 (MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::UASGAL1-TATAGAL1-HIS3 URA3::UASGAL4 17mers(×3)-TATACYC1-lacZ; Stratagene) and the vectors pBD-GAL4 Cam (Stratagene) and pAD-GAL4-2.1 (Stratagene) were used for yeast two-hybrid tests. When exon0 is cloned into the binding domain vector pBD-GAL4 Cam, EXON0 trans-activates the histidine reporter gene (data not shown). Therefore, for all analyses, exon0 was cloned into the activation domain vector (pAD-GAL4-2.1). Eight known nucleocapsid proteins previously identified in both BV and ODV and the nonnucleocapsid protein ME53 as a control were analyzed as candidate interaction partners of EXON0, and the corresponding genes were fused with the binding domain vector pBD-GAL4. The following primers were used for amplifying these genes: primers 1175 (5′-GCGGGAATTCATGACGAATCGTAGATATG-3′) and 1176 (5′-GCGGCTGCAGTTAAGCGCTAGATTCTGTG-3′) for p78/83, primers 1177 (5′-GCGGCTCGAGCATGTGTTCGACCAAGAAAC-3′) and 1178 (5′-GCGGCTGCAGCTACACGTTGTGTGCGTGC-3′) for vp1054, primers 1179 (5′-GCGGGAATTCATGGATCAATTTGAACAGTT-3′) and 1180 (5′-GCGGCTGCAGTTAAATTAAATTTTGAAGCATTT-3′) for fp25, primers 1181 (5′-GCGGGAATTCATGAACGGTTTTAATGTTCG-3′) and 1182 (5′-GCGGCTGCAGCTATTCGTTGCGATAGTAC-3′) for vlf-1, primers 1183 (5′-GCGGGAATTCATGGCGCTAGTGCCCGT-3′) and 1184 (5′-GCGGCTGCAGTTAGACGGCTATTCCTCC-3′) for vp39, primers 1185 (5′-GCGGGAATTCATGAGCGCTATCGCGTTG-3′) and 1186 (5′-GCGGCTGCAGTTAATATTTTTTACGCTTTGCA-3′) for the BV/ODV-C42 gene, primers 1187 (5′-GCGGCTCGAGCATGAACGATTCCAATTCTCT-3′) and 1188 (5′-GCGGCTGCAGTTATATAACATTGTAGTTTGCG-3′) for p87, primers 1189 (5′-GCGGGAATTCATGAACACGGACGCTCAG-3′) and 1190 (5′-GCGGCTGCAGTTATTTATTCAGGCACATTAAA-3′) for p24, and primers 1195 (5′-GCGGGAATTCATGAACCGTTTTTTTCGAGA-3′) and 1196 (5′-GCGGCTGCAGTTAGACATTGTTATTTACAATA-3′) for me53. The resulting activation domain and binding domain fusion plasmids were introduced into the YRG-2 yeast strain by the lithium acetate method according to the instructions of the manufacturer (Stratagene). Transformants were screened on medium lacking the appropriate amino acids, and selection for histidine reporter gene activation was performed on histidine- and tryptophan-deficient or histidine-, tryptophan-, and leucine-deficient agar plates.
Transmission electron microscopy (TEM).
Sf9 cells (2.0 × 106 cells/60-mm-diameter plate) were transfected with 1.0 μg of either exon0 KO or exon0 KO-HA-EXON0 virus. At 24, 36, 48, and 96 hours posttransfection (hpt), the supernatant was removed and the cells were washed once with PBS (pH 7.2) and then fixed in 2.5% glutaradehyde in PBS for 2 h. Cells were then dislodged with a rubber policeman, transferred into Eppendorf tubes, and washed twice in PBS for 15 min each time. Cells were then immobilized with 50 μl of 3% low-melting-point agarose and placed on ice for 30 min. The agarose fraction was then removed, and the cells were fixed with 1% osmium for 1 h, followed by staining with 2% uranyl acetate for 1 h. After dehydration through a series of 30 to 100% ethanol washes, cells were embedded in Spur resin. Ultrathin sections were obtained and subsequently stained with a mixture of 1% uranium acetate and Sato's lead (51). Images were obtained using a Hitachi transmission electron microscope.
RESULTS
Construction of exon0 KO bacmid and growth curve of the repaired virus.
The exon0 gene is essential for the efficient production of BV, as was previously shown using an exon0 KO bacmid that retained 142 and 261 bp of the 5′ and 3′ ends of the exon0 coding region, respectively, in order to preserve the ie0 splice site and the orf142 promoter (8). With this previous construct, intragenomic recombination between the exon0 KO locus and the repair locus could occur. To prevent any possible intragenomic homologous recombination between the exon0 locus and repair constructs, a new exon0 KO bacmid in which the complete ORF of exon0 and the promoters of both exon0 and ie0 were deleted was constructed. This process resulted in a bacmid construct (bMON14272 exon0 KO) that has both exon0 and ie0 knocked out but in which the orf142 promoter has been repaired using the gp64 late promoter (Fig. 1A). It has been shown previously that ie0 is not required for WT levels of BV production (25, 50); therefore, bMON14272 exon0 KO permitted the analysis of exon0 function without the possibility of intragenomic homologous recombination. The correctness of the deletion and repair constructs was confirmed by PCR (Fig. 1B).
The bMON14272 exon0 KO bacmid was repaired by Tn7-mediated transposition at the polyhedrin locus with either the polyhedrin gene alone or the polyhedrin gene and the gene encoding N-terminally HA epitope-tagged EXON0, generating the exon0 KO and exon0 KO-HA-EXON0 viruses, respectively (Fig. 1). To confirm the WT levels of BV production by exon0 KO-HA-EXON0 and the reduction in BV production by the exon0 KO virus, the viruses were compared to the ie0 KO virus ie1 KO-IE1 (50) by analyzing growth curves (Fig. 1C). As expected, the exon0 KO-HA-EXON0 virus and the control virus ie1 KO-IE1 exhibited WT levels of BV production reaching 1.0 × 106 50% tissue culture infective doses (TCID50)/ml by 24 hpt and 1.0 × 108 TCID50/ml by 72 hpt. The exon0 KO virus showed significantly lower levels of BV production, producing fewer than 200 TCID50/ml by 24 hpt, with a peak level of 104 TCID50/ml by 72 hpt. These results correspond to a 99.99% reduction in BV production compared to that by exon0 KO-HA-EXON0 and ie1 KO-IE1. The growth curves confirmed that EXON0 was required for the efficient production of BVs, as previously reported (8), and that the HA tag on the repair construct did not affect the function of EXON0.
Subcellular localization of EXON0 during infection.
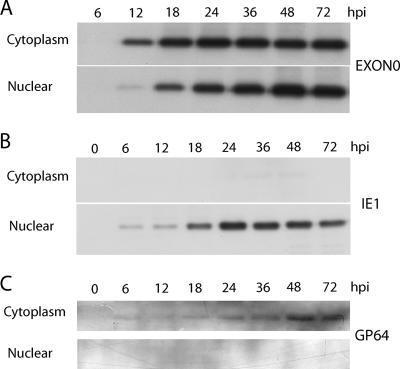
In order to determine the function of EXON0, it was necessary to establish its temporal expression pattern and cellular localization. An initial time course analysis of EXON0 expression was performed using exon0 KO-HA-EXON0. The infected Sf9 cells were collected at various time points pi, and the nuclear and cytoplasmic fractions were separated. Western blot analysis showed that EXON0 was translated from 12 to 72 hpi and was distributed in both the nucleus and the cytoplasm. From 12 to 24 hpi, during peak BV production, EXON0 localized to a greater extent in the cytoplasm (Fig. 2). As a control to test the efficiency of the fractionation process, the same samples were probed with an IE1 antibody and a GP64 antibody. The presence of IE1 and GP64 exclusively in the nuclear and cytoplasmic fractions, respectively, confirmed the efficiency of the cell fractionation.
FIG. 2.
Cellular localization of EXON0 as determined by fractionation of cytoplasm and nuclei and Western blotting. Sf9 cells were infected with exon0 KO-HA-EXON0 virus at an MOI of 10. Cells were harvested and separated into the nuclear and cytoplasmic fractions. Fractions were separated by SDS-PAGE and probed with anti-HA, anti-IE1, and anti-GP64 monoclonal antibodies to detect HA-tagged EXON0, IE1, and GP64, respectively.
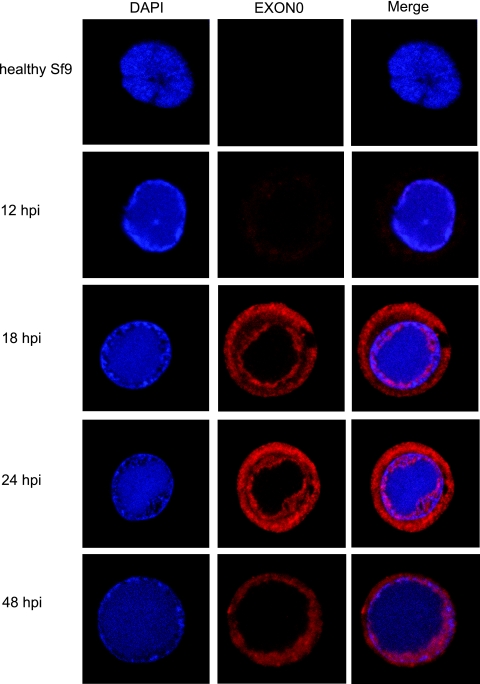
The cellular localization of EXON0 was also analyzed by immunofluorescence and confocal microscopy. As shown in Fig. 3, at 12 hpi more EXON0 could be detected in the cytoplasm than in the nucleus, where it was present at a low level and unevenly distributed. At 18 hpi, EXON0 exhibited a ring pattern concentrating towards the cytoplasmic membrane, and more EXON0 had localized in the nucleus, at the periphery of the virogenic stroma (stained blue by DAPI) that corresponded to the ring zone of the virogenic stroma (7, 59). This double ring localization pattern continued at 24 hpi but with greater levels of expression. By 48 hpi, the EXON0 fluorescence signal had become diffuse and showed slightly greater intensity at the ring zone of the virogenic stroma. The EXON0 signal in both the cytoplasm and the nucleus decreased after 48 hpi (data not shown). The EXON0 localization pattern indicated by immunofluorescence was consistent with the results obtained from the fractionation and Western blot analyses.
FIG. 3.
Localization of EXON0 as demonstrated by immunofluorescence. Sf9 cells were infected with exon0 KO-HA-EXON0 virus at an MOI of 10. At different time points pi, cells were fixed, probed with mouse monoclonal HA antibody to detect HA-tagged EXON0, and visualized by Alexa 635-conjugated goat anti-mouse IgG (red). Additionally, cells were stained with DAPI to directly visualize nuclear DNA (blue). The healthy Sf9 cells probed with the Alexa 635 produced very low levels of background fluorescence, whereas HA-EXON0 was detected in the cytoplasm and nuclei of exon0 KO-HA-EXON0-infected cells from 12 hpi.
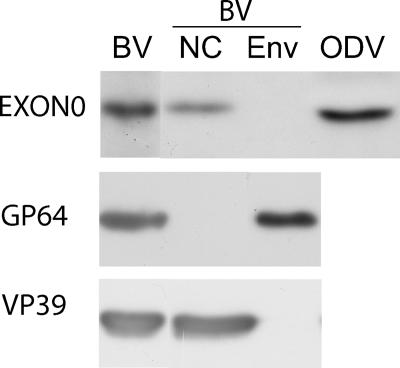
Western blot analysis of EXON0 association with BV and ODV.
As EXON0 affects the production of BV, it was necessary to determine if EXON0 was associated with the viral particles. Therefore, BV and ODV were purified and analyzed by Western blotting for the presence of EXON0. The results (Fig. 4) showed that EXON0 could be detected in both BV and ODV (lanes BV and ODV). The BV particles were also biochemically fractionated into both nucleocapsid and envelope fractions (lanes NC and ENV), and the Western blot analysis indicated that EXON0 was localized in the nucleocapsid fraction. As a control to confirm the efficiency of the fractionation, the known nucleocapsid protein VP39 and the BV envelope-specific protein GP64 were analyzed by Western blotting. Both VP39 and GP64 were observed in the expected fractions. These results showed that EXON0 is associated with both BV and ODV virions and localizes in the nucleocapsids of BV.
FIG. 4.
Analysis of EXON0 in purified and fractionated virions. BV and ODV were purified by using a sucrose gradient and analyzed by SDS-PAGE and Western blotting. Duplicate blots were probed with an anti-HA antibody to detect HA-tagged EXON0, A. californica V5 monoclonal antibody to detect the BV envelope protein GP64, and anti-VP39 to detect the nucleocapsid protein VP39. NC, nucleocapsid fraction; ENV, envelope fraction.
Interaction of EXON0 with nucleocapsid proteins.
The Western blotting results described above suggest that EXON0 is a structural component of the nucleocapsid. It is therefore possible that EXON0 may physically interact with known nucleocapsid proteins. To test this idea, we initially used yeast two-hybrid screening to determine if other nucleocapsid proteins interacted with EXON0. Eight nucleocapsid proteins, P78/83, VP1054, FP25, VLF-1, VP39, BV/ODV-C42, P87, and P24, and, as a control, the nonstructural protein ME53 were selected. Each corresponding gene was cloned into the GAL4 binding domain vector, and an initial control experiment was performed to determine whether these nucleocapsid proteins trans-activated the histidine reporter in the absence of EXON0. exon0 was cloned into the activation domain vector, and the vector was transformed with the nucleocapsid protein fusion constructs as well as an empty binding domain control vector (pBD-Gal4). Based on comparison with the empty vector, medium colony growth was observed for FP25 and BV/ODV-C42 (Table 1).
TABLE 1.
Yeast two-hybrid screening of the nucleocapsid proteins that interact with EXON0
| Construct | Growtha of colonies on:
|
|
|---|---|---|
| His- and Trp-deficient platesb | His-, Trp-, and Leu-deficient platesc | |
| pBD-Gal4 | − | − |
| pBD-Gal4-P78/83 | − | + |
| pBD-Gal4-VP1054 | − | − |
| pBD-Gal4-FP25 | − | ++ |
| pBD-Gal4-VLF-1 | − | + |
| pBD-Gal4-VP39 | − | + |
| pBD-Gal4-C42 | − | ++ |
| pBD-Gal4-P87 | − | + |
| pBD-Gal4-P24 | − | + |
| pBD-Gal4-ME53 | − | + |
−, no growth; +, weak growth; ++, medium growth.
Yeast cells were transformed with the binding domain vector and its fusion constructs and selected on histidine- and tryptophan-deficient plates.
Yeast cells were transformed with the binding domain vector and its fusion constructs along with pAD-Gal4-EXON0 and selected on histidine-, tryptophan-, and leucine-deficient plates.
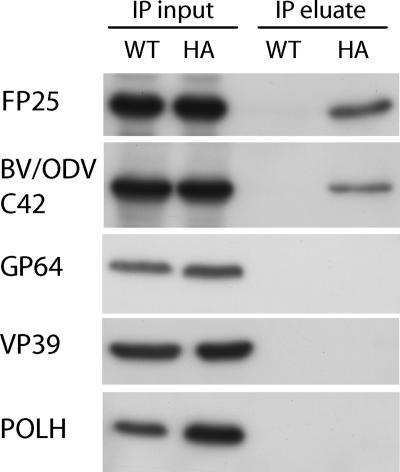
Coimmunoprecipitation assays were performed to confirm the in vivo interaction between EXON0 and FP25 and BV/ODV-C42. Sf9 cells were infected with either WT AcMNPV E2, which does not have HA-tagged EXON0, or exon0 KO-HA-EXON0 virus. At 24 hpi, cells were collected and lysed for immunoprecipitation. Lysates were immunoprecipitated with HA antibody, and the precipitates were analyzed by Western blotting and probed with FP25 and BV/ODV-C42 antibodies. The results showed that FP25 and BV/ODV-C42 coimmunoprecipitated from cells infected with the HA-EXON0 repaired virus but not from those infected with the WT AcMNPV E2 virus. Controls with the major nucleocapsid protein VP39, the BV envelope protein GP64, and the hyperexpressed protein polyhedrin were performed, and all gave negative results (Fig. 5). These results confirmed that EXON0 interacted specifically with FP25 and BV/ODV-C42 in vivo.
FIG. 5.
Interactions of EXON0 with FP25 and BV/ODV-C42 were confirmed by coimmunoprecipitation. Sf9 cells were infected with either WT AcMNPV E2 (WT) or exon0 KO-HA-EXON0 virus (HA) at an MOI of 10, harvested at 24 hpi, and lysed for coimmunoprecipitation. Samples were immunoprecipitated with anti-HA antibody. The total protein from the lysed cells (immunoprecipitation [IP] input) and the immunoprecipitate (IP eluate) were separated by SDS-PAGE, and duplicate blots were probed with antibodies against FP25, BV/ODV-C42, GP64, VP39, and polyhedrin (POLH).
Localization of EXON0 with FP25 and BV/ODV-C42.
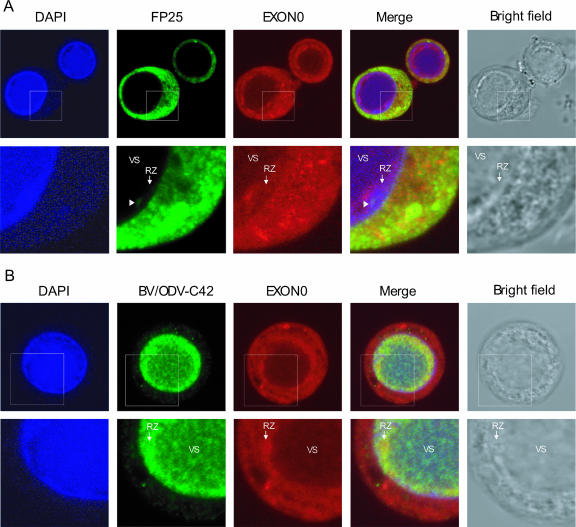
To determine if EXON0 localizes with the nucleocapsid proteins FP25 and BV/ODV-C42, immunofluorescence was used to analyze their patterns of localization in infected Sf9 cells (Fig. 6). At 24 and 48 hpi, FP25 was detected predominantly in the cytoplasm, as reported previously, with very low levels detectable in the nuclei in the ring zone of the virogenic stroma (Fig. 6A, lower panels) (3, 45). An analysis of the merged micrograph images showed that FP25 localized with EXON0 in the cytoplasm, as demonstrated by yellow fluorescence (Fig. 6A). At 24 hpi, BV/ODV-C42 was detected either in the virogenic stroma or at the ring zone of the virogenic stroma, while at 48 hpi, it localized principally at the edge of the virogenic stroma, in agreement with findings in a previous report (Fig. 6B) (4). EXON0 showed strong localization with BV/ODV-C42 mainly at the edge of the virogenic stroma (yellow fluorescence) in the ring zone.
FIG. 6.
Localization of EXON0 with FP25 and BV/ODV-C42. Sf9 cells were infected with exon0 KO-HA-EXON0 virus. Cells were fixed at 24 hpi and probed with anti-HA mouse monoclonal antibody (A and B) followed by anti-FP25 rabbit polyclonal antibody (A) or anti-BV/ODV-C42 rabbit polyclonal antibody (B). EXON0, displayed in red, was visualized with Alexa 635-conjugated goat anti-mouse IgG. FP25 and BV/ODV-C42, displayed in green, were visualized with Alexa 488-conjugated goat anti-rabbit IgG. Cells were also stained with DAPI to visualize the nuclei. In the merged micrographs, yellow indicates colocalization. The lower panels in A and B are enlargements of the regions highlighted by the square boxes in the corresponding upper panels. The white arrowheads in panel A point to the locations of small amounts of nuclear FP25 localized to the ring zone (RZ) of the virogenic stroma (VS), where nucleocapsid assembly occurs. (B) EXON0 and BV/ODV-C42 colocalized predominately in the nuclear ring zone of the virogenic stroma.
Cellular localization of nucleocapsids in Sf9 cells transfected with exon0 KO virus and the repaired virus.
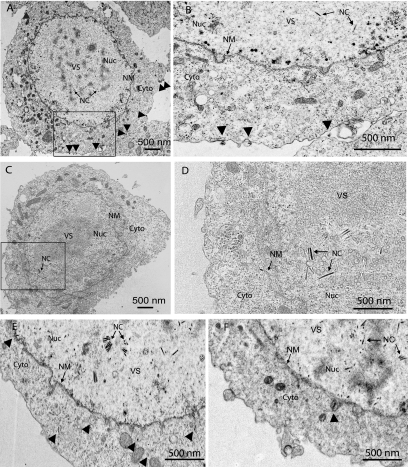
The last essential steps in the AcMNPV BV life cycle are the assembly of nucleocapsids and egress from the nucleus, followed by transport through the cytoplasm and budding from the plasma membrane, where the BV gains its envelope. In order to determine whether the deletion of EXON0 interferes with the assembly and egress of nucleocapsids, Sf9 cells were transfected with either exon0 KO or exon0 KO-HA-EXON0 virus and examined by TEM at 24, 36, 48, and 96 hpt. The TEM analysis showed that the nuclei of cells transfected with either exon0 KO or exon0 KO-HA-EXON0 virus exhibited the typical baculovirus infection symptoms, namely, an extensive number of nucleocapsids, enlargement, and reorganized, electron-dense virogenic stroma (Fig. 7). No physical difference between the nucleocapsids of exon0 KO and exon0 KO-HA-EXON0 viruses was observed, indicating that EXON0 is not required for the assembly of nucleocapsids.
FIG. 7.
Localization of nucleocapsids in Sf9 cells transfected with either exon0 KO-HA-EXON0 (A, B, and E) or exon0 KO (C, D, and F) virus at 24 hpt (A, B, C, and D) or 36 hpt (E and F) as shown by TEM. Panels B and D are higher-magnification images of the boxed regions in panels A and C, respectively. Labels and arrows indicate the virogenic stroma (VS), nucleocapsids (NC), and nuclei (Nuc). Nucleocapsids in the process of egressing that are passing through the nuclear membrane (NM), residing in the cytoplasm (Cyto), or budding at the cytoplasmic membrane are indicated with arrowheads.
In most cells transfected with the exon0 KO virus, nucleocapsids were found in the nucleus but very few nucleocapsids were found to be closely associated with or passing through the nuclear membrane. In addition, very few were observed residing in the cytoplasm or budding at the cytoplasmic membrane. In contrast, cells transfected with exon0 KO-HA-EXON0 virus showed many nucleocapsids penetrating the nuclear membrane, residing in the cytoplasm, and budding at the cytoplasmic membrane. To quantify this observation, 20 Sf9 cells transfected with exon0 KO virus and 20 Sf9 cells transfected with exon0 KO-HA-EXON0 were randomly chosen and the total numbers of nucleocapsids in the nuclei, in the process of egress from the nuclei, in the cytoplasm, or in the process of budding from the cytoplasmic membrane were determined and summarized in Table 2. This analysis showed a total of 2,868 nucleocapsids in the nuclei and 390 nucleocapsids in the BV pathway (inner nuclear membrane, cytoplasm, and cytoplasmic membrane) in the 20 Sf9 cells transfected with exon0 KO-HA-EXON0. In contrast, the 20 Sf9 cells transfected with exon0 KO virus presented a total of 2,957 nucleocapsids in the nuclei and only 25 nucleocapsids in the process of budding. The similar numbers of nucleocapsids found in the nuclei of Sf9 cells transfected with exon0 KO virus and those transfected with exon0 KO-HA-EXON0 would indicate that exon0 does not have an impact on the production or assembly of nucleocapsids. For the exon0 KO virus, the lower numbers of nucleocapsids observed at all steps of the budding process suggest that EXON0 is required for the efficient egress of nucleocapsids from the nucleus.
TABLE 2.
Summary of numbers of nucleocapsids in the nuclei of and in the process of budding from 20 Sf9 cells transfected with the exon0 KO virus and 20 Sf9 cells transfected with exon0 KO-HA-EXON0a
| Location | No. of nucleocapsids at indicated location in cells transfected with:
|
|
|---|---|---|
| exon0 KO virus | exon0 repair virus | |
| Nucleus | 2,957 | 2,868 |
| Nuclear membrane | 4 | 35 |
| Cytoplasm | 19 | 193 |
| Cytoplasmic membrane | 2 | 162 |
| Total no. of budding nucleocapsidsb | 25 | 390 |
The number of cells transfected with the exon0 KO virus was equal to the number of cells transfected with exon0 KO-HA-EXON0 at 24, 36, 48, and 96 hpt.
The total number of budding nucleocapsids comprises nucleocapsids at any stage in the budding process, including those associated with or penetrating the nuclear membrane, those in the cytoplasm, and those budding from the cytoplasmic membrane.
DISCUSSION
AcMNPV EXON0 (ORF141) is required for the efficient production of BV, but its role in the BV pathway was previously unknown. In this study, EXON0 was shown to be a structural protein of both BV and ODV. Western blot analysis indicated that it was localized in the nucleocapsid fraction of BVs (Fig. 4) and was the expected size (30 kDa), but it was a minor component of nucleocapsids compared to VP39, as shown by Coomassie staining of purified virions (data not shown). Therefore, EXON0 is the ninth nucleocapsid protein to be identified, along with P78/83 (46), VP1054 (36), FP25 (3), VLF-1 (60), VP39 (53), BV/ODV-C42 (4), P87 (24), and P24 (57), which have been found to associate with both AcMNPV BV and ODV. Braunagel et al. (5) performed a very detailed proteomic analysis of ODV structural proteins but surprisingly did not identify EXON0. However, they did isolate a protein of the same size as EXON0 from ODV, but they were unable to identify this protein by the methods they used.
EXON0 was shown to interact with the nucleocapsid proteins FP25 and BV/ODV-C42 by yeast two-hybrid screening and coimmunoprecipitation (Table 1 and Fig. 5). It was also shown to localize with FP25 in the cytoplasm and with BV/ODV-C42 in the nucleus (Fig. 6). Previous reports have shown that an FP25 mutant produces fewer occlusion bodies and fewer virions per occlusion body (11, 29, 40, 41) but releases more BVs into the medium (11, 15, 40). The association between FP25 and EXON0 identified in this study indicates that these two proteins may be jointly involved in regulating the BV and ODV nucleocapsid ratio and, ultimately, BV production. Nevertheless, the significance of these interactions with respect to EXON0 functionality remains to be fully determined.
The nucleocapsid proteins P78/83, VP1054, and VLF-1 have previously been shown to be required for nucleocapsid assembly, and in their absence there is either no nucleocapsid production or nucleocapsids are malformed (36, 39, 54). EXON0 as a nucleocapsid protein present in both BV and ODV may potentially affect the efficiency of nucleocapsid assembly. However, this does not seem to be the case, as the nuclei of cells transfected with exon0 KO virus produced normal-length nucleocapsids with an electron-dense nucleoprotein core as determined by TEM. Sections of cells transfected with exon0 KO virus showed that the nucleocapsids were packaged in polyhedra at 96 hpt, but further studies are being done to determine if there is a quantitative difference in packaging efficiency. Finally, there was no significant difference in the number of nucleocapsids found in the nuclei of Sf9 cells transfected with exon0 KO virus and those transfected with exon0 KO-HA-EXON0. These data suggest that EXON0 is not required for the assembly of nucleocapsids.
Interestingly, confocal microscopy shows that EXON0 has distinct cellular localization patterns in both the cytoplasm and the nucleus. The nuclear association of EXON0 occurs at the ring zone of the virogenic stroma, where nucleocapsid morphogenesis and polyhedron development occur (7, 59). This finding agrees with our results showing that EXON0 is a structural component of virions. It is not yet clear what function EXON0 has in the second ring pattern that concentrates towards the cytoplasmic membrane. It is possible that EXON0 has multiple roles during infection, and the cytoplasmic ring pattern may facilitate the transport or budding of nucleocapsids to the cell surface.
The TEM results provide evidence that EXON0 is required for the efficient egress of nucleocapsids from the nucleus to the cytoplasm. The conservation of EXON0 in all lepidopteran nucleopolyhedroviruses suggests that it may play a role in facilitating a common transport pathway for nucleocapsid egress for this group of baculoviruses. However, exon0 is not strictly essential for the production of BV, since a few nucleocapsids in the cells transfected with exon0 KO virus did pass through the nuclear membrane, followed by transport through the cytoplasm and budding at the plasma membrane. Two possible explanations are that the EXON0-facilitated nucleocapsid transport process can be bypassed and that other viral and cellular proteins can replace EXON0, although extremely inefficiently. Granulovirus genomes that have been sequenced to date do not contain an exon0 homolog. Therefore, it is interesting that the characterized Cydia pomonella granulovirus tissue culture system produces very low titers of extracellular virus at very different times pi compared to lepidopteran nucleopolyhedroviruses (56). Therefore, EXON0 may be a relatively recent acquisition by the lepidopteran nucleopolyhedroviruses for the rapid and high-level production of BV.
How nucleocapsids egress and get transported is an important question, as it represents an essential step in the viral life cycle. For other large DNA animal viruses, it has been shown that egress is a complex procedure that involves many viral and cellular proteins. Previous studies showed that the deletion of herpesvirus UL34 produces virus deficient in plaque formation and leads to a 100-fold reduction in viral titer, similar to the AcMNPV exon0 KO phenotype (44). UL34 has been shown to recruit the cellular protein kinase C to phosphorylate the lamin and cause the local dissolution of nuclear lamin for the access of capsids to the nuclear membrane (34, 47, 48). The UL34 and UL31 gene products have been shown to be constituents of primary enveloped virions but not mature virus particles, and the absence of either protein causes the accumulation of capsids in the nucleus (12). Both UL34 and UL31 are required for the formation of the herpesvirus primary envelope acquired by budding through the inner nuclear membrane (20, 42). Nuclear egress is also affected by the US3 kinase, which appears to trigger the de-envelopment of primary enveloped capsids in the perinuclear region by fusion with the outer nuclear membrane (19, 43). Once herpesvirus capsids have egressed from the nucleus, further movement may involve the endoplasmic reticulum and Golgi secretory pathways and interactions with microtubules (9, 28, 30-32, 49, 61). Similarly, EXON0 as a component of nucleocapsids required for the efficient egress of nucleocapsids from the nucleus may interact with cellular proteins to facilitate transport similar to that of herpesvirus capsids. In addition, EXON0 is expressed in specific ring patterns in both the cytoplasm and the nucleus, suggesting that it may have multiple functions in the BV pathway. The elucidation of additional proteins that interact with EXON0 is presently under way and will help to shed light on how BV nucleocapsids are transported during egress.
Acknowledgments
We thank Les Willis, Yingchao Nie, and Christina McCarthy for their critical readings of the manuscript; Michael Weis for his expertise on the TEM; and Sharon Braunagel and George Rohrmann for kindly providing antibodies.
This study was supported in part by funding from the Natural Science and Engineering Research Council and the Biocontrol Network.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Braunagel, S. C., J. K. Burks, G. Rosas-Acosta, R. L. Harrison, H. Ma, and M. D. Summers. 1999. Mutations within the Autographa californica nucleopolyhedrovirus FP25K gene decrease the accumulation of ODV-E66 and alter its intranuclear transport. J. Virol. 73:8559-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunagel, S. C., P. A. Guidry, G. Rosas-Acosta, L. Engelking, and M. D. Summers. 2001. Identification of BV/ODV-C42, an Autographa californica nucleopolyhedrovirus orf101-encoded structural protein detected in infected-cell complexes with ODV-EC27 and p78/83. J. Virol. 75:12331-12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunagel, S. C., W. K. Russell, G. Rosas-Acosta, D. H. Russell, and M. D. Summers. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. USA 100:9797-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunagel, S. C., and M. D. Summers. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315-328. [DOI] [PubMed] [Google Scholar]
- 7.Charlton, C. A., and L. E. Volkman. 1991. Sequential rearrangement and nuclear polymerization of actin in baculovirus-infected Spodoptera frugiperda cells. J. Virol. 65:1219-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, X., T. M. Stewart, J. A. Pathakamuri, Q. Li, and D. A. Theilmann. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J. Virol. 78:9633-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diefenbach, R. J., M. Miranda-Saksena, E. Diefenbach, D. J. Holland, R. A. Boadle, P. J. Armati, and A. L. Cunningham. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, M. J. 1986. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J. Ultrastruct. Mol. Struct. Res. 95:189-195. [Google Scholar]
- 11.Fraser, M. J., and W. F. Hink. 1982. The isolation and characterization of the MP and FP plaque variants of Galleria mellonella nuclear polyhedrosis virus. Virology 117:366-378. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not of mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funk, C. J., S. C. Braunagel, and G. F. Rohrmann. 1997. Baculovirus structure, p. 7-32. In L. K. Miller (ed.), The baculoviruses. Plenum, New York, NY.
- 14.Garrity, D. B., M. Chang, and G. W. Blissard. 1997. Late promoter selection in the baculovirus gp64 envelope fusion protein gene. Virology 231:167-181. [DOI] [PubMed] [Google Scholar]
- 15.Harrison, R. L., and M. D. Summers. 1995. Mutations in the Autographa californica multinucleocapsid nuclear polyhedrosis virus 25 kDa protein gene result in reduced virion occlusion, altered intranuclear envelopment and enhanced virus production. J. Gen. Virol. 76:1451-1459. [DOI] [PubMed] [Google Scholar]
- 16.Hefferon, K. L., A. G. Oomens, S. A. Monsma, C. M. Finnerty, and G. W. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455-468. [DOI] [PubMed] [Google Scholar]
- 17.Hou, S., X. Chen, H. Wang, M. Tao, and Z. Hu. 2002. Efficient method to generate homologous recombinant baculovirus genomes in E. coli. BioTechniques 32:783-784. 786:788. [DOI] [PubMed] [Google Scholar]
- 18.Kingsley, D. H., A. Behbahani, A. Rashtian, G. W. Blissard, and J. Zimmerberg. 1999. A discrete stage of baculovirus GP64-mediated membrane fusion. Mol. Biol. Cell 10:4191-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 20.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leikina, E., H. O. Onaran, and J. Zimmerberg. 1992. Acidic pH induces fusion of cells infected with baculovirus to form syncytia. FEBS Lett. 304:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica multicapsid nucleopolyhedrovirus lef-6-null virus: LEF-6 is not essential for viral replication but appears to accelerate late gene transcription. J. Virol. 76:5503-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long, G., X. Pan, R. Kormelink, and J. M. Vlak. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80:8830-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, A., and E. B. Carstens. 1992. Nucleotide sequence and transcriptional analysis of the p80 gene of Autographa californica nuclear polyhedrosis virus: a homologue of the Orgyia pseudotsugata nuclear polyhedrosis virus capsid-associated gene. Virology 190:201-209. [DOI] [PubMed] [Google Scholar]
- 25.Lu, L., H. Rivkin, and N. Chejanovsky. 2005. The immediate-early protein IE0 of the Autographa californica nucleopolyhedrovirus is not essential for viral replication. J. Virol. 79:10077-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lung, O. Y., M. Cruz-Alvarez, and G. W. Blissard. 2003. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J. Virol. 77:328-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mabit, H., M. Y. Nakano, U. Prank, B. Saam, K. Dohner, B. Sodeik, and U. F. Greber. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 76:9962-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacKinnon, E. A., J. F. Henderson, D. B. Stoltz, and P. Faulkner. 1974. Morphogenesis of nuclear polyhedrosis virus under conditions of prolonged passage in vitro. J. Ultrastruct. Res. 49:419-435. [DOI] [PubMed] [Google Scholar]
- 30.Martin, A., P. O'Hare, J. McLauchlan, and G. Elliott. 2002. Herpes simplex virus tegument protein VP22 contains overlapping domains for cytoplasmic localization, microtubule interaction, and chromatin binding. J. Virol. 76:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monsma, S. A., A. G. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 35.Okano, K., A. L. Vanarsdall, V. S. Mikhailov, and G. F. Rohrmann. 2006. Conserved molecular systems of the Baculoviridae. Virology 344:77-87. [DOI] [PubMed] [Google Scholar]
- 36.Olszewski, J., and L. K. Miller. 1997. Identification and characterization of a baculovirus structural protein, VP1054, required for nucleocapsid formation. J. Virol. 71:5040-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olszewski, J., and L. K. Miller. 1997. A role for baculovirus GP41 in budded virus production. Virology 233:292-301. [DOI] [PubMed] [Google Scholar]
- 38.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297-314. [DOI] [PubMed] [Google Scholar]
- 39.Possee, R. D., T. P. Sun, S. C. Howard, M. D. Ayres, M. Hill-Perkins, and K. L. Gearing. 1991. Nucleotide sequence of the Autographa californica nuclear polyhedrosis 9.4 kbp EcoRI-I and -R (polyhedrin gene) region. Virology 185:229-241. [DOI] [PubMed] [Google Scholar]
- 40.Potter, K. N., P. Faulkner, and E. A. MacKinnon. 1976. Strain selection during serial passage of Trichoplusia ni nuclear polyhedrosis virus. J. Virol. 18:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramoska, W. A., and W. F. Hink. 1974. Electron microscope examination of two plaque variants from a nuclear polyhedrosis virus of the alfalfa looper, Autographa californica. J. Invertebr. Pathol. 23:197-201. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds, A., B. Ryckman, J. Baines, Y. Zhou, L. Liang, and R. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds, A., E. G. Wills, R. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roller, R., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosas-Acosta, G., S. C. Braunagel, and M. D. Summers. 2001. Effects of deletion and overexpression of the Autographa californica nuclear polyhedrosis virus FP25K gene on synthesis of two occlusion-derived virus envelope proteins and their transport into virus-induced intranuclear membranes. J. Virol. 75:10829-10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell, R. L., C. J. Funk, and G. F. Rohrmann. 1997. Association of a baculovirus-encoded protein with the capsid basal region. Virology 227:142-152. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez, V., and D. H. Spector. 2002. CMV makes a timely exit. Science 297:778-779. [DOI] [PubMed] [Google Scholar]
- 48.Scott, E., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart, T. M., I. Huijskens, L. G. Willis, and D. A. Theilmann. 2005. The Autographa californica multiple nucleopolyhedrovirus ie0-ie1 gene complex is essential for wild-type virus replication, but either IE0 or IE1 can support virus growth. J. Virol. 79:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takagi, I., K. Yamada, T. Sato, T. Hanaichi, T. Iwamoto, and L. Jin. 1990. Penetration and stainability of modified Sato's lead staining solution. J. Electron Microsc. 39:67-68. [PubMed] [Google Scholar]
- 52.Theilmann, D. A., G. W. Blissard, B. Bonning, J. A. Jehle, D. R. O'Reilly, G. F. Rohrmann, S. Thiem, and J. M. Vlak. 2005. Baculoviridae, p. 177-185. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Springer, New York, NY.
- 53.Thiem, S. M., and L. K. Miller. 1989. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 63:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2006. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J. Virol. 80:1724-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkman, L. E., and P. A. Goldsmith. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143:143-185. [DOI] [PubMed] [Google Scholar]
- 56.Winstanley, D., and N. E. Crook. 1993. Replication of Cydia pomonella granulosis virus in cell cultures. J. Gen. Virol. 74:1599-1609. [DOI] [PubMed] [Google Scholar]
- 57.Wolgamot, G. M., C. H. Gross, R. L. Russell, and G. F. Rohrmann. 1993. Immunocytochemical characterization of P24, a baculovirus capsid-associated protein. J. Gen. Virol. 74:103-107. [DOI] [PubMed] [Google Scholar]
- 58.Wu, W., T. Lin, L. Pan, M. Yu, Z. Li, Y. Pang, and K. Yang. 2006. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J. Virol. 80:11475-11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xeros, N. 1956. The virogenic stroma in nuclear and cytoplasmic polyhedrosis. Nature 178:412-413. [DOI] [PubMed] [Google Scholar]
- 60.Yang, S., and L. K. Miller. 1998. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 245:99-109. [DOI] [PubMed] [Google Scholar]
- 61.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]