Abstract
Dominant, constitutively expressed antiretroviral factors, including TRIM5α and APOBEC3 proteins, are distinguished from the conventional innate immune systems and are classified as intrinsic immunity factors. Here, we demonstrate that interferon alpha (IFN-α) treatment upregulates TRIM5α mRNA in rhesus monkey cells, which correlates with the enhanced TRIM5α-mediated pre- and postintegration blocks of human immunodeficiency virus replication. In human cells, IFN-α increases the levels of TRIM5α mRNA, resulting in enhanced antiviral activity against N-tropic murine leukemia virus infection. These observations indicate that the TRIM5α-mediated antiviral effects can be orchestrated by the conventional innate immune response. It is conceivable that TRIM5α plays an essential role in controlling both the initial retroviral exposure and the subsequent viral dissemination in vivo.
Mammalian cells have developed diverse strategies to counteract viral infection and replication. The main immune response to viral infection consists of an innate defense and an adaptive defense. The innate defense includes cytokines, complements, local sentinel cells, and natural killer cells and is essential in antiviral defense because it can be activated quickly without any prior exposure to the invading virus (39). In addition to the conventional immune response, several nonimmune strategies which target distinct steps in the retroviral life cycle have been identified (5, 9, 15, 32, 33, 40). Since these antiretroviral factors are constitutively expressed in an immunologically naive host, their antiviral activities are considered to play a crucial role in host defense.
TRIM5α is a restriction factor that blocks retrovirus infection at a postentry, preintegration stage of the viral life cycle (20, 23, 29, 37, 41). Human TRIM5α (TRIM5αhu) potently restricts N-tropic murine leukemia virus (N-MLV) (20, 23, 29, 41) but not human immunodeficiency virus type 1 (HIV-1). Rhesus monkey TRIM5α (TRIM5αrh) expression in normally HIV-1-permissive cells confers strong restriction of HIV-1 but not of simian immunodeficiency virus from macaques (SIVMAC) (20, 23, 29, 37, 41). Recently, we have found that TRIM5αrh also blocks the late phase of HIV-1 replication (31). Similar to the well-characterized postentry restriction activities, TRIM5αrh is active against HIV-1 but not SIVMAC production, whereas TRIM5αhu does not strongly block the late phase of HIV-1 replication (31).
The TRIM5 gene shares the enhancer region with a well-characterized interferon (IFN)-responsive gene, TRIM22 (38). Moreover, the consensus IFN-stimulated response element (TTTCACTTTC) is located within 40 bp upstream of the putative transcription start sites of human and rhesus monkey TRIM5 genes (NCBI Entrez Gene analysis), suggesting that TRIM5α expression is IFN responsive. Indeed, upregulation of TRIM5α mRNA following type I IFN treatment was noted in human HeLa and HepG2 cell lines (3), although its influence on TRIM5α-mediated antiviral activities remains to be determined. In this study, we examined the levels of TRIM5α mRNA and the TRIM5α-mediated antiviral activities in IFN-treated human and rhesus monkey cells.
IFN-α increases the levels of TRIM5αrh transcript in rhesus monkey cells.
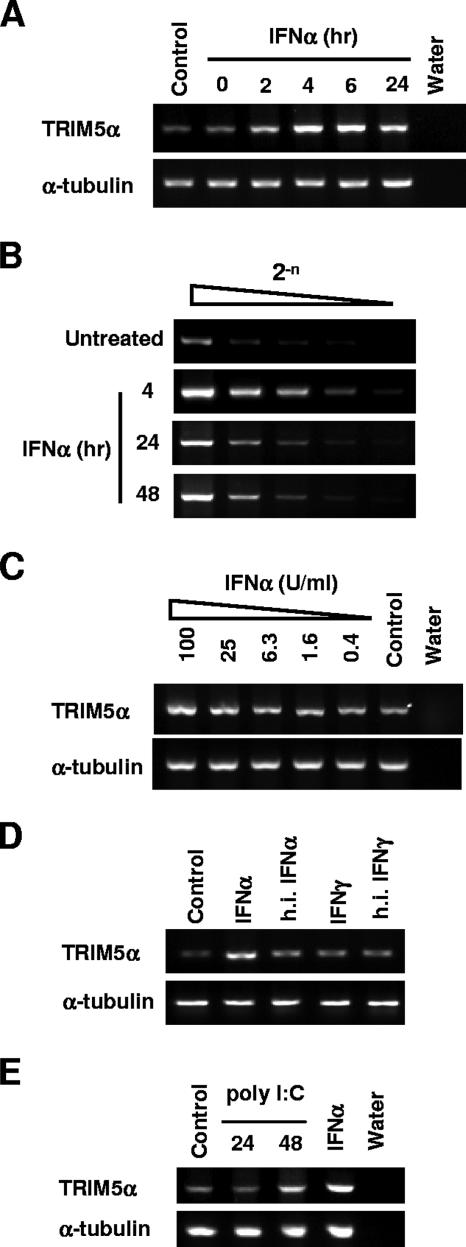
First, we examined the influence of alpha IFN (IFN-α) treatment on the levels of TRIM5αrh transcript in rhesus monkey FrhK4 cells. We used universal IFN-α, a human IFN-αA and -αD hybrid, which is active on most mammalian cells (PBL Biomedical Laboratories). FrhK4 cells were treated with 100 U/ml of IFN-α for 0, 2, 4, 6, 24, and 48 h. Total RNA was extracted by using TRIzol reagent (Invitrogen), and 5.0 μg of RNA was used to synthesize the first-strand DNA with an oligo(dT) primer (SuperScript III first-strand synthesis system; Invitrogen). One-tenth of the reverse transcription (RT) products were used to amplify TRIM5αrh-specific cDNA with primers 5′-GCAGGTACCATGCTCATGGAGGAGGTTGCCCA-3′ and 5′-ACGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATAAGAGCTTGGTGAGCACAGAG TCATG-3′. After IFN-α treatment, notable increases in the levels of TRIM5αrh transcript were observed at 4 and 6 h, while increases to a lesser degree were observed at 24 h (Fig. 1A). In order to estimate the degree of TRIM5αrh induction upon IFN-α treatment, the RT products were serially diluted and used as templates to amplify the TRIM5αrh-specific cDNA. As shown in Fig. 1B, the undiluted RT product of the control cells showed a signal level similar to that of the eightfold dilution of the IFN-α-treated sample. IFN-α treatment for 24 and 48 h resulted in approximate two- to fourfold increases in the TRIM5αrh mRNA levels. The induction of TRIM5αrh mRNA was dose dependent in that the highest concentration of IFN-α (100 U/ml) showed the strongest signal (Fig. 1C). On the other hand, treatments with heat-inactivated IFN-α or 1,000 U/ml of human gamma IFN (IFN-γ; PBL Laboratories) for 6 h did not increase the TRIM5αrh mRNA levels (Fig. 1D). We thus concluded that IFN-α treatment specifically increased the levels of TRIM5αrh mRNA in rhesus monkey cells. Moreover, transfection of double-stranded RNA poly(I:C) resulted in a mild increase in the TRIM5αrh mRNA level (Fig. 1E).
FIG. 1.
IFN-α increases the levels of TRIM5αrh transcript in rhesus monkey cells. (A) FrhK4 cells were treated with IFN-α (100 U/ml) for the indicated periods of time. As a control, untreated FrhK4 cells were used. After treatment, total cellular RNA was isolated and TRIM5αrh mRNA was detected by RT-PCR (upper panel), along with α-tubulin mRNA as a control (lower panel). (B) FrhK4 cells were treated with IFN-α (100 U/ml) for the indicated periods of time. Serially diluted RT products (equivalent to 0.5, 0.25, 0.125, 0.063, and 0.031 μg of total RNA samples) were used to detect the TRIM5αrh-specific transcript. (C) Frhk4 cells were treated with various concentrations of IFN-α, and the TRIM5αrh mRNA was detected by RT-PCR. Untreated cells were used as a control. (D) FrhK4 cells were treated with IFN-α (100 U/ml), heat-inactivated (h.i.) IFN-α (100 U/ml), IFN-γ (1,000 U/ml), and heat-inactivated IFN-γ (1,000 U/ml) for 6 h, and the TRIM5αrh mRNA was detected by RT-PCR. (E) FrhK4 cells were transfected with double-stranded RNA poly(I:C) by using Oligofectamine (Invitrogen). As a positive control, Frhk4 cells were treated with IFN-α (100 U/ml) for 4 h. Total RNA was isolated from the treated or untreated cells (control).
IFN-α enhances the TRIM5αrh-mediated antiviral activities on HIV-1 replication.
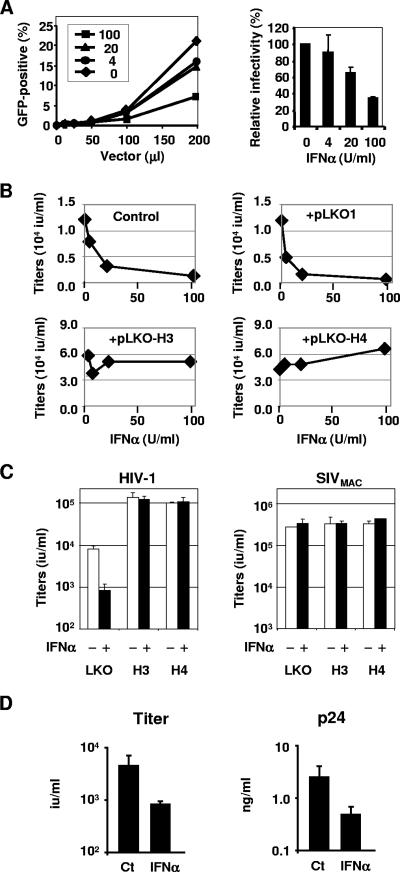
To address the influence of IFN-α treatment on the TRIM5αrh-mediated postentry restriction of HIV-1 infection, we first examined whether IFN-α treatment could render FrhK4 cells less permissive to a vesicular stomatitis virus G protein (VSV-G)-pseudotyped HIV-1 vector infection. A green fluorescent protein (GFP)-carrying HIV-1 vector was produced as described previously (28). We pretreated FrhK4 cells with 0, 10, 20, 50, and 100 U/ml of IFN-α for 6 h, followed by infection with increasing amounts of HIV-1 vectors. The IFN-α treatment marginally enhanced (up to threefold) the postentry restriction in FrhK4 cells (Fig. 2A).
FIG. 2.
IFN-α enhances the TRIM5αrh-mediated blocks of HIV-1 replication. (A) FrhK4 cells were pretreated with 0, 4, 20, or 100 U/ml of IFN-α for 6 h and then infected with increasing amounts of GFP-expressing HIV-1 vector. Two days after infection, the GFP-positive cell populations were determined by flow cytometry (left panel). The relative HIV-1 vector infectivity in IFN-α-treated cells (mean ± standard deviation [SD]) is shown in the right panel. (B) FrhK4 cells (1.0 × 105) were transfected with 0.2 μg of pNL4-3 along with 1.0 μg of either pBlueScriptII (control), a parental shRNA plasmid (pLKO1), or TRIM5α-specific shRNA constructs (pLKO-H3 and pLKO-H4). After overnight transfection, the culture supernatants were replaced with fresh growth media containing various concentrations of IFN-α. Two days after transfection, culture supernatants were harvested to determine HIV-1 production. Viral titers were shown as infectious units per milliliter in GHOST(3)R3/X4/R5 cells. (C) FrhK4 cells (1.0 × 105) were transfected with 0.2 μg of pNL4-3 or pSIVMAC1A11 along with 1.0 μg of pLKO1, pLKO-H3, or pLKO-H4. After overnight transfection, the culture supernatants were replaced with fresh growth media containing 100 U/ml of IFN-α. Two days after transfection, culture supernatants were harvested and viral titers were determined in GHOST(3)R3/X4/R5 cells (mean ± SD). (D) FrhK4 cells (1.0 × 105) were transfected with 1.0 μg of pNL4-3. After overnight transfection, the culture supernatants were replaced with fresh growth media containing 100 U/ml of IFN-α. Two days after IFN-α treatment, culture supernatants were harvested to determine the viral titers and p24 concentrations (mean ± SD).
We then examined the influence of TRIM5αrh upregulation on the TRIM5αrh-mediated block of the late phase of HIV-1 replication. FrhK4 cells were transfected with an HIV-1 infectious molecular clone, pNL4-3 (2), by using Fugene6 (Roche). After overnight transfection, the culture supernatants were replaced with fresh growth media containing various concentrations of IFN-α. Two days after transfection, the culture supernatants were harvested to determine the viral titers in GHOST(3)R3/X4/R5 cells (26). Increasing concentrations of the IFN-α treatments correlated with reduced HIV-1 titers from the treated cells (Fig. 2B), and treatment with 100 U/ml of IFN-α led to a sevenfold reduction in HIV-1 titers.
Since IFN-α induces a series of antiviral factors, we tested whether the IFN-α-mediated antiviral effect on HIV-1 production was mediated by the upregulated TRIM5αrh or by other antiviral factors. In order to dissect the influence of TRIM5αrh upregulation from the other antiviral factors, we used two anti-TRIM5α-short-hairpin-RNA (shRNA) constructs, pLKO1-H3 and pLKO1-H4 (OpenbioSystems), which were used to disrupt TRIM5αrh mRNA in our previous study (31). FrhK4 cells were transfected with 0.2 μg of pNL4-3 along with 1.0 μg of the shRNA-expressing plasmids or their parental plasmid, pLKO1. After overnight transfection, the culture supernatants were replaced with fresh growth media containing various concentrations of IFN-α. Two days after transfection, viral titers in the supernatants were determined. TRIM5αrh knockdown by pLKO1-H3 and -H4 abrogated the effects of IFN-α on HIV-1 production, whereas the parental pLKO1 vector did not affect the IFN-α-mediated antiviral activities (Fig. 2B and C). In contrast, production of SIVMAC1A11 (24), which is resistant to the TRIM5αrh-mediated postintegration block (31), was not affected by the IFN-α or shRNA treatment (Fig. 2C). These observations indicate that TRIM5αrh plays a central role in the IFN-α-mediated block of HIV-1 production in rhesus monkey cells.
We then compared viral titers and the levels of p24 release from the IFN-α-treated and untreated FrhK4 cells. p24 concentrations in the supernatants were analyzed with a p24 enzyme-linked immunosorbent assay kit (Zeptmatrix). The reduction of viral titers upon IFN-α treatment correlated well with the reduced p24 level in the supernatant (Fig. 2D), suggesting that IFN-α reduced the HIV-1 titers by reducing the number of virions in the supernatants.
IFN-α upregulates TRIM5αhu mRNA and enhances postentry restriction against N-MLV.
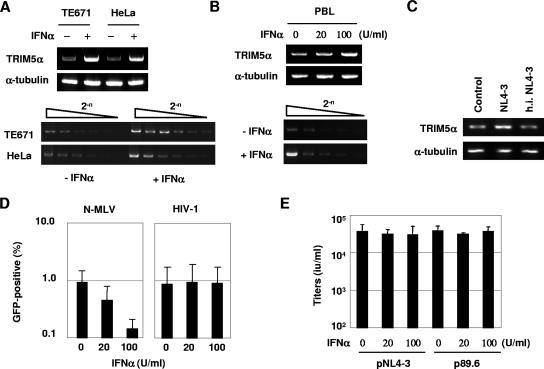
We next examined the influence of IFN-α treatment on the levels of TRIM5αhu mRNA. The human cell lines TE671 and HeLa were treated with 100 U/ml of IFN-α for 4 h. The total cellular RNA was extracted, and the TRIM5αhu-specific mRNA was detected by RT-PCR. An oligo(dT) primer was used for the first-strand synthesis, while TRIM5αhu-specific primers, 5′-GCGAATTCCACCATGGCTTCTGGAATCCTG-3′ and 5′-GGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATAAGAGCTTGGTG AGCACAG-3′, were used for PCR. As shown in Fig. 3A, IFN-α treatment increased the levels of TRIM5αhu mRNA approximately eight- and fourfold in TE671 and HeLa cells, respectively. Similarly, approximately fourfold increases in the TRIM5αhu mRNA were observed in phytohemagglutinin-stimulated peripheral blood lymphocytes (PBL) after treatment with IFN-α for 4 h (Fig. 3B). A marginal increase in the TRIM5αhu mRNA was also noted when the PBL were infected with HIV-1 NL4-3 at a multiplicity of infection of 0.5 (Fig. 3C). Since TRIM5αhu is known to block infection of N-MLV, we tested the influence of IFN-α-mediated TRIM5αhu upregulation on N-MLV infectivity. pCIG-N, which expresses N-MLV Gag-Pol, was used to produce VSV-G-pseudotyped, GFP-carrying N-MLV vector (7). IFN-α treatment rendered TE671 cells approximately fivefold less permissive to a VSV-G-pseudotyped N-MLV vector (Fig. 3D). In contrast, IFN-α-mediated TRIM5αhu upregulation in TE671 cells did not affect infection of a VSV-G-pseudotyped HIV-1 vector (Fig. 3D) or HIV-1 production from infectious molecular clones pNL4-3 and p89.6 (10) (Fig. 3E).
FIG. 3.
IFN-α upregulates TRIM5αhu mRNA and enhances postentry restriction against N-MLV in human cells. (A) TE671 and HeLa cells were treated with 100 U/ml of IFN-α for 4 h. A 1.0-μg amount of total RNA was used to synthesize the first-strand DNA, and one-tenth of the RT products were used to detect TRIM5αhu and α-tubulin transcripts. Serially diluted RT products (equivalent to 0.1, 0.05, 0.025, 0.0125, 0.0063, and 0.0031 μg of total RNA samples) were used to detect the TRIM5αhu-specific transcript. (B) Activated PBL from a healthy donor were treated the same as described forTE671 and HeLa cells in panel A. (C) Activated PBL from a healthy donor were infected with HIV-1 NL4-3 at a multiplicity of infection of 0.5 for 48 h. Heat-inactivated (h.i.) virus (NL4-3, preincubated at 56°C for 1 h) was also used as a noninfectious control. The TRIM5αrh-specific transcript was detected by RT-PCR. (D) TE671 cells were pretreated with 0, 20, or 100 U/ml of IFN-α for 4 h and then infected with GFP-expressing N-MLV and HIV-1 vectors. Three days after infection, the GFP-positive cell populations were determined by flow cytometry (mean ± SD). (E) TE671 cells (1.0 × 105) were transfected with 0.2 μg of pNL4-3 or p89.6 by using Fugene6 (Roche). After overnight transfection, the culture supernatants were replaced with fresh growth media containing 0, 20, or 100 U/ml of IFN-α. Two days after transfection, culture supernatants were harvested to determine HIV-1 production. Viral titers were determined in GHOST(3)R3/X4/R5 cells (mean ± SD).
Dominant, constitutively expressed antiviral factors, including TRIM5α and APOBEC3G, are distinguished from the conventional innate immune systems and are often classified as intrinsic immunity factors (1, 6, 25). Here, we showed that TRIM5α mRNA was upregulated by IFN-α treatment, which correlated with the enhanced antiviral activities in rhesus and human cells. These observations indicate that the TRIM5α expression can be orchestrated by the conventional innate immune response. TRIM5α has been thought to be a constitutively expressed factor targeting an incoming retroviral capsid. This notion has led to an image of TRIM5α as a factor which protects a host from an initial retroviral exposure. However, considering the antiviral activity against the late stage of HIV-1 replication and the IFN-α responsiveness, TRIM5α is more likely an antiviral factor which controls both initial retroviral exposure and subsequent viral dissemination in the exposed host. It is plausible that cells constitutively express low levels of TRIM5α to block possible retroviral infection but that they upregulate TRIM5α expression upon viral invasion for better control of retroviral replication. Recently, Chen et al. (8) reported that IFN-α can enhance APOBEC3G-mediated antiviral activity in resting T cells. These observations may offer an attractive possibility that the other antiviral factors of intrinsic immunity, such as Fv1 (5), cross talk with the innate immunity in response to viral infection.
Primate TRIM5α proteins have distinct pre- and postintegration restriction activities against a wide range of retroviruses and lentiviruses, yet generally lack the activity against their own host-specific viruses (20, 23, 29, 31, 37, 41). Intriguingly, even IFN-α-mediated upregulation of TRIM5α failed to restrict SIVMAC and HIV-1 in rhesus and human cell lines, respectively (Fig. 2 and 3). These findings suggest that retroviruses and lentiviruses had to evade TRIM5α-mediated restriction in order to colonize certain species. Lentivirus infection is known to trigger IFN-α production from several cell types, especially from plasmacytoid dendritic cells (17, 42). Considering that TRIM5α can block two distinct steps of retroviral/lentiviral replication and is IFN-α responsive, crossing a species barrier seems to be a very difficult task for a lentivirus; the virus needs to overcome the two TRIM5α-mediated restrictions, which are likely to be upregulated after viral infection, as well as the other potent antiviral factors such as APOBEC3 proteins (33, 40). This is probably the reason for the narrow host ranges of lentiviruses and why no group has managed to adapt HIV-1 to replicate in rhesus monkey cells. Indeed, recently developed simian-tropic HIV-1 strains were generated by introducing partial gag or vif sequences from SIVMAC before serial passages in simian cells (21, 22). Further understanding of the TRIM5αrh-mediated blocks of HIV-1 replication will be necessary to develop a rhesus monkey-tropic HIV-1 without using SIV-derived sequences.
In clinical trials, systemic administration of IFN-α in HIV-1-infected patients showed little or no clinical benefit with notable toxicity, suggesting that HIV-1 is not particularly sensitive to IFN-α treatment in vivo (11, 13, 18, 34). However, in some human cell types and at relatively high concentrations (100 to 10,000 U/ml), type I IFN is known to inhibit HIV-1 replication at early and late steps (12, 35, 36). Among them, the observations that IFN-α (i) inhibits HIV-1 reverse transcription in infected monocytes (4), (ii) reduces the virion infectivity (19, 30), and (iii) induces a defect in a late stage of virus maturation (30) are particularly interesting in that these phenotypes are reminiscent of the TRIM5αrh-mediated antiviral activities in rhesus monkey cells. In this study, we did not use high doses of IFN-α treatment to avoid possible artifacts resulting from the IFN-α-mediated global host translational shutoff. Further careful studies will be necessary to address whether TRIM5αhu is involved in the IFN-α-mediated blocks of HIV-1 replication in certain human cell types.
Recently, we have shown that TRIM5αrh blocks the late phase of HIV-1 replication. When 293T cells were transfected with 1.0 μg of TRIM5αrh-expressing plasmid and 0.1 μg of pNL4-3, TRIM5αrh protein reduced both the progeny virus titer and the p24 concentration in the supernatant approximately 100-fold (31). Under this condition, the virion infectivity (6,302 infectious units [IU]/ng p24) was similar to that of the control (7,490 IU/ng p24). In contrast, when 293T cells were transfected with 0.25 μg of TRIM5αrh plasmid and 0.1 μg of pNL4-3, TRIM5αrh expression reduced the viral titer and the p24 concentration 10- and 2-fold, respectively (31), resulting in reduced virion infectivity (1,460 IU/ng p24). These observations suggest that high levels of TRIM5αrh expression block the late phase of HIV-1 replication predominantly by reducing the number of HIV-1 virions but that modest TRIM5αrh expression blocks HIV-1 production by reducing the virion infectivity as well as the virion numbers. This appears to be the case with endogenous TRIM5αrh in rhesus monkey cells. Disruption of TRIM5αrh mRNA in FrhK4 cells enhanced the HIV-1 titer and the p24 concentration in the supernatants approximately 10- and 2-fold, respectively. Upregulation of TRIM5αrh upon IFN-α treatment reduced both the HIV-1 titer and the p24 release (Fig. 2D), while the virion infectivity was unchanged (1,770 IU/ng, control; 1,760 IU/ng, with IFN-α treatment). Thus, the primary mode of TRIM5αrh-mediated late restriction upon IFN-α treatment is the reduction in the number of HIV-1 virions in the supernatants.
TRIM5α is a member of the large family of TRIM proteins. Like TRIM5α, several TRIM proteins, such as TRIM14, -21, -22, -25, and -34, are upregulated following IFN treatment or viral infection (16, 27, 38). Although their biological properties are poorly characterized, some of them are reported to exhibit antiviral activities or to play a critical role in eliciting host antiviral innate immunity (14, 27). We are currently examining whether these factors have any antiviral effects on the late stages of the retroviral life cycle.
Acknowledgments
We thank J. Stoye for pCIG-N. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: GHOST(3)R3/X4/R5 cells from V. N. KewalRamani and D. R. Littman, pNL4-3 from M. Martin, p89.6 from R. G. Collman, and pSIVMAC1A11 from P. Luciw.
This work was supported by the Mayo Foundation (Y.I.).
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Abudu, A., A. Takaori-Kondo, T. Izumi, K. Shirakawa, M. Kobayashi, A. Sasada, K. Fukunaga, and T. Uchiyama. 2006. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 16:1565-1570. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asaoka, K., K. Ikeda, T. Hishinuma, K. Horie-Inoue, S. Takeda, and S. Inoue. 2005. A retrovirus restriction factor TRIM5alpha is transcriptionally regulated by interferons. Biochem. Biophys. Res. Commun. 338:1950-1956. [DOI] [PubMed] [Google Scholar]
- 4.Baca-Regen, L., N. Heinzinger, M. Stevenson, and H. E. Gendelman. 1994. Alpha interferon-induced antiretroviral activities: restriction of viral nucleic acid synthesis and progeny virion production in human immunodeficiency virus type 1-infected monocytes. J. Virol. 68:7559-7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, S., P. Le Tissier, G. Towers, and J. P. Stoye. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826-829. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 7.Bock, M., K. N. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, K., J. Huang, C. Zhang, S. Huang, G. Nunnari, F. X. Wang, X. Tong, L. Gao, K. Nikisher, and H. Zhang. 2006. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 80:7645-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 10.Collman, R., J. W. Balliet, S. A. Gregory, H. Friedman, D. L. Kolson, N. Nathanson, and A. Srinivasan. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Cruz, E., J. M. Lang, J. Frissen, V. Furner, M. Chateauvert, C. A. Boucher, P. Dowd, J. Stevens, et al. 1995. Zidovudine plus interferon-alpha versus zidovudine alone in HIV-infected symptomatic or asymptomatic persons with CD4+ cell counts > 150 × 10(6)/L: results of the Zidon trial. AIDS 9:1025-1035. [PubMed] [Google Scholar]
- 12.Fernie, B. F., G. Poli, and A. S. Fauci. 1991. Alpha interferon suppresses virion but not soluble human immunodeficiency virus antigen production in chronically infected T-lymphocytic cells. J. Virol. 65:3968-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischl, M. A., D. D. Richman, M. Saag, T. C. Meng, K. E. Squires, J. Holden-Wiltse, and P. M. Meehan. 1997. Safety and antiviral activity of combination therapy with zidovudine, zalcitabine, and two doses of interferon-α2a in patients with HIV: AIDS Clinical Trials Group study 197. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 16:247-253. [DOI] [PubMed] [Google Scholar]
- 14.Gack, M. U., Y. C. Shin, C. H. Joo, T. Urano, C. Liang, L. Sun, O. Takeuchi, S. Akira, Z. Chen, S. Inoue, and J. U. Jung. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916-920. [DOI] [PubMed] [Google Scholar]
- 15.Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703-1706. [DOI] [PubMed] [Google Scholar]
- 16.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurney, K. B., A. D. Colantonio, B. Blom, H. Spits, and C. H. Uittenbogaart. 2004. Endogenous IFN-alpha production by plasmacytoid dendritic cells exerts an antiviral effect on thymic HIV-1 infection. J. Immunol. 173:7269-7276. [DOI] [PubMed] [Google Scholar]
- 18.Haas, D. W., J. Lavelle, J. P. Nadler, S. B. Greenberg, P. Frame, N. Mustafa, M. St. Clair, R. McKinnis, L. Dix, M. Elkins, and J. Rooney. 2000. A randomized trial of interferon alpha therapy for HIV type 1 infection. AIDS Res. Hum. Retrovir. 16:183-190. [DOI] [PubMed] [Google Scholar]
- 19.Hansen, B. D., P. L. Nara, R. K. Maheshwari, G. S. Sidhu, J. G. Bernbaum, D. Hoekzema, M. S. Meltzer, and H. E. Gendelman. 1992. Loss of infectivity by progeny virus from alpha interferon-treated human immunodeficiency virus type 1-infected T cells is associated with defective assembly of envelope gp120. J. Virol. 66:7543-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatziioannou, T., M. Princiotta, M. Piatak, Jr., F. Yuan, F. Zhang, J. D. Lifson, and P. D. Bieniasz. 2006. Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314:95. [DOI] [PubMed] [Google Scholar]
- 22.Kamada, K., T. Igarashi, M. A. Martin, B. Khamsri, K. Hatcho, T. Yamashita, M. Fujita, T. Uchiyama, and A. Adachi. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. USA 103:16959-16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luciw, P. A., K. E. Shaw, R. E. Unger, V. Planelles, M. W. Stout, J. E. Lackner, E. Pratt-Lowe, N. J. Leung, B. Banapour, and M. L. Marthas. 1992. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac). AIDS Res. Hum. Retrovir. 8:395-402. [DOI] [PubMed] [Google Scholar]
- 25.Mangeat, B., and D. Trono. 2005. Lentiviral vectors and antiretroviral intrinsic immunity. Hum. Gene Ther. 16:913-920. [DOI] [PubMed] [Google Scholar]
- 26.Mörner, A., Å. Bjorndal, J. Albert, V. N. KewalRamani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyö, and E. Björling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nisole, S., J. P. Stoye, and A. Saib. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 28.Noser, J. A., G. J. Towers, R. Sakuma, J.-M. Dumont, M. K. L. Collins, and Y. Ikeda. 2006. Cyclosporine increases human immunodeficiency virus type 1 vector transduction of primary mouse cells. J. Virol. 80:7769-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poli, G., J. M. Orenstein, A. Kinter, T. M. Folks, and A. S. Fauci. 1989. Interferon-alpha but not AZT suppresses HIV expression in chronically infected cell lines. Science 244:575-577. [DOI] [PubMed] [Google Scholar]
- 31.Sakuma, R., J. A. Noser, S. Ohmine, and Y. Ikeda. 2007. Rhesus monkey TRIM5alpha restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat. Med. 13:631-635. [DOI] [PubMed] [Google Scholar]
- 32.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 33.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 34.Shepherd, F. A., R. Beaulieu, K. Gelmon, C. A. Thuot, C. Sawka, S. Read, and J. Singer. 1998. Prospective randomized trial of two dose levels of interferon alfa with zidovudine for the treatment of Kaposi's sarcoma associated with human immunodeficiency virus infection: a Canadian HIV Clinical Trials Network study. J. Clin. Oncol. 16:1736-1742. [DOI] [PubMed] [Google Scholar]
- 35.Shirazi, Y., and P. M. Pitha. 1992. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 66:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirazi, Y., and P. M. Pitha. 1998. Interferon downregulates CXCR4 (fusin) gene expression in peripheral blood mononuclear cells. J. Hum. Virol. 1: 69-76. [PubMed] [Google Scholar]
- 37.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 38.Tissot, C., S. A. Taviaux, S. Diriong, and N. Mechti. 1996. Localization of Staf50, a member of the Ring finger family, to 11p15 by fluorescence in situ hybridization. Genomics 34:151-153. [DOI] [PubMed] [Google Scholar]
- 39.Tosi, M. F. 2005. Innate immune responses to infection. J. Allergy Clin. Immunol. 116:241-250. [DOI] [PubMed] [Google Scholar]
- 40.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonezawa, A., R. Morita, A. Takaori-Kondo, N. Kadowaki, T. Kitawaki, T. Hori, and T. Uchiyama. 2003. Natural alpha interferon-producing cells respond to human immunodeficiency virus type 1 with alpha interferon production and maturation into dendritic cells. J. Virol. 77:3777-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]