Abstract
Human immunodeficiency virus type 1 (HIV-1) Gag is expressed as a polyprotein that is cleaved into six proteins by the viral protease in a maturation process that begins during assembly and budding. While processing of the N terminus of Gag is strictly required for virion maturation and infectivity, the necessity for the C-terminal cleavages of Gag is less well defined. To examine the importance of this process, we introduced a series of mutations into the C terminus of Gag that interrupted the cleavage sites that normally produce in the nucleocapsid (NC), spacer 2 (SP2), or p6Gag proteins. Protein analysis showed that all of the mutant constructs produced virions efficiently upon transfection of cells and appropriately processed Gag polyprotein at the nonmutated sites. Mutants that produced a p9NC/SP2 protein exhibited only minor effects on HIV-1 infectivity and replication. In contrast, mutants that produced only the p8SP2/p6 or p15NC/SP2/p6 protein had severe defects in infectivity and replication. To identify the key defective step, we quantified reverse transcription and integration products isolated from infected cells by PCR. All mutants tested produced levels of reverse transcription products either similar to or only somewhat lower than that of wild type. In contrast, mutants that failed to cleave the SP2-p6Gag site produced drastically less provirus than the wild type. Together, our results show that processing of the SP2-p6Gag and not the NC-SP2 cleavage site is important for efficient viral DNA integration during infection in vitro. In turn, this finding suggests an important role for the p9NC/SP2 species in some aspect of integration.
The Gag polyprotein is the major structural component of retroviruses and is the only viral protein strictly required for particle formation by orthoretroviruses (reviewed in references 8 and 13). During assembly and budding, human immunodeficiency virus type 1 (HIV-1) Gag is cleaved by the viral protease, liberating six mature proteins from the Gag polyprotein (13, 45), p15MA, p24CA, SP1 (spacer peptide 1, also known as p2), p7NC, SP2 (also known as p1), and p6Gag. The organization of HIV-1 Gag and its mature cleavage products is presented in Fig. 1. The newly liberated mature Gag proteins then complete the maturation process by rearranging the interior core structure of the virion, observable by electron microscopy as a transition from the doughnut-shaped immature core morphology to a cone-shaped core of the mature virus (13, 15, 42, 45). By this maturation process, the proteins within Gag and Gag-Pol shift roles from parts of a polyprotein that is dedicated to assembly to individual proteins that function in the infection process.
FIG. 1.
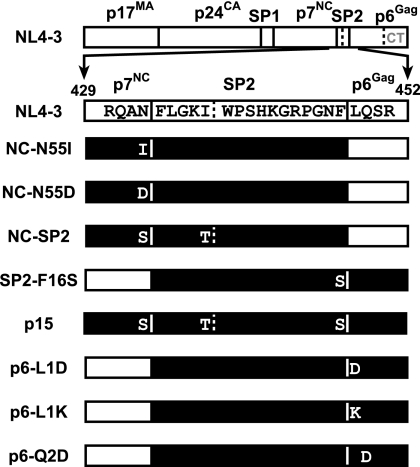
Proviral constructs. Diagrams of the Gag regions of the various pNL4-3 constructs used in this study are presented. The mature proteins within Gag and the minor cleavage sites (denoted by dotted lines in SP2 and p6Gag) are indicated on the NL4-3 wild-type construct map. The region mutated is presented for each mutant with the expected product highlighted in black. A dotted line indicates the C-terminal partial protease cleavage site (after position 36) in p6Gag.
Gag processing occurs in an ordered fashion (37, 41, 48). The initial cleavage of Gag appears to take place between SP1 and NC, liberating MA-CA-SP1 (p40Gag) and NC-SP2-p6Gag (p15NC/SP2/p6) as intermediary products. Processing at this site and at the others in the p40Gag partial cleavage product is essential for proper maturation and virion infectivity (19, 26, 36). In contrast, the importance of processing at the C-terminal sites in Gag, NC-SP2-p6Gag, is not altogether clear. In vitro, processing at the SP2-p6Gag site can be dependent on the RNA-binding sequences in the NC protein (44). Alteration of the SP2-p6Gag site by mutating both the P1 and P1′ positions produced virions with mostly p8SP2/p6 with some p15NC/SP2/p6 that exhibited a replication defect in T cells (54). Mutations altering the two proline residues in the middle of SP2 eliminated viral replication (23). Nevertheless, p9NC/SP2 has been found to be more efficient than p7NC at promoting reverse transcriptase (RT)-associated functions in vitro (3, 27). Still, it is not clear how important the processing of NC, SP2, and p6Gag is for the infection process.
To better understand the requirement for proteolytic processing of the C-terminal p15NC/SP2/p6 region of Gag, we produced a series of NL4-3 constructs with mutations designed to block processing at these cleavage sites. The results showed that while processing of the NC-SP2 site is dispensable for infectivity, cleavage at the SP2-p6Gag site is required for efficient integration by the infecting virus.
MATERIALS AND METHODS
DNA mutagenesis.
The pNL4-3 infectious molecular clone of HIV-1 (1) (GenBank accession no. AF324493) was used for these studies and altered by site-directed mutagenesis using PCR-based methods, either by direct amplification with a mutagenic primer or by two rounds of amplification using the overlap extension procedure (24). The NL4-3 mutants constructed included the following: NC-N55D, with nucleotide 2083 changed from A to G, resulting in an asparagine-to-aspartic acid substitution at NC position 55; NC-N55I, with nucleotide 2084 changed from A to T, resulting in an asparagine-to-isoleucine substitution at NC position 55; NC-SP2, with nucleotide 2084 changed from A to G, resulting in an asparagine-to-serine substitution at NC position 55 that was combined with another mutation, nucleotide 2089 changed from T to C, resulting in an isoleucine-to-threonine substitution at SP2 position 5; SP2-F16S, with nucleotide 2131 changed from T to C, resulting in a phenylalanine-to-serine substitution at SP2 position 16; p15, a combination of the NC-SP2 and SP2-p6Gag mutants; p6-L1D, with nucleotides 2133 to 2136 changed from TCTT to CGAT, resulting in an aspartic acid-to-leucine substitution at p6Gag position 1 and a serine-to-arginine substitution at polymerase (Pol) position 17; p6-L1K, with nucleotides 2133 to 2136 changed from TCTT to CAAA, resulting in an aspartic acid-to-lysine substitution at p6Gag position 1 and an alteration of Pol codons serine 17 to glutamine and serine 18 to threonine; p6-Q2D, with nucleotides 2136 to 2139 changed from TCAG to CGAC, resulting in a glutamine-to-aspartic acid substitution at p6Gag position 2 and an alteration of Pol serine 17 to leucine and serine 18 to arginine. After construction, the regions of DNA that were PCR amplified were sequenced to confirm the mutations and to rule out the possibility of any additional changes introduced during the PCR-mediated mutagenesis process.
Cell culture.
293T human embryonic kidney, HeLa-CD4-LTR-lacZ (HCLZ) (18), JC53BL-13 (also known as TZM-bl) (49), and HOS human osteosarcoma cell line cells were cultured in Dulbecco's modified Eagle's medium, while H9 human T-cell leukemia cells were cultured in RPMI 1640. All media were supplemented with 2 mM l-glutamine, 100 U per ml penicillin, 100 μg per ml streptomycin, and either 10% (vol/vol) fetal bovine serum (293T, JC53BL-13, HCLZ, and H9) or 5% (vol/vol) fetal bovine serum and 5% (vol/vol) calf serum (HOS). All cell culture products were obtained from Invitrogen, Inc. (Carlsbad, CA). Transient transfections of 293T cells were carried out by using calcium phosphate transfection (20) or TransIT-293 (Mirus Bio Corp., Madison, WI). The HIV-1 infection assays using either HCLZ or JC53BL-13 cells were carried out as previously described (18). Briefly, the cells were infected with dilutions of virus and the assay was developed for β-galactosidase activity by staining with 5-bromo-4-chloro-3-indolyl-β-galactoside 48 h postinfection. Infected cells, those staining blue, were observed by light microscopy and counted to score infection events. The ability of viruses to replicate on H9 cells was carried out as previously described (18). Briefly, H9 cells were exposed to 10-fold dilutions of virus and the cultures were monitored periodically for infection by the presence of and increase in RT activity in the culture medium over a 6-week culture period. Samples that were 10 times the background level and greater than 1/100 of the peak level of the undiluted wild-type control were considered positive for infection.
Protein analysis.
Isolation of virions by density centrifugation and immunoblot analysis were performed as previously described (33). Primary goat antiserum against p7NC (goat 77), p24CA (goat 81), and p17MA (goat 83), as well as rabbit antiserum against p6Gag (DJ-30552), were obtained from the AIDS Vaccine Program, NCI-Frederick, Frederick, MD. Mouse antibody against RT, NEA-9304001EA, was obtained from Perkin-Elmer (Wellesley, MA). Proteins were detected by developing blots with either horseradish peroxidase-conjugated anti-goat, anti-rabbit (both from Biochain Institute, Hayward, CA), or anti-mouse (Life Technologies, Inc., Gaithersburg, MD) secondary antibody and the Immun-star horseradish peroxidase substrate kit (Bio-Rad, Hercules, CA) on LumiFilm (Roche Applied Science, Indianapolis, IN). Reversed-phase high-pressure liquid chromatography (HPLC) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry were carried out as previously described (33).
Real-time PCR of reverse transcription products and proviral DNA.
Assays for reverse transcription and integration products within HOS cells after infection were carried out as previously described (25). Briefly, wild-type and mutant viruses pseudotyped with the vesicular stomatitis virus G surface glycoprotein (VSV-G) were generated by calcium phosphate cotransfection of 293T cells followed by a repetitive washing procedure to remove plasmid DNA. The virus stocks were then used to infect HOS cells, and total DNA was isolated from the cells 24 h postinfection. For HIV-1 Env-mediated infections, virus stocks were prepared by calcium phosphate transfection and then treated with DNase to remove carryover plasmid from transfections: supernatants were exposed to 10 U/ml of DNase (Sigma-Aldrich, St. Louis, MO) in 4 mM MgCl2 at 37°C for 1 h. H9 cells (3 × 106) were infected in a 24-well plate using Virofect R/L Magnetofection reagent (OZ Biosciences, Inc., Marseilles, France) according to the manufacturer's instructions using the recommended magnetic plate (OZ Biosciences). Analyses of early (strong stop; R-U5) and late reverse transcription DNA products (plus-strand transfer; R-5′ untranslated region [UTR]) and 2-LTR junction products were carried out as previously described (5). The proviral DNA assay (Alu-LTR) was performed as described elsewhere (46). Negative control infections with heat-inactivated (65°C for 20 min) aliquots of the virus stocks were used to assess the levels of carryover plasmid DNA from the transfection. The quantities of carryover plasmid DNA were typically less than 0.1% of that obtained from live virus.
RESULTS
To examine the importance of the processing of the p15NC/SP2/p6 precursor, we produced a series of mutant constructs in full-length pNL4-3 designed to eliminate Gag proteolytic processing in the C terminus of p7NC and the N terminus of SP2 to generate viruses that contain p9NC/SP2, p8SP2/p6, or p15NC/SP2/p6 (Fig. 1). One potential complication to this approach is that, in addition to these primary cleavage sites, there are minor secondary sites in this region of Gag (Fig. 1) that can be cleaved in mature virions (21, 22, 33). One of these minor cleavages occurs after SP2 amino acid 5, potentially cleaving within a p9NC/SP2 or p15NC/SP2/p6 species produced by our cleavage site mutants. To eliminate this possibility, we mutated the P1 amino acid of this site (SP2 amino acid 5, Gag position 437) (Fig. 1) in constructs that had mutations either at the NC-SP2 site (NC-SP2 in Fig. 1) or at both the NC-SP2 and SP2-p6Gag sites (p15 in Fig. 1).
A complication in genetically blocking protease cleavage is the difficulty in predicting what sequence substitutions at the protease cleavage site will eliminate protease recognition in the viral mutants. General selection rules based on protease crystal structure and studies with peptides suggest that insertion of hydrophilic and charged residues at the cleavage site interrupts processing (39, 52, 53). Nevertheless, our experience has shown that some of the mutations suggested by these rules do not always prevent cleavage when placed in the context of the virus. Using these rules as well as our previous observations, we introduced a series of mutations into the C terminus of Gag. Contrary to expectations, some of the mutated cleavage sites were either completely or mostly cleaved in virions despite a poor predicted substrate site: preliminary immunoblotting with p7NC antiserum showed that NC-SP2 site mutants containing isoleucine, histidine, and lysine in the P1 position were processed by protease, while those containing serine and aspartic acid were not cleaved (data not shown). Also, an initial p6Gag immunoblot assay revealed that a serine in the P1′ position of SP2-p6Gag was also processed, while aspartic acid and lysine substitutions were not cleaved.
The NL4-3-based constructs selected for further study are displayed in Fig. 1. All of the NC and SP2 mutants were constructed so that the Gag mutations did not alter the overlapping Pol reading frame or alter the frameshift slippery site (which overlaps the wobble codon of the asparagine at P1 of the NC-SP2 site). The p6Gag mutants had changes in the Pol frame at the beginning of the p6Pol protein (also referred to as the preprotease or transframe protein). These secondary mutations did not alter processing of Gag at other sites or reverse transcriptase incorporation (presented below).
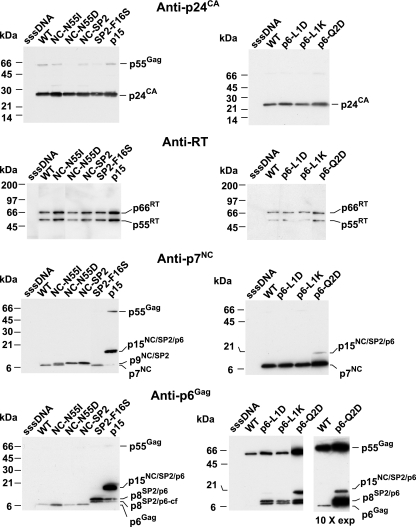
To study the mutant virions, preparations were produced by transfecting each molecular clone into 293T cells and then isolating virions by density centrifugation for protein analyses. To examine any impact the mutations might have on virus production and processing of the N-terminal Gag proteins (i.e., those in the p40Gag partial cleavage product), equal amounts of virion preparations (by volume) were analyzed by immunoblotting with p24CA antiserum. The results showed that the mutants produced nearly equivalent levels of virions, indicating that they exhibit no overt assembly defects. The virion samples also did not contain appreciable amounts of p55Gag or partially processed products; thus, the mutations in the C terminus of Gag did not alter the overall processing of p24CA or p17MA. Since changes to the middle of SP2 can alter Gag processing and Gag-Pol incorporation (23), we detected the Pol protein product in the samples by stripping the blot and exposing it to an antibody against RT. The blot showed that the relative intensities of the RT signals present in the mutant samples compared to those of Gag were similar to the wild-type sample (Fig. 2). Hence, the amount of Pol incorporation does not appear to be significantly altered by these mutations.
FIG. 2.
Immunoblots of mutant virion preparations. p24CA, RT, p7NC, and p6Gag immunoblots of equal volumes of virion preparations are presented. Samples are identified above the respective lanes. sssDNA, a virus preparation produced from transfected, sheared salmon sperm DNA as a mock control. Molecular masses, as calculated by relative mobilities, and identities of bands are indicated at the margins of the blots. Due to the poor retention of wild-type p6Gag, a 10-fold-longer exposure is provided to the right of the second p6Gag immunoblot to show the migration of wild-type p6Gag.
A p7NC immunoblot analysis of the virions revealed that mutating the NC-SP2 cleavage site had various effects on processing. The NC-N55I sample contained a band that was similar to the wild type, p7NC, as well as a readily detectable minor band that migrated higher in the gel, likely p9NC+SP2. Therefore, processing apparently still occurred, though inefficiently, at the NC-SP2 site in this mutant. Only one band was present in the NC-N55D and NC-SP2 virion samples, migrating at a position higher than the wild-type band, consistent with the presence of the p9NC/SP2 species that would be produced by blocking the NC-SP2 cleavage site. As expected, the SP2-F16S, p6-L1D, p6-L1K, and p6-Q2D mutants contained a normal-sized p7NC band (Fig. 2), as the NC-SP2 site was not mutated in these constructs. The p15 mutant sample contained a band that migrated much higher, consistent with the expected p15NC/SP2/p6 product.
To examine the status of p6Gag in the mutants, the blot was stripped and exposed to p6Gag antiserum. This revealed that the samples of the NC-N55I, NC-N55D, and NC-SP2 mutants contained a wild-type-sized p6Gag band. (The wild-type p6 bands are less intense due to the nature of p6Gag, a protein that is relatively hydrophilic and does not efficiently transfer from the gel onto polyvinylidene difluoride membranes [33].) The SP2-F16S, p6-L1D, p6-L1K, and p6-Q2D mutant samples contained two higher-migrating bands on the blots. Since the C terminus of p6Gag is alternatively processed after p6Gag residue 36 (Gag position 483) (Fig. 1) (21, 22, 33), the former is likely an SP2-p6Gag product while the latter is an apparent SP2-p6Gag product missing its extreme C terminus. The p15 mutant contained a much higher migrating band that coincided with the band in the p7NC immunoblot, confirming the presence of both p7NC and p6Gag in this species. In addition to the p15NC/SP2/p6 product, there were weaker bands that corresponded to those observed for the SP2-p6Gag mutants, indicating that there was a small amount of processing at the NC-SP2 site. Together, these results indicate that we have produced a functional series of cleavage site mutants.
HPLC and mass spectrometry analysis.
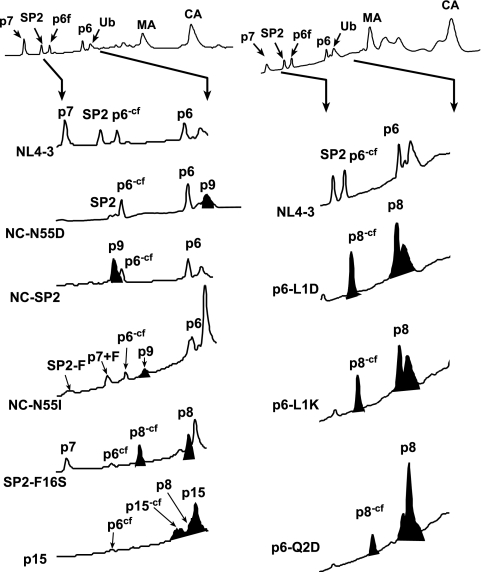
Considering the relatively small size of the mutant proteins and the potential for minor cleavages in this region, we confirmed the identities of the p7NC, SP2, and p6Gag products by isolating them from virion preparations with reversed-phase HPLC and identified them with a subsequent MALDI-TOF mass spectrometry analysis (Fig. 3; Table 1). These data generally confirmed the immunoblot data (Fig. 2). Nearly all of the NC in the NC-N55D mutant was present as a p9NC/SP2 product (Fig. 3; Table 1), though these virions did contain a small amount of p7NC (less than approximately 5%, based on relative absorbance at 206 nm) (Fig. 3; Table 1). (There is also another peak in the HPLC that elutes at a similar time as the p9NC/SP2 peak that corresponds to ubiquitin [Ub] [Fig. 3], which is incorporated into virions [34, 35, 43].) The NC-SP2 mutant contained only p9NC/SP2 with no detectable p7NC (Fig. 3; Table 1). Thus, NC-SP2 processing appears to be completely blocked in this mutant. NC-N55I virions displayed an intermediate phenotype (Fig. 3), containing a mixture of p7NC and p9NC/SP2 (approximately 40% based on relative absorbance) (Fig. 3; Table 1), in agreement with our prior blotting data (Fig. 2). Mass spectrometry revealed that the p7NC in this mutant virus, while appearing normal-sized by immunoblotting, contained additional mass that is consistent with the addition of a phenylalanine from the N terminus of SP2. Likewise, the SP2 protein was missing mass that would account for this residue. Thus, the mutation appears to have altered processing at this site in two ways, either moving the cleavage site over one residue, making p7NC+F, or preventing cleavage entirely (Fig. 3). This underscores the important of biochemical analyses of cleavage site mutants.
FIG. 3.
HPLC analysis of mutants. The complete HPLC chromatogram for wild-type NL4-3 virions is shown at the top of the figure with the region that is presented for the mutants indicated by arrows. The identities of the Gag proteins, determined by Coomassie brilliant blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotting, protein sequencing, and mass spectrometry, are identified above each respective peak. The mutant proteins as identified by MALDI-TOF mass spectrometry are shaded in black. The p6Gag-containing species that are missing the C-terminal 16 amino acids due to cleavage after residue 36 are indicated by a “-cf” notation, e.g., p6-cf, in the identifying superscript. The C-terminal fragment itself was labeled p6cf when observed.
TABLE 1.
Mass spectrometry data for cleavage site products
| Virus/protein detected | Observed mass (Da) | % Difference from theoretical |
|---|---|---|
| NL4-3/NC | 6,352.1 | 0.01 |
| NL4-3/SP2 | 1,841.9 | 0.04 |
| NL4-3/p6Gag | 5,803.6 | 0.06 |
| NL4-3/p6Gag-cf | 4,135.7 | 0.04 |
| NC-N55D/p9NC/SP2 | 8,178.2 | 0.03 |
| NC-N55D/p7NC | 6,354.7 | 0.05 |
| NC-N55I/p9NC/SP2 | 8,199.5 | 0.3 |
| NC-N55I/p7NC+F | 6,497.4 | 0.3 |
| NC-N55I/SP2-F | 1,693.93 | 0.1 |
| NC-SP/p9NC/SP2 | 8,140.5 | 0.06 |
| SP2-F16S/p8SP2/p6 | 7,577.5 | 0.09 |
| SP2-F16S/p8SP2/p6-cf | 5,927.5 | 0.1 |
| p6-L1D/p8SP2/p6 | 7,640.4 | 0.1 |
| p6-L1D/p8SP2/p6-cf | 5,977.2 | 0.1 |
| p6-L1K/p8SP2/p6 | 7,653.5 | 0.1 |
| p6-L1K/p8SP2/p6-cf | 6,002.9 | 0.02 |
| p6-Q2D/p8SP2/p6 | 7,617.39 | 0.05 |
| p6-Q2D/p8SP2/p6-cf | 5,975.0 | 0.02 |
| p15 | 13,908.4 | 0.3 |
As expected, the SP2-p6Gag cleavage site mutants, SP2-F16S, p6-L1D, p6-L1K, and p6-Q2D, contained only the p8SP2/p6 product with the expected mass (Table 1), though we did observe a truncated form (Fig. 3, p8-cf) that matched the expected size for a p8SP2/p6 product with a C-terminal cleavage at p6 residue 36. This species was previously observed in our immunoblot data (Fig. 2). Additionally, we also could detect the C-terminal 16-amino-acid protein of p6Gag in this sample (Fig. 3, cf).
Analysis of the p15 mutant virions detected the p15NC/SP2/p6 species (Fig. 3; Table 1) and two proteins with masses consistent with the expected N- and C-terminal fragments for a cleavage in the C-terminal tail (Fig. 3, p15-cf and cf, respectively). In addition, we did find a slight amount of p8SP2/p6 (Fig. 3). Together, these data confirm the immunoblot data and further demonstrate that there was no unexpected processing at any minor sites.
Mutant infectivity and replication.
The ability of the HIV-1 mutants examined above to carry out a single round of infection was tested by using JC53bl-13 cells in an LTR-lacZ Tat complementation assay (18). Their ability to perform multiple rounds of replication in H9 cells was determined using an endpoint titer method. The results (Table 2) showed that the NC mutants NC-N55D, NC-N55I, and NC-SP2 produced single-round titers and H9 endpoint tissue culture infective doses that were only somewhat lower than those containing wild-type NC. Thus, even though both the NC-N55D and NC-SP2 mutants contain mostly uncleaved p9NC+SP2, they had only modest reductions in the infectivity and replication assays. Also, the NC-N55I virions, which contain partially or miscleaved p7NC, maintained high relative infectivities. These results show that the cleavage of p7NC from SP2 is not essential for replication.
TABLE 2.
Infectivity and replication of mutants
| Virus | Single-round titera | Relative titerb | H9 replicationc |
|---|---|---|---|
| sssDNA | 4 | 2 × 10−6 | <1 |
| NL4-3 | 3.6 × 107 | 1 | 106 |
| NC-N55D | 6.9 × 107 | 2 | 105 |
| NC-N55I | 1.1 × 107d | 0.3 | 104 |
| NC-SP2 | 3.9 × 107 | 1 | 106 |
| SP2-F16S | 4.0 × 105 | 0.01 | 1 |
| P6-L1D | 7.5 × 104 | 2 × 10−3 | 1 |
| P6-L1K | 2.9 × 105 | 8 × 10−3 | <1 |
| P6-Q2D | 5.9 × 104 | 2 × 10−3 | <1 |
| p15 | 8.1 × 104 | 2 × 10−3 | <1 |
Single-round infectivity was based on β-galactosidase activity and is reported as blue cell-forming units per ml.
Relative to wild-type titer.
Tissue culture infective dose per milliliter.
Value normalized from HCLZ data.
In contrast, the single-round infectivities of the SP2-p6Gag cleavage site mutants, SP2-F16S, p6-L1D, p6-L1K, and p6-Q2D, were at least 2 logs lower than wild-type levels. The effect on replication was even more dramatic: the titers for these mutants were ≤10−6 of the titer of the wild type. Similar to the SP2-p6Gag cleavage site mutants, the infectivity and replication competence of the p15 mutant were also reduced to levels near those of the SP2-p6Gag mutants. Therefore, these data indicate that cleavage of SP2 from p6Gag is important in HIV-1 replication.
Analysis of infection products.
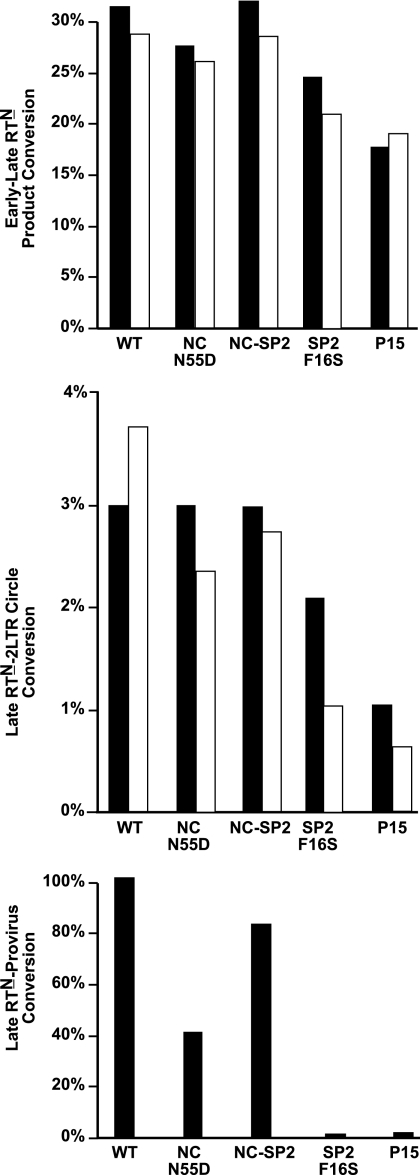
To determine the replication block for the SP2-p6Gag and p15 mutants, we examined the ability of VSV-G-pseudotyped mutants (NC-N55D, NC-SP2, SP2-F16S, and p15) to carry out reverse transcription and integration steps in HOS cells by detecting key DNA products inside the cell with real-time PCR 24 h postinfection. Processivity of reverse transcription, measured by conversion of early (strong stop; R-U5) to late (plus-strand transfer; R-5′ untranslated region [UTR]) reverse transcription products (RTN), was only slightly reduced for the NC-ND, SP2-F16S, and p15 mutants when compared to the wild-type samples (a representative experiment is shown in Fig. 4, early-late RTN; full data sets are presented in Tables S1 to S3 of the supplemental material). The NC-SP2 mutant produced reverse transcriptase products essentially at a wild-type efficiency, consistent with its infectivity phenotype (Fig. 4; see also Tables S1 to S3). Therefore, a gross failure in carrying out reverse transcription was not the major defect in the SP2-F16S and p15 mutants.
FIG. 4.
Reverse transcription and integration products assayed by real-time PCR. Representative results of a quantitative real-time PCR analysis are presented as percent conversion between two representative steps in the infection process. Results for VSV-G-mediated infection are presented as black bars, and data from HIV-1 Env-mediated infections are in white. Parameters presented are indicated next to each graph. Typical samples analyzed contained extract from about 1.1 × 104 cells. For the wild type, the target quantities detected were 3.2 × 105 copies of R-U5, 1.33 × 105 copies of R-5′UTR, 3.3 × 104 copies of provirus, and 4.8 × 103 copies of 2-LTR circles. The amounts of provirus in H9 cells infected with HIV-1 Env-containing virus were below the level of detection and thus were omitted. Full data sets of experiments are presented in Tables S1 to S6 in the supplemental material.
Another aspect assayed was the production of 2-LTR circles (representative data shown in Fig. 4, late RTN-2-LTR circles). A small amount of the newly reverse transcribed DNA forms 2-LTR circles as a dead-end by-product that is produced during infection. This is commonly used as an indicator that plus-strand synthesis was completed and that the resulting viral DNA was exposed to nuclear enzymes. The PCR analysis showed that 2-LTR formation for the SP2-F16S and the p15 mutants was one-half to one-third of the wild-type value and that of the NC cleavage site mutants (Fig. 4; see also Tables S1 to S3 in the supplemental material). While this is somewhat reduced, the differences do not account for the greater than 6-log difference in replication observed between these viruses.
Unlike the two other steps, integration, as measured by an Alu-LTR-based PCR assay, was drastically reduced in the cells infected with the SP2-F16S and p15 mutants (representative data shown in Fig. 4, late RTN-provirus). The analysis showed that the conversion of late reverse transcription product (R-5′UTR) to provirus (Alu-LTR) was greatly reduced for these mutants, to less than 3% of the wild-type levels (Fig. 4; see also Table S1 in the supplemental material). Thus, the primary defect in the SP2-F16S and p15 mutants appears to be a severe defect in integration.
Even though both VSV-G- and HIV-1 Env-mediated infection of cells with NL4-3 produces reverse transcription products at the same efficiency in unperturbed cells (47), VSV-G pseudotyping can overcome various blocks to HIV reverse transcription during infection that are induced by genetic mutants and cellular inhibitors (2, 40). Thus, the use of VSV-G could possibly mask a reverse transcription defect in our mutants. To address this possibility, we infected H9 cells using our mutants without VSV-G pseudotyping (NC-N55D, NC-SP2, SP2-F16S, p6-Q2D, and p15), thereby relying on CD4/gp120-mediated infection. The results essentially mirrored those obtained from the VSV-G/HOS cell experiments (Fig. 4; see also Tables S4 to S6 in the supplemental material): the mutants had only minor decreases in reverse transcription (early-late RTN) and a modest defect in nuclear accessibility (late RTN-2 LTR circle). Neither defect accounts for the gross defect in replication. The amounts of provirus for these experiments were below the level of detection due to the reduced number of infection events with the HIV-1 Env-mediated infections. Overall, these results demonstrated that the mutants carry out reverse transcription relatively efficiently. Thus, the defect in infection is at a later step, immediately prior to or during integration.
DISCUSSION
Our results show that removal of SP2 from p7NC is not required for efficient HIV-1 replication. In contrast, processing at the SP2-p6Gag cleavage site is required for efficient HIV-1 replication. These latter results are consistent with a replication defect observed by Yu et al. with a different SP2-p6Gag cleavage mutant (54). Our real-time PCR data revealed that the primary defect caused by blocking SP2-p6Gag processing was at or just before the integration step. It is unclear how blocking the SP2-p6Gag site could affect integration. The loss of free SP2 does not appear to play a direct role, as our p9NC/SP2 mutants were infectious despite containing no detectable SP2. The Pol polyproteins appear to be incorporated into virions at near-wild-type levels (Fig. 2; immunoblotting results for protease are not shown), and so it is unlikely that there is a simple IN-related explanation. Therefore, the production of either p6Gag or p9NC-SP2, both of which are not produced by the SP2-p6Gag or p15 mutants, could be required for integration. The p6Gag protein has not been implicated in any postentry/infection events and is located outside the mature core in virions; thus, it is unlikely to interact with the RNA or integrase (50). In contrast, the NC protein has already been implicated in promoting efficient integration (5, 16, 17, 46). Thus, it is possible that a block in the production of p9NC-SP2 is responsible for the defect in integration. Similar to the results presented here, our previous results from genetic studies support some role for NC at or just before the integration step: HIV-1 mutants with CCHC NC zinc finger substitutions to other nonretroviral motifs, CCCC and CCHH, maintain genomic RNA incorporation and exhibit only somewhat reduced levels of reverse transcription, yet have a severe block in integration (5, 16, 17, 46).
How NC, potentially as a p9NC/SP2 species, could be required for integration is not clear. NC could be acting as a somewhat labile component of the integration complex or as a protective factor for the cDNA ends, presumably by binding to them (5, 17, 46). In vitro, p7NC and especially p9NC/SP2 have been found to increase the efficiency of concerted integration (7, 14, 38). Interestingly, p9NC/SP2 can condense double-stranded DNA into dense protein/DNA complexes much more efficiently than either p7NC or p15NC/SP2/p6 (32), suggesting that p9NC/SP2 might specifically associate with the viral cDNA. Alternatively, p9NC/SP2 could play a role in nuclear localization of the preintegration complex, although our experiments were conducted on dividing cell cultures. Despite all of these indirect supporting data, neither NC nor p9NC/SP2, also called NCp7 or NC[1-72] (11), is routinely observed in isolated preintegration complexes (30). Detectable amounts of p9NC/SP2 do not appear to be present in HIV-1 cores (50): nearly all of the NC appears to be p7NC with a very minute amount of p15NC/SP2/p6. Also, in our years of biochemical analyses of wild-type HIV-1 virions, we have not found much evidence for p9NC/SP2 in virions (21) (data not shown). However, it is possible that only a small amount, two to four molecules per virion bound to the cDNA ends, might be present, levels below the sensitivity of our past and current protein analyses.
Alternatively, a p9NC/SP2 species could act transiently at the assembly/maturation step by assisting the correct formation of the mature virus, perhaps placing IN in the correct position within the viral core. Recent reports of in vitro experiments have implicated p9NC/SP2 in the maturation process (31, 32). Theoretically, p9NC/SP2 would not have to be present in the core to assist the integration process in this way.
From our data and other processing site mutation studies (19, 26, 42, 51), it appears that the only Gag proteolyic cleavage site that is not strictly required for replication in vitro is the NC-SP2 junction. Unlike NC, the proteins in the N-terminal region of Gag play mostly structural roles. Therefore, having additional sequences attached to them due to incomplete processing likely causes defects in their fairly constrained protein-protein interactions. However, NC primarily functions in RNA binding and chaperone activity (4, 10, 28), roles that apparently can tolerate additional sequences attached to p7NC.
One question that remains unanswered is the following: why is p7NC produced if p9NC/SP2 is sufficient for replication? It is important to consider that in cell culture with limited rounds of replication, we may not detect minor replication inefficiencies that would apply a selective pressure for the maintenance of this site over the many cycles of replication that occur in an infected individual (9). Indeed, sequential passaging of the NC-N55D and NC-SP2 mutants results in a restoration of the NC-SP2 site within four passages (4 weeks) (data not shown), supporting this hypothesis. Furthermore, some protease inhibitor-resistant viruses isolated from patients have sequence changes at both the NC-SP2 and the SP2-p6Gag sites that appear to restore normal levels of processing by the mutant protease (6, 12, 29, 55). Thus, processing at both sites appears to be important for long-term viral fitness in vivo.
Overall, our findings are consistent with a role for NC in promoting the integration process, possibly as a p9NC/SP2 species. The potential involvement of NC in integration suggests further studies to clarify its role in this process, e.g., either a direct or indirect interaction of p9NC/SP2 with the preintegration complex or its providing an organizing function during the maturation phase of assembly.
Supplementary Material
Acknowledgments
We thank Laurie Queen and Teresa Shatzer for technical assistance and Lou Henderson for the inspiration to examine this topic.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 18 July 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, A., S. Koenig, H. E. Gendelman, D. Daugherty, S. Gattoni-Celli, A. S. Fauci, and M. A. Martin. 1987. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 61:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bampi, C., A. Bibillo, M. Wendeler, G. Divita, R. J. Gorelick, S. F. Le Grice, and J. L. Darlix. 2006. Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J. Biol. Chem. 281:11736-11743. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 5.Buckman, J. S., W. J. Bosche, and R. J. Gorelick. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrillo, A., K. D. Stewart, H. L. Sham, D. W. Norbeck, W. E. Kohlbrenner, J. M. Leonard, D. J. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J., S. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Press, Plainview, NY. [PubMed]
- 9.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 10.Cruceanu, M., M. A. Urbaneja, C. V. Hixson, D. G. Johnson, S. A. Datta, M. J. Fivash, A. G. Stephen, R. J. Fisher, R. J. Gorelick, J. R. Casas-Finet, A. Rein, I. Rouzina, and M. C. Williams. 2006. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 34:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rocquigny, H., D. Ficheux, C. Gabus, M. C. Fournie-Zaluski, J. L. Darlix, and B. P. Roques. 1991. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 180:1010-1018. [DOI] [PubMed] [Google Scholar]
- 12.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 14.Gao, K., R. J. Gorelick, D. G. Johnson, and F. Bushman. 2003. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol. 77:1598-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelderblom, H. 1997. Fine structure of HIV and SIV. Los Alamos National Laboratory, Los Alamos, NM.
- 16.Gorelick, R. J., L. O. Arthur, A. Rein, L. E. Henderson, and S. Oroszlan. October 1997. Design and construction of noninfectious human retroviral mutants deficient in genomic RNA. U.S. patent 5,674,720.
- 17.Gorelick, R. J., T. D. Gagliardi, W. J. Bosche, T. A. Wiltrout, L. V. Coren, D. J. Chabot, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92-104. [DOI] [PubMed] [Google Scholar]
- 18.Gorelick, R. J., S. M. Nigida, J. W. Bess, Jr., L. E. Henderson, L. O. Arthur, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 21.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson, L. E., T. D. Copeland, R. C. Sowder, A. M. Schultz, and S. Oroszlan. 1988. Analysis of proteins and peptides purified from sucrose gradient banded HTLV-III, p. 135-147. In D. Bolognesi (ed.), Human retroviruses, cancer and AIDS: approaches to prevention and therapy. Alan R. Liss, Inc., New York, NY.
- 23.Hill, M. K., M. Shehu-Xhilaga, S. M. Crowe, and J. Mak. 2002. Proline residues within spacer peptide P1 are important for human immunodeficiency virus type 1 infectivity, protein processing, and genomic RNA dimer stability. J. Virol. 76:11245-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 25.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 99:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krausslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lener, D., V. Tanchou, B. P. Roques, S. F. Le Grice, and J. L. Darlix. 1998. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 273:33781-33786. [DOI] [PubMed] [Google Scholar]
- 28.Levin, J. G., J. Guo, I. Rouzina, and K. Musier-Forsyth. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217-286. [DOI] [PubMed] [Google Scholar]
- 29.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and Gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirambeau, G., S. Lyonnais, D. Coulaud, L. Hameau, S. Lafosse, J. Jeusset, I. Borde, M. Reboud-Ravaux, T. Restle, R. J. Gorelick, and E. Le Cam. HIV-1 protease and reverse transcriptase control the architecture of their nucleocapsid partner. PLoS One, in press. [DOI] [PMC free article] [PubMed]
- 32.Mirambeau, G., S. Lyonnais, D. Coulaud, L. Hameau, S. Lafosse, J. Jeusset, A. Justome, E. Delain, R. J. Gorelick, and E. Le Cam. 2006. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 364:496-511. [DOI] [PubMed] [Google Scholar]
- 33.Ott, D. E., E. N. Chertova, L. K. Busch, L. V. Coren, T. D. Gagliardi, and D. G. Johnson. 1999. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6Gag protein produces a mutant that fails to package its envelope protein. J. Virol. 73:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 35.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, I. Sowder, R. C. Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettit, S. C., N. Sheng, R. Tritch, S. Erickson-Viitanen, and R. Swanstrom. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 436:15-25. [DOI] [PubMed] [Google Scholar]
- 38.Poljak, L., S. M. Batson, D. Ficheux, B. P. Roques, J. L. Darlix, and E. Kas. 2003. Analysis of NCp7-dependent activation of HIV-1 cDNA integration and its conservation among retroviral nucleocapsid proteins. J. Mol. Biol. 329:411-421. [DOI] [PubMed] [Google Scholar]
- 39.Poorman, R. A., A. G. Tomasselli, R. L. Heinrikson, and F. J. Kezdy. 1991. A cumulative specificity model for proteases from human immunodeficiency virus types 1 and 2, inferred from statistical analysis of an extended substrate data base. J. Biol. Chem. 266:14554-14561. [PubMed] [Google Scholar]
- 40.Qi, M., and C. Aiken. 2007. Selective restriction of Nef-defective human immunodeficiency virus type 1 by a proteasome-dependent mechanism. J. Virol. 81:1534-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasnick, D. 1997. Kinetics analysis of consecutive HIV proteolytic cleavages of the Gag- Pol polyprotein. J. Biol. Chem. 272:6348-6353. [PubMed] [Google Scholar]
- 42.Ross, E. K., T. R. Fuerst, J. M. Orenstein, T. O'Neill, M. A. Martin, and S. Venkatesan. 1991. Maturation of human immunodeficiency virus particles assembled from the gag precursor protein requires in situ processing by gag-pol protease. AIDS Res. Hum. Retrovir. 7:475-483. [DOI] [PubMed] [Google Scholar]
- 43.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheng, N., S. C. Pettit, R. J. Tritch, D. H. Ozturk, M. M. Rayner, R. Swanstrom, and S. Erickson-Viitanen. 1997. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 71:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- 46.Thomas, J. A., T. D. Gagliardi, W. G. Alvord, M. Lubomirski, W. J. Bosche, and R. J. Gorelick. 2006. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology 353:41-51. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, J. A., D. E. Ott, and R. J. Gorelick. 2007. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J. Virol. 81:4367-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tozser, J., I. Blaha, T. D. Copeland, E. M. Wondrak, and S. Oroszlan. 1991. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 281:77-80. [DOI] [PubMed] [Google Scholar]
- 49.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welker, R., H. Hohenberg, U. Tessmer, C. Huckhagel, and H. G. Krausslich. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 74:1168-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:453-585. [DOI] [PubMed] [Google Scholar]
- 53.You, L., D. Garwicz, and T. Rognvaldsson. 2005. Comprehensive bioinformatic analysis of the specificity of human immunodeficiency virus type 1 protease. J. Virol. 79:12477-12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, X. F., Z. Matsuda, Q. C. Yu, T. H. Lee, and M. Essex. 1995. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J. Gen. Virol. 76:3171-3179. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.