Abstract
Human papillomavirus type 16 (HPV16) and other high-risk HPVs are etiologically linked to the development of cervical carcinomas and contribute to a number of other tumors of the anogenital tract, as well as oral cancers. The high-risk HPV E6 and E7 oncoproteins are consistently expressed in cervical cancer cells and are necessary for the induction and maintenance of the transformed phenotype. An important aspect of HPV16 E7's oncogenic activities is destabilization of the retinoblastoma tumor suppressor (pRB) through a ubiquitin/proteasome-dependent mechanism, although the exact molecular mechanism is unknown. Here, we report that HPV16 E7 is associated with an enzymatically active cullin 2 ubiquitin ligase complex and that the HPV16 E7/pRB complex contains cullin 2. Depletion of cullin 2 by RNA interference causes increased steady-state levels and stability of pRB in HPV16 E7-expressing cells, and ectopic expression of HPV16 E7 and the cullin 2 complex leads to pRB ubiquitination in vivo. Hence, we propose that the HPV16 E7-associated cullin 2 ubiquitin ligase complex contributes to aberrant degradation of the pRB tumor suppressor in HPV16 E7-expressing cells.
Human papillomaviruses (HPVs) are small DNA viruses that infect epithelial cells. Approximately 200 HPV types have been identified. The ∼30 HPVs that are associated with mucosal infections are classified into high- and low-risk groups according to the relative malignant potential of the lesions that they cause. Low-risk HPVs, such as HPV type 6 (HPV6) and HPV11, cause benign genital warts, whereas high-risk HPVs, such as HPV16 and HPV18, cause premalignant squamous intraepithelial neoplasia. Infections with high-risk HPVs are associated with more than 95% of cervical cancer cases, the second most common cancer affecting young women worldwide (reviewed in reference 69). A number of other anogenital tract carcinomas, as well as approximately 20% of oral cancers, are caused by high-risk HPV infections (23). Hence, these viruses have been classified as carcinogenic agents in humans (33). Integration of viral DNA into the host genome is a consistent hallmark of malignant progression and leads to persistent, dysregulated expression of the high-risk HPV E6 and E7 oncoproteins, which is necessary for the induction and maintenance of the transformed phenotype (reviewed in reference 44).
HPV16 E7 is sufficient for oncogenic transformation of established rodent fibroblasts, cooperates with the ras oncogene to transform primary baby rat kidney cells, and in combination with E6 facilitates immortalization of primary human foreskin keratinocytes. High-risk HPV E7 proteins also dysregulate apoptosis, inhibit cell cycle arrest in response to differentiation or DNA damage, and contribute to genomic instability through induction of centrosome duplication errors, as well as other mechanisms (reviewed in reference 44). Expression of HPV16 E7 in basal epithelial cells, in conjunction with low-dose estrogen treatment, was sufficient for cervical cancer development in a transgenic-mouse model (55).
The major transforming activities of high-risk HPV E6 and E7 proteins have been linked to inactivation of the p53 and retinoblastoma (pRB) tumor suppressors, respectively (reviewed in reference 44). Mutational inactivation of these two tumor suppressor pathways is an almost universal hallmark of human solid-tumor development (reviewed in reference 29). The pRB tumor suppressor and the related p107 and p130 “pocket proteins,” associate with E2F transcription factors to inhibit S-phase entry and have also been implicated in regulating other cellular processes, including apoptosis and differentiation (reviewed in reference 15). The paradigm of pRB inactivation by viral oncoproteins was initially established by studies with adenovirus (Ad) E1A, simian virus 40 (SV40) large tumor antigen, and HPV16 E7 (17, 19, 63). Each of these viral oncoproteins contains conserved sequence domains, including the LXCXE motif, which constitutes the core pRB-binding site of HPV16 E7 (45, 62). HPV16 E7 binding causes disruption of pRB/E2F complexes (13, 65), thereby subverting G1/S cell cycle control. Unlike Ad E1A and SV40 large tumor antigen, which inactivate pRB through stoichiometric association, HPV16 E7 causes ubiquitin-dependent proteasomal pRB degradation (9, 11, 34). The >9-h half-life of pRB in normal cells is decreased to approximately 3 h in HPV16 E7-expressing cells (11, 24). In addition to the LXCXE motif, amino-terminal HPV16 E7 sequences that do not directly contribute to pRB binding are necessary for pRB degradation (24, 34, 35). The molecular mechanisms by which HPV16 E7 induces degradation of the pRB tumor suppressor, however, are not yet mechanistically defined.
The ubiquitin-proteasome pathway importantly contributes to regulated turnover of many cellular proteins. Conjugation of ubiquitin to a target protein involves at least three enzymatic reactions. The ubiquitin-activating enzyme (E1) activates and transfers ubiquitin to a ubiquitin-conjugating enzyme (E2). E2 then cooperates with a specific ubiquitin ligase (E3) to transfer ubiquitin to a substrate (reviewed in references 28 and 53). Several viruses target cellular ubiquitin ligases to induce degradation of a specific cellular protein(s) to alter host cellular pathways (reviewed in reference 6). Most importantly for the biology of HPVs, high-risk HPV E6 proteins associate with a cellular ubiquitin ligase, E6AP, to induce polyubiquitination, followed by proteasomal degradation of the p53 tumor suppressor (56). E6AP is the prototype of the HECT domain ubiquitin ligase family. Cullins act as central scaffolding modules for the assembly of a large family of multisubunit ubiquitin ligases (reviewed in reference 49). Seven family members, cullins 1, 2, 3, 4A, 4B, 5, and 7, have been identified in humans thus far (reviewed in reference 49). They bind to the ring finger protein Rbx1, as well as a diverse set of specific substrate adaptors, and consequently mediate the ubiquitination of a large number of proteins that regulate many different cellular signaling pathways (reviewed in references 25 and 49).
We identified cullin 2 by mass spectrometric analysis of HPV16 E7-associated cellular proteins that were isolated by tandem-affinity purification. We show that HPV16 E7 binds to an active cullin 2 ubiquitin ligase complex and that association correlates with the ability of HPV16 E7 to transform cells and to dysregulate G1/S cell cycle checkpoints. Lastly, we provide evidence that the HPV16 E7-associated cullin 2 ubiquitin ligase complex also contains pRB and that this complex contributes to HPV16 E7-mediated pRB destabilization.
MATERIALS AND METHODS
Cell lines and expression vectors.
RKO and RKO E7 cells were maintained in modified McCoy's 5A medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum, penicillin/streptomycin, and 500 μg/ml G418. HeLa S3 cells expressing the wild-type and mutant HPV16 E7 proteins (31) were grown in Dulbecco's modified Eagle medium, 10% fetal bovine serum, and penicillin/streptomycin. HPV16-positive CaSki human cervical carcinoma and 293 cells were cultured in Dulbecco's modified Eagle medium, 10% fetal bovine serum, and penicillin/streptomycin. Normal oral keratinocytes (NOK) immortalized by human telomerase (hTERT) and NOK hTERT/E7 (52) were maintained in keratinocyte-serum free medium (Invitrogen). SF9 insect cells were grown in TNM-FH medium (BD Biosciences, San Jose, CA) at 27°C.
Plasmids pcDNA3 T7 elongin C, pSP72 elongin B, and pcDNA3 Zeo HA elongin B were obtained from W. G. Kaelin, Jr. (Dana-Farber Cancer Institute). pcDNA3 HA2 Rbx1 was obtained from Y. Xiong (Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill). pCMV HA- ubiquitin was obtained from Li-Huei Tsai (The Picower Institute, Massachusetts Institute of Technology). pCMV RB and pCMV HPV-16 E7 were previously described (24), and pCMV myc cullin 2 was obtained from Christopher L. Carpenter (Beth Israel Deaconess Medical Center/Harvard Medical School).
Recombinant baculoviruses for expression of hemagglutinin (HA)-cullin 2 (Autographa californica multiple nucleopolyhedrosis virus [AcNPV] HA Cul2), elongin B (AcNPV elongin B), elongin C (AcNPV elongin C), and Rbx1 (AcNPV his-myc-Rbx1) were kind gifts from J. Conaway (Stowers Institute for Medical Research, Kansas City, MO). pVL1393 HPV16 E7 FLAG/HA (C-E7) was constructed by PCR amplification of a cDNA fragment corresponding to C-E7 from pOZ-CE7 (31). The amplified cDNA fragment was inserted into BamHI and EcoRI sites of pVL1393 (BD Biosciences) and confirmed by DNA sequencing. C-E7-expressing baculovirus was generated by transfecting SF9 cells with pVL1393 C-E7 according to the manufacturer's instructions (BD Transduction Laboratories, BD Biosciences, San Jose, CA).
Ubiquitination experiments.
For in vitro ubiquitination experiments, recombinant proteins were produced in Sf9 insect cells. At ∼72 h after infection with recombinant baculoviruses, the cells were harvested and lysed in 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, EDTA-free protease inhibitor cocktail (Roche) for 10 min at 4°C. The cell extracts were centrifuged at 15,700 × g in a microcentrifuge for 30 min, and the supernatants were recentrifuged for an additional 10 min. The cell extracts were immunoprecipitated with M2 FLAG antibody beads (Sigma, St. Louis, MO). After 45 min, the immunoprecipitates were washed three times with lysis buffer and eluted with FLAG peptide (1 mg/ml). The eluted HPV16 E7/cullin 2 complex was added to a ubiquitination reaction mixture (1) (50 mM Tris-HCl, 5 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, EDTA-free protease inhibitor cocktail [Roche], 2 mM ATP, 1× Energy Regeneration Mix, 1 mM ubiquitin aldehyde, 12 nM NEDD8, 6 nM UbcH12, 6 nM NEDD-activating enzyme [NAE1], 200 ng/μl UbcH5a, 250 ng/μl E1, 5 μg ubiquitin [all from Boston Biochem, Cambridge MA]) in a volume of 15 μl. The reaction mixtures were incubated at 30°C for 1 h, and the reaction was terminated with sample loading buffer, followed by analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Ubiquitin conjugates were visualized by Western blotting using ubiquitin antibody.
In vivo ubiquitination experiments were performed by transfecting 293 cells with the appropriate expression vectors using Fugene 6 (Roche) in six-well plates according to the manufacturer's protocol. pCMV hRB (1 μg), pCMV HPV16 E7 (1 μg), pCMV myc-cullin 2 (0.8 μg), pcDNA3 HA2-Rbx1 (0.4 μg), pcDNA3 T7 elongin C (0.3 μg), pcDNA3 Zeo HA elongin B (0.3 μg), and pCMV HA ubiquitin (0.8 μg) were cotransfected as indicated in Fig. 5C. Approximately 24 h after transfection, the cells were washed twice with phosphate-buffered saline (PBS) and lysed in 50 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 25 mM N-ethylmaleimide (Sigma), and protease inhibitors (Roche) on ice for ∼20 min. The cell lysates were cleared by centrifugation for 2 min at 15,700 × g in a microcentrifuge at 4°C. The supernatants were incubated for 2 h at 4°C with 25 μl HA beads (HA probe F-7; Santa Cruz) to immunoprecipitate the ubiquitinated proteins. The immunoprecipitates were washed three times with lysis buffer, boiled in SDS sample buffer for 5 min, and analyzed by SDS-PAGE.
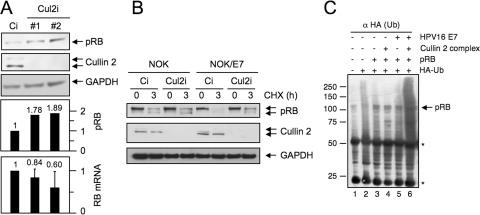
FIG. 5.
The cullin 2 ubiquitin ligase contributes to HPV16 E7-mediated pRB degradation. (A) Cullin 2 depletion by transfection of two siRNA oligonucleotide duplex populations results in pRB accumulation in the HPV16-positive CaSki cervical cancer cell line. Transfection of a control siRNA duplex (Ci) was used as a control. The concentration of the siRNA duplexes in the transfection mixture was 47.5 nM. Bar graphs with quantification of pRB (after normalization to GAPDH [glyceraldehyde-3-phosphate dehydrogenase] levels) and RB mRNA levels as determined by real-time PCR analysis from the same cells are shown below the blots. The RB mRNA levels represent averages and standard deviations of a single analysis performed in duplicate. Two additional experiments with higher siRNA concentrations (225 nM) yielded similar results. (B) Inhibition of pRB degradation in HPV16 E7-expressing cells upon cullin 2 depletion by siRNA. Control and HPV16 E7-expressing hTERT-immortalized NOK were transfected with the cullin 2-specific siRNA oligonucleotide duplex no. 1 (Cul2i) or a control siRNA (Ci), followed by treatment with cycloheximide to inhibit new protein synthesis. The cells were harvested at 0 and 3 h after cycloheximide treatment, and pRB levels were assessed by Western blotting. Cullin 2 and GAPDH levels are shown as controls. The concentration of the siRNA duplexes in the transfection mixture was 225 nM. (C) The HPV16 E7/cullin 2 ubiquitin ligase complex enhances pRB ubiquitination. 293 cells were cotransfected with the indicated plasmids. At 24 h after transfection, the ubiquitinated (Ub) proteins were immunoprecipitated with HA antibody beads, and ubiquitinated pRB products were detected by Western blotting with pRB antibody. Immunoglobulin heavy and light chains are denoted by asterisks.
Immunoprecipitation and glutathione S-transferase (GST) pull-down experiments.
Cells were washed twice with PBS and collected by scraping them in PBS. The cells were transferred into an Eppendorf tube, pelleted at 100 × g at 4°C, and rinsed once with PBS. The cell pellets were resuspended in 3 volumes of 0.3B buffer (20 mM Tris-HCl, pH 8.0, 0.3 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 10 mM β-mercaptoethanol, 0.2 mM phenylmethanesulfonyl fluoride). The cells were lysed by two freeze-thaw cycles (liquid nitrogen; 37°C), followed by rotation at 4°C for 30 min. The lysates were cleared by centrifugation at 15,700 × g at 4°C for 30 min. The supernatants were recentrifuged at 15,700 × g at 4°C for 10 min and precleared with protein G Sepharose for 30 min at 4°C with rotation. Monoclonal antibody to HPV-16 E7 (ED17; Santa Cruz, Santa Cruz, CA) was added, and the mixture was incubated for 3 to 4 h with rotation at 4°C. Protein G-Sepharose was added, and the mixture was incubated for 1 h at 4°C with rotation and washed three times with 1 ml of 0.3B buffer. Bound proteins were eluted with 2× SDS-PAGE sample buffer and analyzed on a 4 to 12% bis-Tris gradient gel (Invitrogen), followed by Western blotting with the indicated antibodies.
For double-immunoprecipitation experiments, HeLa S3 cells expressing C-E7 or control plasmid were treated with 5 μM of lactacystin (Biomol) for 12 h. The cells were harvested and lysed as described above. The cell extracts were immunoprecipitated with M2 FLAG antibody gel (Sigma). Immune complexes were eluted with FLAG peptide (Sigma; 1 mg/ml) at room temperature for 30 min. The eluates were resuspended in 0.3B buffer and immunoprecipitated with pRB antibody (Oncogene). The immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blotting with the appropriate antibodies.
For gradient centrifugation, 200 μl of FLAG-immunopurified C-E7-associated protein complexes isolated from HeLa S3 cells were loaded onto a 10 to 40% glycerol gradient and centrifuged for 16 h at 368,000 × g (Beckman; SW55Ti). After centrifugation, 23 fractions (200 μl each) were collected from the top, and a 10-μl aliquot was analyzed by silver staining and Western blotting with the indicated antibodies.
For GST pull-down experiments, the indicated proteins were generated by in vitro transcription/in vitro translation using rabbit reticulocyte lysates (TNT kit; Promega, Madison, WI), and 2 to 3 μl of each protein was used for in vitro binding in ELB buffer (250 mM NaCl, 50 mM HEPES, pH 7.0, 0.1% NP-40) for 6 h at 4°C. The bound proteins were washed three times with ELB buffer and subjected to SDS-PAGE.
Antibodies.
The E7 antibodies were 8C9 (Zymed; Invitrogen, Carlsbad, CA) and ED17 (Santa Cruz, Santa Cruz, CA). Rabbit cullin 1 and cullin 2 antibodies were from Zymed and Cul-4 (H-66) and Cul-5 (H-300) antibodies from Santa Cruz, polyclonal rabbit elongin B antibody was generously provided by J. W. Conaway (Stowers Institute for Medical Research, Kansas City, MO), elongin C/SIII p15 antibody was from BD Transduction Laboratories (BD Biosciences, San Jose, CA), Roc1/Rbx1 Ab-1 (rabbit polyclonal antibody) was from Lab Vision (Fremont, CA), anti-FLAG M2 affinity gel was from Sigma (Saint Louis, MO), HA antibody (Y11) was from Santa Cruz, pRB antibody (Ab-5) was from Oncogene (Cambridge, MA), rabbit anti-ubiquitin antiserum was from Sigma, and NEDD8 antibody was from Biomol (Plymouth Meeting, PA).
RNA interference (RNAi).
CaSki cells or NOK (2 × 105) were seeded onto six-well plates 1 day before transfection with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The small interfering RNA (siRNA) duplexes for cullin 2 (no. 1, D-007277-03; no. 2, a pool of no. 1 and D-007277-01) and a 21-nucleotide nonspecific siRNA duplex (D-001210-02) were obtained from Dharmacon Inc. (Chicago, IL). The concentrations of the siRNA duplexes in the transfection reactions ranged from 47.5 to 225 nM and are indicated in the figure legends.
To assess pRB stability after RNAi, NOK and NOK-E7 (52) were treated for 3 h with 30 μg/ml cycloheximide (Sigma, St. Louis, MO) at 48 h after RNAi transfection, followed by lysis in 150 mM NaCl, 1% NP-40, 50 mM Tri-HCl, pH 8.0, containing 45 nM sodium orthovanadate and protease inhibitor cocktail (Roche, Indianapolis, IN).
Quantitative reverse transcription-PCR.
Total RNA was extracted from CaSki cells using a commercial kit (Agilent Total RNA Isolation Mini Kit; Agilent Technologies, Inc., Santa Clara, CA) according to the manufacturer's instructions. Real-time PCR was performed at the TaqMan Core at Beth Israel Deaconess Medical Center. Primer hRB(b)-63F (TGGTGAATCATTCGGGACTTC), primer hRB(b)-146R (GCACTTCTTTTGAGCACACGG), and the TaqMan probe hRB(b)-86T (AGAAGTTCCAGAAAATAAATCAGATGGTATGTAACAGCG) were used.
RESULTS
The HPV16 E7 oncoprotein associates with cullin 2.
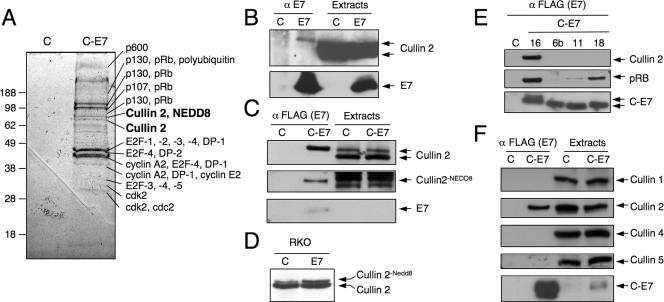
To identify cellular protein complexes associated with the HPV16 E7 oncoprotein, we expressed carboxyl terminally FLAG/HA epitope-tagged HPV16 E7 (C-E7) in HeLa cervical carcinoma cells and performed tandem-affinity purification, followed by mass spectrometric identification of individual components (31). In addition to known cellular HPV16 E7 targets (pRB, p107, p130, E2F family members, cdk2, cyclin A, and cyclin E), we isolated additional novel targets, including p600 (31) and steroid receptor coactivator 1 (SRC-1) (3). These analyses also revealed the presence of cullin 2-derived tryptic peptides in two closely spaced, adjacent bands migrating at an apparent molecular mass of approximately 75 kDa. The faster-migrating band contained five cullin 2-derived peptides, whereas the slower-migrating band contained 21 peptides. The slower-migrating band also contained NEDD8-derived tryptic peptides (Fig. 1A). Association of untagged HPV16 E7 with cullin 2 was confirmed by coprecipitation experiments with HPV16 E7-expressing RKO human colon cancer cells (Fig. 1B). These experiments revealed that HPV16 E7 preferentially associates with the slower-migrating form of cullin 2. Cullins are modified by covalent modification with the ubiquitin-related protein NEDD8, and only the NEDD8-bound form of cullin 2 was found to be active (14, 22). To determine whether HPV16 E6 preferentially associates with active, NEDD8-conjugated cullin 2, we performed FLAG immunoprecipitations from C-E7-expressing HeLa cells. Consistent with the results obtained with RKO E7 cells, C-E7 was preferentially associated with the slower-migrating cullin 2 band (Fig. 1C, top). Reprobing the same membrane with a NEDD8-specific antibody confirmed that the slower-migrating C-E7-associated cullin 2 band contained NEDD8 (Fig. 1C, middle). These results suggested that HPV16 E7 preferentially associates with NEDD8-modified cullin 2. Whereas the overall steady-state level of cullin 2 in HPV16 E7-expressing cells was not significantly different from that of control cells, the relative amount of neddylated cullin 2 was consistently increased in cells highly expressing HPV16 E7 (Fig. 1D). These results suggested that HPV16 E7 expression and/or association might induce cullin 2 neddylation and/or stabilize neddylated cullin 2.
FIG. 1.
HPV16 E7 associates with cullin 2. (A) Tandem-affinity purification of cellular protein complexes associated with C-E7 expressed in HeLa cells. Representative HPV16 E7-associated cellular proteins isolated by this procedure are indicated (31). Vector-transfected HeLa cells (lanes C) were used as a control. (B) Coprecipitation of cullin 2 with E7 antibodies in HPV16 E7-expressing RKO colon carcinoma cells. Parental RKO cells (lanes C) were used as a control. (C) E7 preferentially associates with the slower-migrating form of cullin 2 (top). The blot was stripped and reprobed with NEDD8-specific antibody to demonstrate that this band corresponds to NEDD8-modified cullin 2 (middle). A blot demonstrating immunoprecipitation of E7 is also shown (bottom). Lysates of HeLa cells with stable expression of C-E7 were immunoprecipitated with FLAG antibody, followed by Western blotting with the corresponding antibodies. Vector-transfected HeLa cells (lanes C) were used as a control. (D) Relative levels of NEDD8-conjugated cullin 2 are increased in HPV16 E7-expressing RKO cells (lane E7) compared to parental RKO cells (lane C). (E) Low-risk HPV11 and HPV6b and high-risk HPV18 E7 do not form a detectable complex with cullin 2. Lysates of HeLa cells with stable expression of the corresponding FLAG/HA-tagged E7 proteins were analyzed by immunoprecipitation with FLAG antibody, followed by Western blotting with the indicated antibodies. Vector-transfected HeLa cells (lane C) were used as a control. (F) HPV16 E7 binds to cullin 2, but not to cullin 1, 4, or 5. Lysates of HeLa cells with stable expression of C-E7 were immunoprecipitated with FLAG antibody, followed by Western blotting with the corresponding cullin antibodies. Vector-transfected HeLa cells (lanes C) were used as a control.
We next investigated whether E7 proteins encoded by other HPVs were also associated with cullin 2. We performed FLAG immunoprecipitations from HeLa cell lines expressing carboxyl-terminally FLAG/HA-tagged versions of HPV18, HPV6b, and HPV11 (31), followed by Western blotting for cullin 2 and pRB as a control. Consistent with published results, the low-risk HPV6 and HPV11 E7 proteins associated with pRB at reduced levels (45) but did not associate with cullin 2 at detectable levels (Fig. 1E). Somewhat unexpectedly, we did not detect cullin 2 in association with HPV18 E7 (Fig. 1E).
It was previously reported that HPV16 E7 associates with the cullin 1 ubiquitin ligase complex (47). Even though our tandem-affinity purification/mass spectrometry analyses did not provide any evidence for other cullin family members in complex with HPV16 E7 (data not shown), we performed coimmunoprecipitation experiments with C-E7-expressing HeLa cells, followed by Western blotting with antibodies specific for different cullins. These experiments revealed that HPV16 E7 specifically associates with cullin 2, but not with the closely related cullin 5, cullin 1, or cullin 4 (Fig. 1F).
HPV16 E7 is associated with an active cullin 2 ubiquitin ligase complex.
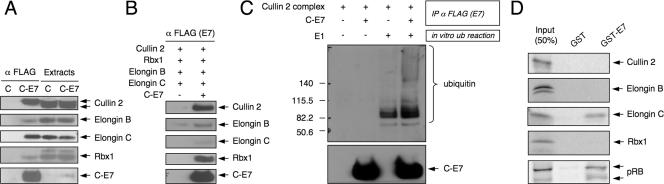
Cullin 2 acts as a scaffold for the formation of a multisubunit ubiquitin ligase complex with elongin B, elongin C, Rbx1, and BC box-containing cellular proteins, such as the von Hippel Lindau (VHL) tumor suppressor, mMED8, or elongin A (49). We next examined whether HPV16 E7 associates with the cullin 2 complex or solely with the cullin 2 subunit and performed immunoprecipitation/Western blot experiments with lysates from C-E7-expressing HeLa cells. These experiments revealed that HPV16 E7 coimmunoprecipitates not only cullin 2, but also elongin B, elongin C, and Rbx1 (Fig. 2A). The VHL tumor suppressor protein, a BC box protein that acts a substrate adaptor for the cullin 2 ubiquitin ligase complex, however, was not detected in these experiments, even though it was readily identified in the HeLa cell extracts (data not shown). These data suggest that HPV16 E7 forms a complex with the cullin 2 ubiquitin ligase complex and that this complex most likely does not contain VHL.
FIG. 2.
HPV16 E7 associates with an active cullin 2 ubiquitin ligase complex. (A) HPV16 E7 associates with cullin 2, elongin B, elongin C, and Rbx1. Lysates of HeLa cells with stable expression of C-E7 were immunoprecipitated with FLAG antibodies, followed by Western blotting with the corresponding cullin 2 ubiquitin ligase components. Vector-transfected HeLa cells (lanes C) were used as a control. (B) HPV16 E7 interacts with the cullin 2 ubiquitin ligase complex produced in insect cells. Sf9 insect cells were coinfected with recombinant baculoviruses encoding the various components of the cullin 2 ubiquitin ligase complex and C-E7 as indicated. E7 immunoprecipitations were performed with FLAG antibody, and coprecipitated components of the cullin 2 complex were detected by immunoblotting. (C) The HPV16 E7/cullin 2 complex isolated from insect cells (panel B) was subjected to an in vitro ubiquitination reaction including ubiquitin and UbcH5a (E2) with or without E1. Ubiquitin polymerization was analyzed by Western blotting using ubiquitin antibody. (D) HPV16 E7 binds to elongin C in vitro. [35S]methionine/cysteine-labeled cullin 2, Rbx1, elongin C, elongin B, and pRB were synthesized by in vitro transcription/in vitro translation using rabbit reticulocyte lysates. The in vitro-translated proteins were incubated with bacterially produced GST-HPV16 E7 fusion protein (GST-E7) or GST in a buffer containing 0.1% NP-40, followed by affinity chromatography on glutathione-Sepharose.
Based on these results, we examined whether HPV16 E7 forms an enzymatically active cullin 2 ubiquitin ligase. SF9 insect cells were infected with recombinant baculoviruses encoding HA-tagged cullin 2, elongin B, and elongin C and myc-tagged Rbx1 with or without a baculovirus encoding C-E7. C-E7 was immunoprecipitated from coinfected insect cells using FLAG antibody beads, and coprecipitation of cullin 2, elongin B, elongin C, and Myc-Rbx1 was evaluated by Western blotting. Consistent with the results obtained with HeLa cells (Fig. 2A), HPV16 E7 was found associated with cullin 2, elongin B, elongin C, and Rbx1 in insect cells (Fig. 2B). Similarly, immunoprecipitation experiments with insect cells coinfected with the different baculoviruses with a cullin 2 antibody resulted in coprecipitation of HPV16 E7, elongin B, elongin C, and Rbx1 (data not shown). To assess the enzymatic activity of the HPV16 E7-associated cullin 2 complex, purified UbcH 5a (E1) and ubiquitin were added, and conjugation of ubiquitin to larger-molecular-size structures was assessed by immunoblotting (Fig. 2C). It has been reported that the cullin complexes can undergo autoubiquitination (66). Thus, the bands observed may represent ubiquitinated forms of the cullin 2 complex. These experiments showed that HPV16 E7 associates with an enzymatically active cullin 2 complex.
HPV16 E7 associates with the elongin C subunit of the cullin 2 ubiquitin ligase complex in vitro.
To identify the subunit of the cullin 2 ubiquitin ligase complex that is directly targeted by HPV16 E7, we performed pull-down assays with GST-HPV16 E7 and individual cullin 2 complex components generated by in vitro transcription/in vitro translation. These experiments revealed that HPV16 E7 associates with elongin C but not significantly with other components of the cullin 2 complex (Fig. 2D). Since cullin 2 generated by in vitro translation is not efficiently neddylated (data not shown), we cannot rule out the possibility that HPV16 E7 may also interact with neddylated cullin 2. Nevertheless, these results suggest that HPV16 E7 associates with elongin C of the cullin 2 ubiquitin ligase complex.
Association with the cullin 2 ubiquitin ligase complex requires the conserved region 1 (CR1) homology domain, as well as carboxyl-terminal sequences of HPV16 E7.
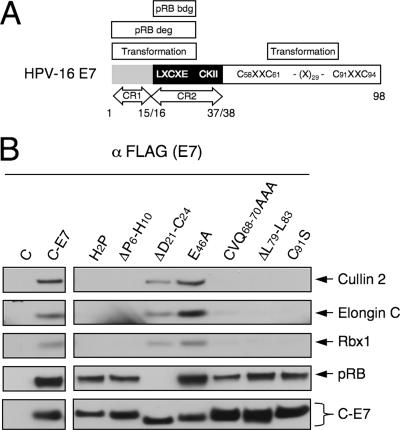
To gain insight into the potential functional significance of the association of HPV16 E7 with the cullin 2 complex, we examined the association of HPV16 E7 mutants with the cullin 2 complex in vivo. HPV16 E7 consists of three domains: the extreme amino terminus is homologous to a small portion of CR1 of Ad E1A, followed by a region homologous to Ad E1A CR2 and a carboxyl-terminal domain that contains a unique cysteine-rich zinc-binding domain (20, 51) (Fig. 3A). HeLa cells with stable expression of various carboxyl-terminally FLAG/HA epitope-tagged HPV16 E7 mutants were generated (31), and associated cellular proteins were isolated by immunoprecipitation with FLAG antibody. Since these HPV16 E7 mutants were expressed at somewhat different levels, the relative amounts of cell extracts used for the experiments were normalized for C-E7 expression. Coprecipitation of cullin 2 complex components, as well as pRB, was assessed by Western blotting (Fig. 3B). The two HPV16 E7 mutants within CR1, H2P and ΔP6-H10, were negative for association with the cullin 2 complex. As expected, these mutants efficiently associated with pRB, even though they are defective for cellular transformation and pRB degradation (5, 34, 35, 50). In contrast, the pRB-binding- and transformation-defective HPV16 E7 mutant in CR2, ΔD21-C24 (45, 50), retained the capacity to associate with the cullin 2 complex. Of the four HPV16 E7 mutants within the carboxyl terminus of HPV16 E7, only the E46A mutant efficiently associated with the cullin 2 complex, whereas the CVQ68-70AAA, ΔL79-L83, and C91S mutants each displayed a dramatically decreased capacity to bind to the cullin 2 ubiquitin ligase complex. As expected, each of these mutants was able to efficiently associate with pRB (Fig. 3B). The E46A mutant displayed no apparent transformation defects (50), whereas the CVQ68-70AAA and ΔL79-L83 mutants were defective in overriding a G1 cell cycle arrest in response to DNA damage (27) and the C91S mutant was defective for association with the S4 subunit of the proteasome (8).
FIG. 3.
HPV16 E7 sequences in CR1 and the carboxyl-terminal region contribute to association with the cullin 2 complex. (A) Schematic representation of HPV16 E7 functional regions. (B) Association of the cullin2 ubiquitin ligase complex with different HPV16 E7 mutants. Lysates of HeLa cells with stable expression of the corresponding C-E7 proteins were immunoprecipitated with FLAG antibody, followed by Western blotting with the corresponding antibodies; pRB and E7 blots (using HA antibody) are shown as controls.
Thus, our results demonstrated that the ability of HPV16 E7 to associate with the cullin 2 ubiquitin ligase complex is independent of pRB binding and that sequences within the CR1 homology domain implicated in cellular transformation and pRB degradation are necessary for cullin 2 association. In addition, sequences within the HPV16 E7 carboxyl-terminal domain, which have been implicated in pRB binding/degradation-independent transformation-associated activities of E7, also contribute to efficient association of HPV16 E7 with the cullin 2 ubiquitin ligase complex.
The pRB tumor suppressor is a component of the HPV16 E7-associated cullin 2 ubiquitin ligase complex.
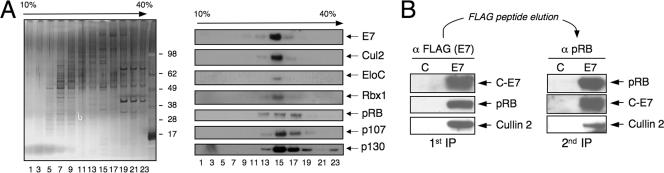
HPV16 E7 interacts with the cullin 2 ubiquitin ligase complex and pRB through different sequences (Fig. 3B). Thus, binding to the cullin 2 ubiquitin complex and pRB may not be mutually exclusive, and pRB may be a component of the HPV16 E7-associated cullin 2 ubiquitin ligase complex. Consistent with this notion, the HPV16 E7-associated cullin 2, pRB, p107, and p130 complexes peak in overlapping fractions upon sucrose gradient centrifugation (Fig. 4A). To definitively determine whether cullin 2 is a component of HPV16 E7-associated pRB complexes, C-E7-associated cellular complexes were isolated by FLAG immunoprecipitation and eluted from the antibody beads with FLAG peptide. The eluted proteins were subjected to a second round of immunoprecipitation with a pRB-specific antibody, followed by Western blot analysis for pRB, C-E7, and cullin 2. These experiments revealed that cullin 2 is indeed a component of HPV16 E7/pRB complexes (Fig. 4B).
FIG. 4.
Cullin 2 is a component of the HPV16 E7/pRB complex. (A) Cosedimentation of the HPV16 E7-associated pRB, p107, p130, and cullin 2 complexes on a glycerol gradient. HPV16 E7-associated protein complexes isolated from HeLa cells expressing C-E7 by immunoprecipitation with FLAG antibody followed by elution with FLAG peptide were resolved on a 10 to 40% glycerol gradient. Representative fractions were analyzed by silver staining (left) and Western blotting with the indicated antibodies (right). (B) The HPV16 E7/pRB complex contains cullin 2. HPV16 E7-associated proteins were isolated from HeLa cells expressing C-E7 by immunoprecipitation (IP) with FLAG antibody, followed by elution with FLAG peptide. The eluted protein complexes were reprecipitated with pRB antibody. The presence of cullin 2 and E7 in pRB-associated protein complexes was assessed by Western blotting. Vector-transfected HeLa cells (lanes C) were used as a control.
The cullin 2 ubiquitin ligase complex contributes to HPV16 E7-mediated pRB destabilization.
Since cullin 2 is a component of the HPV16 E7/pRB complex, we next determined whether the HPV16-associated cullin 2 ubiquitin ligase might be involved in HPV16 E7-mediated pRB degradation. Cullin 2 depletion by two different siRNA populations in the HPV16-positive CaSki human cervical carcinoma cell line caused 1.78- and 1.89-fold increases in pRB steady-state levels, respectively. To ensure that this effect was not a consequence of increased RB mRNA expression, we compared retinoblastoma protein (RB) mRNA levels in these cells by quantitative real-time PCR. These results showed no concomitant increase in RB mRNA levels; indeed RB mRNA levels were decreased by 16 and 40% (Fig. 5A). Thus, the increased pRB levels in HPV16 E7-expressing cells upon cullin 2 depletion are not due to increased transcription or mRNA stability.
To determine whether increased pRB levels in HPV16 E7-expressing cells upon RNAi-mediated cullin 2 depletion were due to protein stabilization rather than increased RB mRNA translation, we performed cycloheximide chase experiments. A matched pair of hTERT-immortalized NOK with or without stable expression of HPV16 E7 (NOK and NOK/E7, respectively) (52) were transfected with cullin 2 siRNA and control siRNA. At 48 h after transfection, protein synthesis was inhibited by cycloheximide and pRB levels were analyzed. Compared to control siRNA-transfected cells, cullin 2 depletion in NOK cells had only a minor effect on pRB levels during the 3-hour chase period. A dramatic decrease in pRB levels was observed in control siRNA-transfected NOK/E7 cells after the 3-h incubation with cycloheximide, consistent with previous reports that HPV16 E7 expression decreases pRB half-life (11, 24). In contrast, pRB levels in cullin 2-depleted NOK E7 were similar to those in control or cullin 2 siRNA-transfected NOK after the 3-hour cycloheximide chase period (Fig. 5B). We also detected increases of pRB levels upon siRNA-mediated HPV16 E7 depletion in these cells or upon treatment with the proteasome inhibitor Lactacystein (data not shown). These results indicate that cullin 2 is necessary for accelerated degradation of pRB in HPV16 E7 cells.
HPV16 E7 and the cullin 2 complex ubiquitinate pRB in vivo.
The detection of cullin 2 in the HPV16 E7/pRB complex (Fig. 4B) and the observed increase in the pRB protein level and half-life upon siRNA-mediated cullin 2 silencing in HPV16 E7-expressing cells (Fig. 5A and B) suggest that pRB may be a substrate of the HPV16 E7/cullin 2 ubiquitin ligase complex. To address this issue, we cotransfected 293 cells with various combinations of expression plasmids for pRB, HPV16 E7, and HA-ubiquitin, as well as the cullin 2 ubiquitin ligase components cullin 2, elongin C, elongin B, and Rbx 1. Ubiquitination of pRB was assessed by immunoprecipitation with HA antibody beads, followed by Western blotting with pRB antibody. Whereas expression of E7 or the cullin 2 complex alone did not significantly enhance pRB ubiquitination in 293 cells (Fig. 5C, lanes 2 to 5), cotransfection of the cullin 2 complex and HPV16 E7 resulted in enhanced pRB ubiquitination (Fig. 5C, lane 6). These experiments indicate that the HPV16 E7/cullin 2 complex can induce ubiquitination of pRB in vivo.
DISCUSSION
The pRB tumor suppressor is a critical regulator of the G1-to-S cell cycle transition. Mutations of the human RB gene are rate limiting for the development of retinoblastomas, and the pRB tumor suppressor pathway is rendered dysfunctional in the majority of human carcinomas (reviewed in reference 15). Previous studies have shown that HPV16 E7 expression causes pRB destabilization (9, 11, 34). Proteasome inhibitors interfere with HPV16 E7-mediated pRB destabilization (9, 11, 35), and HPV16 E7 is incompetent for pRB destabilization at the nonpermissive temperature in a cell line expressing a temperature-sensitive E1 enzyme (11). Destabilization of pRB is necessary for functional inactivation by HPV16 E7 (24) and has been linked to HPV16 E7-mediated transformation (35), although HPV16 E7 exhibits pRB-independent transforming activities, as well (4, 16, 27, 31).
Here, we provide evidence that HPV16 E7 is associated with a cullin 2 ubiquitin ligase complex. HPV16 E7 preferentially binds to the NEDD8-modified form of cullin 2 (Fig. 1C), which is associated with enhanced ubiquitin ligase activity (14, 22). Consistent with this notion, we found that the HPV16 E7-associated cullin 2 complex is enzymatically active in vitro (Fig. 2C). We also observed a consistent increase of NEDD8-modified cullin 2 species in HPV16 E7-expressing cells (Fig. 1D). NEDD8 is a small ubiquitin-like molecule that covalently modifies cullins, leading to an enhancement of their ubiquitin ligase activities (18, 40; reviewed in reference 48). Similar to the ubiquitin system, conjugation of NEDD8 to target proteins is achieved by NEDD8-specific activating (E1) and conjugating (E2) enzymes and the NEDD8-ligase (E3) ROC1. Removal of NEDD8 is achieved by the COP9 signalosome (reviewed in reference 64). The NEDD8 modification pathway is important for viability, cell cycle control, and embryonic development in different organisms (reviewed in reference 48). The molecular mechanisms of HPV16 E7-induced alteration of the cullin 2 neddylation status and its potential biological ramifications remain to be determined.
In vitro binding studies with individual components of the cullin 2 ubiquitin ligase complex revealed binding of HPV16 E7 to the elongin C subunit (Fig. 2D). Since cullin 2 synthesized by in vitro transcription/in vitro translation is not efficiently neddylated, our results do not rule out the possibility that HPV16 E7 may also bind to the NEDD8-modified cullin 2 subunit. This might explain why we did not detect association of HPV16 E7 with cullin 5, which, similar to cullin 2, also associates with elongins B and C (Fig. 1F). Cullin 2 is associated with the HPV16 E7/pRB complex (Fig. 4B), cullin 2 depletion by RNAi causes accumulation and stabilization of pRB in HPV16 E7-expressing cells (Fig. 5A and B), and the HPV16 E7-associated cullin 2 complex can ubiquitinate pRB in vivo (Fig. 5C). Thus, the HPV16 E7-associated active cullin 2 ubiquitin ligase complex significantly contributes to HPV16 E7-mediated pRB degradation.
Association with the cullin 2 ubiquitin ligase complex, however, may not represent a universal mechanism for HPV E7-mediated pRB destabilization. Most remarkably, we did not detect any cullin 2 associated with an HPV18 E7 protein identically HA/FLAG epitope tagged at its carboxyl terminus when expressed in HeLa cells (Fig. 1E). This suggests that HPV18 E7, which is known to destabilize pRB, may utilize other cellular components to destabilize pRB. Alternatively, addition of epitope tags may interfere with its ability to efficiently associate with the cullin 2 ubiquitin ligase. Similarly, it has been recently reported that expression of the low-risk HPV6 E7 protein results in destabilization of the pRB family member p130 (68). Even though our results indicate that the associated cullin 2 complex also contributes to HPV16 E7-mediated p130 destabilization (data not shown), we did not detect association of the cullin 2 ubiquitin ligase complex with carboxyl-terminally FLAG/HA-tagged HPV6 and HPV11 E7 proteins expressed in HeLa cells (Fig. 1E). This suggests that HPV6 and HPV11 E7 proteins may destabilize p130 through a different mechanism. Alternatively, low-risk HPV E7 proteins may decrease p130 levels indirectly. The stability of p130 is regulated by ubiquitination of phospho-p130 through the cullin 1 ubiquitin ligase complex (60). Since expression of low-risk E7 proteins also stimulates G1/S cell cycle progression and thus p130 phosphorylation, it is possible that levels of phospho-p130 are increased in such cells and that the observed destabilization of p130 by HPV6 E7 reflects increased degradation of phospho-p130 through the cullin 1 ubiquitin ligase complex.
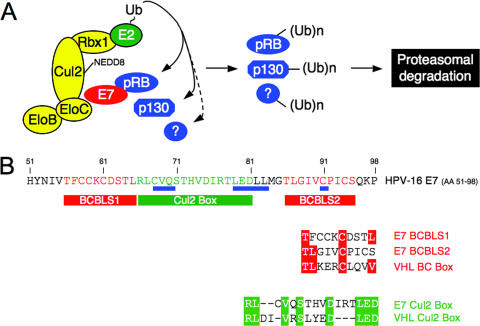
Our results indicate that HPV16 E7 associates with the cullin 2 ligase complex through two distinct sequences. An amino-terminal domain that shares sequence similarity with a portion of CR1 of the Ad E1A oncoprotein (51) is required for association with cullin 2 (Fig. 3B). HPV16 E7 mutants in the CR1 homology domain that are incompetent for association with the cullin 2 complex are also defective for cellular transformation and pRB degradation (35). We have recently identified another HPV E7 cellular target protein, p600, which also associates through sequences within the CR1 homology domain (31). Our data suggest that the E7/cullin 2 and E7/p600 complexes represent independent interactions, since, unlike the cullin 2 complex, p600 can also associate with the low-risk HPV6 and HPV11 E7 proteins (31), as well as with HPV18 E7 (K. W. Huh and K. Münger, unpublished data). Sequences in the HPV16 E7 carboxyl-terminal domain also contribute to efficient association of HPV16 E7 with the cullin 2 ubiquitin ligase complex. The HPV16 E7 CVQ68-70AAA and ΔL79-L83 mutants are defective for overriding DNA damage-induced G1 growth arrest and remain competent for pRB binding and destabilization (27), yet they associate with the cullin 2 complex at severely reduced, barely detectable levels (Fig. 3B). While this remaining level of association with the cullin 2 complex may be sufficient for pRB degradation in vivo, it is possible that an additional E7-associated ubiquitin ligase(s) may also contribute to HPV16 E7-mediated pRB degradation. Indeed, in addition to p600, a ubiquitin ligase of the N-end rule pathway (59) that does not appear to contribute to E7-mediated pRB degradation (31), our tandem-affinity purification data provide evidence for additional ubiquitin ligases among the HPV16 E7 host cell protein complex components (Huh and Münger, unpublished). Studies aimed at confirming these associations and determining their potential biological consequences are currently in progress. In addition, our results are consistent with a model in which the HPV16 E7-associated cullin 2 ubiquitin ligase complex may target additional HPV16 E7-associated proteins for degradation (Fig. 6A).
FIG. 6.
A hypothetical model of the HPV16 E7/cullin 2 complex. (A) HPV16 E7 associates with the cullin 2 complex and recruits cellular proteins, including pRB, p130, and potentially additional, unidentified cellular proteins (?) for ubiquitination [(Ub)n]. In addition, cullin 2-associated ubiquitin ligase activity may be increased in HPV16 E7-expressing cells due to increased cullin 2 neddylation. (B) HPV16 E7 contains two BC box-like sequences (BCBL1 and BCBL2) and a cullin 2 box-like amino acid sequence within the carboxyl-terminal domain. The carboxyl-terminal HPV16 E7 sequence (amino acid residues 51 to 98) is shown, with positions of mutations that are necessary for efficient association with the cullin 2 ubiquitin ligase complex underlined in blue. BC box-like sequences (BCBLS1 and -2) are indicated by red boxes, and a sequence related to the cullin 2 box in the VHL tumor suppressor is shown in green. Detailed sequence comparisons are shown underneath, and identical residues are boxed.
It is interesting that the carboxyl-terminal HPV16 E7 sequences that contribute to efficient association with cullin 2 contain two sequence motifs that share some sequence similarity with BC boxes (Fig. 6B). BC boxes are signature sequences of cellular proteins that act as substrate adaptors for elongin B/elongin C-containing cullin 2 and cullin 5 ubiquitin ligases. In addition, the HPV16 E7 carboxyl terminus contains a cullin 2 box-related sequence (Fig. 6B), which in the VHL tumor suppressor protein determines its specificity to associate with the cullin 2, but not the cullin 5, ubiquitin ligase complex (38). Even though we have not carefully assessed the functionalities of these sequences, HPV16 E7 specifically associates with cullin 2, not cullin 5 (Fig. 1F), and binds to elongin C in vitro (Fig. 2D). Consistent with the notion that HPV16 E7 and the cellular adapter VHL associate with the cullin 2 ubiquitin ligase through similar sequences, we did not detect VHL as a component of the HPV16 E7-associated cullin 2 complex (data not shown). Under conditions of limiting cullin 2 levels, HPV16 E7 may thus displace VHL from cullin 2 ubiquitin ligase complexes, thereby affecting VHL tumor suppressor activity. This possibility is currently being addressed experimentally. Indeed, HPV16 E7 expression has been shown to trigger an angiogenesis-related transcriptional response (61), and there is no correlation between HIF1α expression and tumor oxygenation status in HPV-associated cervical cancers (43).
The paradigm that viral oncoproteins can reprogram the substrate specificities of cellular ubiquitin ligases and alter their substrate specificities was originally established by studies with the HPV16 E6 oncoprotein. HPV16 E6 accelerates the degradation of the p53 tumor suppressor (57) by forming a complex with E6AP (56), the founding member of the HECT domain-containing ubiquitin ligase family (32). In the absence of E6, E6AP does not appear to be involved in the regulated turnover of p53 (7, 58). Similarly, the Ad E1B55K and E4orf6 proteins can induce degradation of p53 (and potentially other proteins) through complex formation by E4orf6 with the cullin 5 ubiquitin ligase complex (26, 54). Similar to what we report here for HPV16 E7, Ad E4orf6 associates with the cullin ubiquitin complex through elongin C, but the substrate p53 is targeted to the complex through the E4orf6-associated Ad E1B 55,000-molecular-weight (55K) protein (10). Various other viral proteins have been reported to associate with cullin-based ubiquitin ligases (reviewed in reference 6). The respiratory syncytial virus NS1 protein has been shown to degrade STAT2 through association with the cullin 2 ubiquitin ligase complex (21). The human immunodeficiency virus viral infectivity factor (Vif) protein abrogates the antiviral activity of the host cytidine deaminase APOBEC3G by targeting it for proteasomal degradation through association with cullin 5 (67). The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen has also been reported to associate with cullin 5 to induce degradation of the p53 and VHL tumor suppressors (12). In addition, the SV40 large tumor antigen, which shares some sequence similarity with HPV16 E7, associates with cullin 7, but the substrates of this complex have yet to be identified (1, 39).
The tumor suppressor pRB has a long half-life, and its biological activity is controlled by cell cycle-dependent phosphorylation/dephosphorylation. However, a number of mechanisms leading to degradation of pRB have been published. Overexpression of the oncogenic ankyrin repeat protein gankyrin in hepatomas has been reported to lead to pRB destabilization and has been correlated with tumor formation (30). A cathepsin B-like protease, termed SPase, that can cleave pRB has been described (46). Similarly, pRB is cleaved by a caspase during apoptosis (2). In addition, a number of other viral proteins have been reported to induce pRB degradation. They include the Epstein-Barr virus nuclear antigen 3C, which degrades pRB through a cullin 1 ubiquitin ligase complex (42), as well as cytomegalovirus pp71 (36) and human T-cell leukemia virus type 1 Tax (41), which induce ubiquitin-independent proteasomal pRB degradation (37, 41).
In summary, we have identified the cullin 2 ubiquitin ligase as a novel cellular target of the HPV16 E7 oncoprotein. HPV16 E7 mutants that are defective for cullin 2 association are impaired for cellular transformation and/or G1/S cell cycle checkpoint engagement in response to DNA damage. Thus, association of HPV16 E7 with the cullin 2 ubiquitin ligase complex is linked to its transforming activities. Our results are consistent with the model in which HPV16 E7 may act as a viral substrate adaptor for the cullin 2 ubiquitin ligase complex, thereby targeting HPV16 E7-associated cellular proteins for ubiquitination and subsequent proteasomal degradation (Fig. 6A). In addition, HPV16 E7 expression correlates with enhanced cullin 2 neddylation. Hence, cullin 2-associated ubiquitin ligase activity may be increased in HPV16 E7-expressing cells. We provided evidence that cullin 2 is associated with the HPV16 E7/pRB complex and that pRB can be a substrate for ubiquitination by the HPV16 E7/cullin 2 ubiquitin ligase complex. Aberrant proteasomal turnover of cellular proteins, including the pRB tumor suppressor, through association with and reprogramming of cellular ubiquitin ligase(s) may be one of the mechanisms through which HPV16 E7 exerts its diverse transforming activities.
Acknowledgments
This work was supported by PHS grants CA066980 (K.M.) and GM54137 (J.W.H.).
We thank the members of Pat Nakatani's laboratory (Dana Farber Cancer Institute, Boston, MA) for their help with glycerol gradient centrifugation; Keiichi I. Nakayama and other members of Takumi Kamura's laboratory (Kyushu University, Fukuoka, Japan), as well as Dana Gabuzda and Andrew Mehle (Dana-Farber Cancer Institute, Boston, MA), for helpful suggestions and advice; Jennifer Altreuter, Brendan N. Lilley, Gustavo Martinez-Noel, and Jianxin You for critical reading of the manuscript and editorial assistance; Joseph DeMasi (Harvard Medical School, Boston, MA) for his help with the Megalign DNASTAR program; W. G. Kaelin, Jr. (Dana-Farber Cancer Institute, Boston, MA) for pcDNA3 T7 elongin C, pSP72 elongin B, and pcDNA3 Zeo HA elongin B; Li-Huei Tsai (The Picower Institute, MIT, Cambridge, MA) for pCMV HA-ubiquitin; Christopher L. Carpenter (Beth Israel Deaconess Medical Center, Boston, MA) for pCMV myc cullin 2; J. W. Conaway (Stowers Institute for Medical Research, Kansas City, MO) for baculoviruses; and Y. Xiong (Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill) for pcDNA3 HA2 Roc I. We thank the TaqMan Core at Beth Israel Deaconess Medical Center and the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for real-time PCR and mass spectrometry, respectively.
This article is dedicated to the memory of Konrad Lerch.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Ali, S. H., J. S. Kasper, T. Arai, and J. A. DeCaprio. 2004. Cul7/p185/p193 binding to simian virus 40 large T antigen has a role in cellular transformation. J. Virol. 78:2749-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, B., and Q. P. Dou. 1996. Cleavage of retinoblastoma protein during apoptosis: an interleukin 1 beta-converting enzyme-like protease as candidate. Cancer Res. 56:438-442. [PubMed] [Google Scholar]
- 3.Baldwin, A., K. W. Huh, and K. Munger. 2006. Human papillomavirus E7 oncoprotein dysregulates steroid receptor coactivator 1 localization and function. J. Virol. 80:6669-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balsitis, S., F. Dick, D. Lee, L. Farrell, R. K. Hyde, A. E. Griep, N. Dyson, and P. F. Lambert. 2005. Examination of the pRb-dependent and pRb-independent functions of E7 in vivo. J. Virol. 79:11392-11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banks, L., C. Edmonds, and K. Vousden. 1990. Ability of the HPV16 E7 protein to bind RB and induce DNA synthesis is not sufficient for efficient transforming activity in NIH 3T3 cells. Oncogene 5:1383-1389. [PubMed] [Google Scholar]
- 6.Barry, M., and K. Fruh. 2006. Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci. STKE 2006:pe21. [DOI] [PubMed] [Google Scholar]
- 7.Beer-Romero, P., S. Glass, and M. Rolfe. 1997. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene 14:595-602. [DOI] [PubMed] [Google Scholar]
- 8.Berezutskaya, E., and S. Bagchi. 1997. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26 S proteasome. J. Biol. Chem. 272:30135-30140. [DOI] [PubMed] [Google Scholar]
- 9.Berezutskaya, E., B. Yu, A. Morozov, P. Raychaudhuri, and S. Bagchi. 1997. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 8:1277-1286. [PubMed] [Google Scholar]
- 10.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 12.Cai, Q. L., J. S. Knight, S. C. Verma, P. Zald, and E. S. Robertson. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiba, T., and K. Tanaka. 2004. Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr. Protein Pept. Sci. 5:177-184. [DOI] [PubMed] [Google Scholar]
- 15.Cobrinik, D. 2005. Pocket proteins and cell cycle control. Oncogene 24:2796-2809. [DOI] [PubMed] [Google Scholar]
- 16.Collins, A. S., T. Nakahara, A. Do, and P. F. Lambert. 2005. Interactions with pocket proteins contribute to the role of human papillomavirus type 16 E7 in the papillomavirus life cycle. J. Virol. 79:14769-14780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeCaprio, J. A., J. W. Ludlow, J. Figge, J. Y. Shew, C. M. Huang, W. H. Lee, E. Marsilio, E. Paucha, and D. M. Livingston. 1988. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275-283. [DOI] [PubMed] [Google Scholar]
- 18.Dharmasiri, S., N. Dharmasiri, H. Hellmann, and M. Estelle. 2003. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 22:1762-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 20.Edmonds, C., and K. H. Vousden. 1989. A point mutational analysis of human papillomavirus type 16 E7 protein. J. Virol. 63:2650-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott, J., O. T. Lynch, Y. Suessmuth, P. Qian, C. R. Boyd, J. F. Burrows, R. Buick, N. J. Stevenson, O. Touzelet, M. Gadina, U. F. Power, and J. A. Johnston. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J. Virol. 81:3428-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furukawa, M., Y. Zhang, J. McCarville, T. Ohta, and Y. Xiong. 2000. The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20:8185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillison, M. L., W. M. Koch, R. B. Capone, M. Spafford, W. H. Westra, L. Wu, M. L. Zahurak, R. W. Daniel, M. Viglione, D. E. Symer, K. V. Shah, and D. Sidransky. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92:709-720. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guardavaccaro, D., and M. Pagano. 2004. Oncogenic aberrations of cullin-dependent ubiquitin ligases. Oncogene 23:2037-2049. [DOI] [PubMed] [Google Scholar]
- 26.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helt, A. M., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75:6737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 29.Hickman, E. S., M. C. Moroni, and K. Helin. 2002. The role of p53 and pRB in apoptosis and cancer. Curr. Opin. Genet. Dev. 12:60-66. [DOI] [PubMed] [Google Scholar]
- 30.Higashitsuji, H., K. Itoh, T. Nagao, S. Dawson, K. Nonoguchi, T. Kido, R. J. Mayer, S. Arii, and J. Fujita. 2000. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat. Med. 6:96-99. [DOI] [PubMed] [Google Scholar]
- 31.Huh, K. W., J. DeMasi, H. Ogawa, Y. Nakatani, P. M. Howley, and K. Munger. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. USA 102:11492-11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin protein ligase. Proc. Natl. Acad. Sci. USA 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Agency for Research on Cancer. 1995. IARC monographs on the evaluation of carcinogenic risks to humans, vol. 64. Human papillomaviruses. International Agency for Research on Cancer, Lyon, France.
- 34.Jones, D. L., and K. Munger. 1997. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J. Virol. 71:2905-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, D. L., D. A. Thompson, and K. Munger. 1997. Destabilization of the RB tumor suppressor protein and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology 239:97-107. [DOI] [PubMed] [Google Scholar]
- 36.Kalejta, R. F., J. T. Bechtel, and T. Shenk. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalejta, R. F., and T. Shenk. 2003. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. USA 100:3263-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamura, T., K. Maenaka, S. Kotoshiba, M. Matsumoto, D. Kohda, R. C. Conaway, J. W. Conaway, and K. I. Nakayama. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasper, J. S., H. Kuwabara, T. Arai, S. H. Ali, and J. A. DeCaprio. 2005. Simian virus 40 large T antigen's association with the CUL7 SCF complex contributes to cellular transformation. J. Virol. 79:11685-11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawakami, T., T. Chiba, T. Suzuki, K. Iwai, K. Yamanaka, N. Minato, H. Suzuki, N. Shimbara, Y. Hidaka, F. Osaka, M. Omata, and K. Tanaka. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kehn, K., L. C. de Fuente, K. Strouss, R. Berro, H. Jiang, J. Brady, R. Mahieux, A. Pumfery, M. E. Bottazzi, and F. Kashanchi. 2005. The HTLV-I Tax oncoprotein targets the retinoblastoma protein for proteasomal degradation. Oncogene 24:525-540. [DOI] [PubMed] [Google Scholar]
- 42.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. USA 102:18562-18566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayer, A., A. Wree, M. Hockel, C. Leo, H. Pilch, and P. Vaupel. 2004. Lack of correlation between expression of HIF-1α protein and oxygenation status in identical tissue areas of squamous cell carcinomas of the uterine cervix. Cancer Res. 64:5876-5881. [DOI] [PubMed] [Google Scholar]
- 44.Munger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78:11451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishinaka, T., Y. H. Fu, L. I. Chen, K. Yokoyama, and R. Chiu. 1997. A unique cathepsin-like protease isolated from CV-1 cells is involved in rapid degradation of retinoblastoma susceptibility gene product, RB, and transcription factor SP1. Biochim. Biophys. Acta 1351:274-286. [DOI] [PubMed] [Google Scholar]
- 47.Oh, K. J., A. Kalinina, J. Wang, K. Nakayama, K. I. Nakayama, and S. Bagchi. 2004. The papillomavirus E7 oncoprotein is ubiquitinated by UbcH7 and Cullin 1- and Skp2-containing E3 ligase. J. Virol. 78:5338-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan, Z. Q., A. Kentsis, D. C. Dias, K. Yamoah, and K. Wu. 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23:1985-1997. [DOI] [PubMed] [Google Scholar]
- 49.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 50.Phelps, W. C., K. Munger, C. L. Yee, J. A. Barnes, and P. M. Howley. 1992. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J. Virol. 66:2418-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phelps, W. C., C. L. Yee, K. Munger, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547. [DOI] [PubMed] [Google Scholar]
- 52.Piboonniyom, S., S. Duensing, N. W. Swilling, P. W. Hinds, and K. Münger. 2003. Abrogation of the retinoblastoma tumor suppressor checkpoint during keratinocyte immortalization is not sufficient for induction of centrosome-mediated genomic instability. Cancer Res. 63:476-483. [PubMed] [Google Scholar]
- 53.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 54.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riley, R. R., S. Duensing, T. Brake, K. Munger, P. F. Lambert, and J. M. Arbeit. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 63:4862-4871. [PubMed] [Google Scholar]
- 56.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-Protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 57.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 58.Talis, A. L., J. M. Huibregtse, and P. M. Howley. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 273:6439-6445. [DOI] [PubMed] [Google Scholar]
- 59.Tasaki, T., L. C. Mulder, A. Iwamatsu, M. J. Lee, I. V. Davydov, A. Varshavsky, M. Muesing, and Y. T. Kwon. 2005. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25:7120-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tedesco, D., J. Lukas, and S. I. Reed. 2002. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toussaint-Smith, E., D. B. Donner, and A. Roman. 2004. Expression of human papillomavirus type 16 E6 and E7 oncoproteins in primary foreskin keratinocytes is sufficient to alter the expression of angiogenic factors. Oncogene 23:2988-2995. [DOI] [PubMed] [Google Scholar]
- 62.Vousden, K. H., and P. S. Jat. 1989. Functional similarity between HPV16E7, SV40 large T and adenovirus E1a proteins. Oncogene 4:153-158. [PubMed] [Google Scholar]
- 63.Whyte, P., K. J. Buchkovich, J. M. Horowitz, S. H. Friend, M. Raybuck, R. A. Weinberg, and E. Harlow. 1988. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334:124-129. [DOI] [PubMed] [Google Scholar]
- 64.Wolf, D. A., C. Zhou, and S. Wee. 2003. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5:1029-1033. [DOI] [PubMed] [Google Scholar]
- 65.Wu, E. W., K. E. Clemens, D. V. Heck, and K. Munger. 1993. The human papillomavirus E7 oncoprotein and the cellular transcription factor E2F bind to separate sites on the retinoblastoma tumor suppressor protein. J. Virol. 67:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, K., A. Chen, and Z. Q. Pan. 2000. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275:32317-32324. [DOI] [PubMed] [Google Scholar]
- 67.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, B., W. Chen, and A. Roman. 2006. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc. Natl. Acad. Sci. USA 103:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]