Abstract
The structure of virus-like particles of the lymphotropic, immunosuppressive strain of minute virus of mice (MVMi) in complex with the neutralizing Fab fragment of the mouse monoclonal antibody (MAb) B7 was determined by cryo-electron microscopy to 7-Å resolution. The Fab molecule recognizes a conformational epitope at the vertex of a three-fold protrusion on the viral surface, thereby simultaneously engaging three symmetry-related viral proteins in binding. The location of the epitope close to the three-fold axis is consistent with the previous analysis of MVMi mutants able to escape from the B7 antibody. The binding site close to the symmetry axes sterically forbids the binding of more than one Fab molecule per spike. MAb as well as the Fab molecules inhibits the binding of the minute virus of mice (MVM) to permissive cells but can also neutralize MVM postattachment. This finding suggests that the interaction of B7 with three symmetry-related viral subunits at each spike hinders structural transitions in the viral capsid essential during viral entry.
A number of reports of structural studies of antibodies or antigen binding fragments (Fab) bound to icosahedral viruses, such as picornaviruses, flaviviruses, and parvoviruses, are available (24, 25, 31, 61, 64, 68, 74). It has previously been shown by mutational analyses that neutralizing antibodies are usually directed to major antigenic sites on the viral surface (54). In most cases, the Fab molecules have been found to extend radially outwards from the surface of the virus, permitting the cross-linking of virions by antibodies. Although structural studies show multiples of 60 Fab molecules bound to the viral surface, a lower level of occupancy may be sufficient for neutralization in many cases, indicating that many factors can be involved in determining neutralization efficiency (34).
Minute virus of mice (MVM) is an autonomous member of the Parvoviridae, a family of small, single-stranded DNA viruses. The two best-characterized variants of MVM, the prototype strain (MVMp) (16) and the immunosuppressive strain (MVMi) (7, 18, 50), are serologically indistinguishable but differ in their in vitro cell tropism (27, 66) and in vivo pathogenicity (10, 33, 58, 59), despite their high degree of sequence identity of about 97%. The MVM capsid is composed of a mixture of the viral proteins VP1 (83 kDa), VP2 (64 kDa), and VP3 (60 kDa) (67). VP2, the major capsid protein, is structurally equivalent to the C-terminal portion of VP1. Although VP2 is sufficient for the formation of icosahedral particles that are able to encapsidate DNA, these particles are not infectious (70). However, the presence of about nine VP1 molecules per 60 subunits renders a virion infectious (67, 70). VP3, a proteolytic cleavage product of VP2, has been observed only in DNA-containing particles (1, 13, 51, 67). Empty, recombinant virus-like particles (VLPs), self-assembled from MVMp VP2, are morphologically and immunologically similar to native, empty MVMp particles (11, 23, 35, 56), as demonstrated previously for other parvoviral VLPs (29, 38, 39, 57).
Similar to that of other autonomous parvoviruses, the icosahedral protein shell of MVM has depressions at the two-fold axes, canyon-like regions around the cylindrical structures at the five-fold axes, and protrusions at the three-fold symmetry axes (1, 40). The protrusions at or around the icosahedral three-fold axes have been implicated in parvoviral tropism (12, 35, 60), pathogenicity (49), antigenicity (6, 9, 36, 65, 76, 77), and receptor attachment (26, 32). Residues defining MVM tissue tropism and pathogenicity have been mapped to the three-dimensional (3D) atomic structures of the viral capsid (23, 35, 45). A major determinant of MVM tissue tropism was found in the area surrounding the depression at the icosahedral two-fold axis of the virion (4, 17, 21, 48). This dimple-like depression has also been identified as a sialic acid binding site of MVM and canine parvovirus (5, 44, 69).
The neutralizing monoclonal antibody (MAb) B7 elicited against MVMp is protective in passive immunotherapy against a lethal leukopenia disease in SCID mice caused by MVMi (43). However, MVMi evades the therapy by in vivo selection of antibody-resistant mutants on the viral surface. Analyses of antibody escape mutants indicated that residues 433 through 439 of VP2, located on the tip of the spike at the three-fold axes of symmetry, are a part of the B7 epitope (43). MAb B7 has also been used as a marker for intact capsids in in vivo studies of MVM infection (41, 42, 46), but the MAb is unable to recognize viral capsid protein (VP) trimers, a major assembly intermediate (52).
The present study examines the interaction of the Fab fragment of B7 with MVMi VLPs by cryo-electron microscopy (cryoEM). The fitting of a B7 Fab homology model into the cryoEM density map demonstrated that only one B7 Fab molecule per spike can be accommodated at the three-fold axes. The structural results, together with the analysis of B7 binding and neutralization activity, suggest that this antibody may hinder conformational transitions in the viral capsid required for the MVMi infection process.
MATERIALS AND METHODS
Production and purification of VLPs of MVMi.
Recombinant baculovirus expressing the VP2 protein of MVMi under the control of the polh promoter was constructed using the Bac-to-Bac baculovirus expression system (Invitrogen). The VP2-encoding sequence of the MVMi genome (2) was amplified by PCR using the primers VP2-Forward (5′ GCA GTG GGA TCC ATG AGT GAT GGC ACC AGC CAA C 3′) and VP2-Reverse (5′ AAG CAT CTC GAG TTA GTA AGT ATT TCT AGC AAC 3′) and inserted between the BamHI and XhoI restriction sites of the pFastBac1 shuttle vector. MVMi VLPs were purified from High Five insect cells by a modification of the procedure described by Hernando et al. (23). Infected cells (multiplicity of infection of 1) were harvested 2 days postinfection and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 100 mM NaCl, 0.2% sodium dodecyl sulfate). VLPs were purified from the lysate by sedimentation through a 20% sucrose cushion in the presence of 0.2% Triton X-100, followed by density gradient purification using a linear 10 to 40% sucrose gradient (68,000 × g for 6.5 h at 5°C). Fractions with hemagglutination activity were pooled and subjected to equilibrium centrifugation in a CsCl gradient at 150,000 × g for 24 h at 10°C. MVMi VLPs banding at a density of 1.32 g/cm3 were harvested and extensively dialyzed against phosphate-buffered saline (PBS). The purified sample was stored at 4°C after the addition of 0.02% sodium azide.
Production of B7 hybridoma cells and purification of Fab fragments from MAb B7.
Mice were subcutaneously immunized with purified and UV radiation (254-nm wavelength)-treated MVMp capsids in Freund's complete adjuvant, followed by two injections with the capsids in Freund's incomplete adjuvant. A final intravenous booster immunization with the same antigen was administered 4 days before splenectomy. Spleen cells were fused to Sp2/0-Ag14 mouse myeloma cells and selected using standard methods (20). Culture supernatants of the hybridomas were tested against purified MVMp capsids in an enzyme-linked immunosorbent assay, and positive cells were cloned by end point dilution. MAb B7, secreted by the hybridoma clone D4H1.B7, was of subtype γ1/κ as determined using the mouse typer subisotyping kit (Bio-Rad).
For B7 Fab production, ascitic fluid was recovered from BALB/c mice injected with 2 × 106 hybridoma D4H1.B7 cells per mouse. MAb B7 was purified by using protein A-Sepharose (Pharmacia) (71) and digested for 5 h at 37°C with soluble papain (Sigma) in digestion buffer (PBS [pH 7.2], 0.8 mM EDTA, 4.2 mM l-cysteine) at an immunoglobulin G (IgG)/enzyme ratio of 104:1 (wt/wt). Upon precipitation with 85% ammonium sulfate, the Fab moiety was purified by protein A-Sepharose and Sephacryl S-200 Spun (Pharmacia) chromatography.
MVM neutralization assays.
MVMi virions (150 PFU) were incubated with purified intact MAb or Fab fragments in 0.4 ml of PBS for 30 min at 37°C, and the remaining infectivity was determined in plaque assays using NB324K cells as described previously (43). One neutralization unit (Nu) was defined as the amount of MAb or Fab required to neutralize 50% of the virus. The inhibition of MVM capsid attachment to cells was tested in binding assays using 35S-labeled capsids (56). To test for neutralization postattachment, NB324K monolayers were inoculated with purified MVMi for 1 h at 4°C. After the removal of unbound virus by repeated washing with PBS, fresh culture medium was added to allow virus internalization at 37°C. At different time points postadsorption, 10 Nu of purified MAb B7/ml, an amount exceeding the concentration necessary for complete neutralization of the inoculum, was added. Incubation at 37°C was continued for 30 min before the replacement of the culture medium with plaque assay medium.
Determination of sequences of variable domains of B7 Fab.
Total RNA was extracted from hybridoma cells by using the RNeasy Midi kit (QIAGEN). The coding regions for the heavy and light (kappa) variable domains were amplified from the total RNA extracts by reverse transcriptase PCR with the Superscript III One-Step reverse transcriptase PCR kit (Invitrogen). For the heavy-chain gene, primer sequences were obtained from the Centre for Protein Research at the University of Cambridge, Cambridge, United Kingdom (http://www.mrc-cpe.cam.ac.uk/phage/mouse_seq.php). The selected primers bind to either the constant (5′ TAR CCY TTG ACM AGG CAT CC 3′) or the variable (5′ GGA ACC CTT TGG CCC AGC CGG CCA TGG CCS AGG TYC AGC TBC AGC AGT C 3′) domain of the rearranged IgG heavy-chain gene. The kappa chain primers bind to the rearranged kappa light-chain gene in either its constant (5′ GCC ATC AAT CTT CCA CTT GAC ACT GAT GTC 3′) or variable (5′ GTG ACA TTG TGA TGA CNC AGT CTC C 3′) domain. The amplification products were cloned into the vector pGEM-T Easy (Promega) and sequenced using T7 or SP6 primers.
Generation of a 3D homology model of B7 Fab.
A 3D homology model of the B7 variable domain (VL and VH) was obtained by submitting the amino acid sequences of the heavy and light chains (Table 1) to Web Antibody Modelling (http://antibody.bath.ac.uk/). Residues corresponding to the complementarity-defining regions (CDRs) in the model Protein Data Bank (PDB) file were as follows: residues 22 to 32 (CDR L1), 48 to 54 (CDR L2), 87 to 95 (CDR L3), 127 to 136 (CDR H1), 151 to 160 (CDR H2), and 200 to 208 (CDR H3). The canonical loops L2, L3, H1, and H2 were modeled with particularly high confidence.
TABLE 1.
Amino acid sequences of the light and heavy variable chains of the B7 Fab fragmenta
| Chain | Sequence |
|---|---|
| 1 11 21 31 41 51 61 | |
| Light (kappa) | ------KFMS TSVGDRVSIT CKASQDVSTT VAWYQEKPGQ SPKLLIYSAS YRFTGVPDRF TGSGFGT VFT |
| L1 L2 | |
| 71 81 91 101 | |
| FTISSVQAED LAVYYCQQHY STPWTFGGGT KLEIKRA | |
| L3 | |
| 101 111 121 131 141 151 161 | |
| Heavy | --------GA ELAKPGASVK MSCKASGYTF TNYWMHWVKQ RPGQGLEWIG YIYPTTGYTE YNQKFND KAT |
| H1H2 | |
| 171 181 191 201 211 | |
| FTADKSSNTA YMQLTSLTSG DSAVYYCARN PGDYYFDYWG QGTTLTVS | |
| H3 |
Sequences are shown as submitted to the Web Antibody Modelling server. The canonical and noncanonical CDR loops are double underlined and underlined, respectively, and labeled 1 through 3.
The B7 sequences were subjected to a BLAST search of transcripts from translated hybridoma open reading frames. A significant alignment with the variable domain of a mouse IgG1 antibody (PDB accession no. 1IGY) was found (22). The Cα atomic coordinates of the constant domain of accession no. 1IGY Fab2 were used for the B7 Fab constant domain (CL and CH1).
Complex formation, cryoEM, image analysis, and 3D image reconstruction.
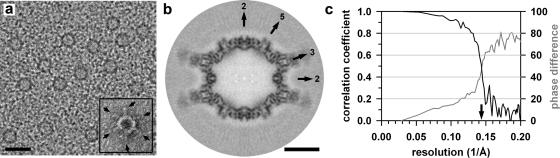
Purified MVMi VLPs were incubated with B7 Fab in the presence of 100 mM NaCl at 4°C overnight at a ratio of about four Fab molecules per VP2 subunit. Small aliquots of the sample were applied to 400-mesh copper grids coated with a holey carbon film and rapidly frozen by plunging into an ethane slush. Micrographs of the freeze hydrated samples were recorded on SO-163 films (Kodak, Rochester, NY) with a CM300 field emission gun transmission electron microscope (Philips, Eindhoven, The Netherlands). Images were taken at a calibrated magnification of ×47,190 and a total electron dose of approximately 22 electrons/Å2. The cryoEM micrographs were digitized at 7-μm intervals by using a Zeiss SCAI scanner (see Fig. 1a).
FIG. 1.
CryoEM analysis of MVMi VLPs in complex with the Fab fragment of the neutralizing anti-VP2 MAb B7. (a) CryoEM micrograph of the vitrified complex. The insert shows an Fab molecule (arrows)-decorated MVMi VLP negatively stained with uranyl acetate at a slightly higher magnification. The scale bar represents 500 Å. (b) Central cross-section of the 3D image reconstruction of the complex viewed down an icosahedral two-fold axis. The positions of icosahedral two-, three-, and five-fold axes are indicated. The scale bar represents 100 Å. (c) Resolution assessment of the 3D reconstruction. Correlation coefficients (black) and phase differences (gray) for different resolution shells were computed from the structure factors derived from the independent reconstructions of two half data sets. These plots demonstrate that the data are reliable to at least 7-Å resolution (arrow) based on a correlation coefficient of 0.5 or a phase difference of 45°.
A cryoEM density map of recombinant B19 VP2 capsids (30) was used to initialize the 3D reconstruction. The orientations and origins of all the particles were determined by comparing the contrast transfer function-corrected particle images against reference projections of the current electron microscopy-reconstructed map by using the programs EMPFT (3) and POR (28). The procedure was repeated at progressively higher resolutions until no further increase in the correlation coefficients could be obtained. To improve the reliability of the orientation refinement, the image data were band-pass filtered to reduce low- and high-frequency noise. The resolution of the resulting map was estimated by comparing structure factors of the virus shell computed from two independent half data sets (Table 2). For the final 3D reconstruction, data were included to a resolution at which the correlation between the two independent data sets was 0.3 (see Fig. 2). The magnification of the cryoEM map was standardized to a map calculated from the atomic coordinates of the MVMi VP2 structure (PDB accession no. 1Z1C) (35), resulting in a final pixel separation of 1.47 Å. A difference map between the cryoEM map of the MVM-Fab complex and the MVMi map derived from the atomic coordinates was calculated after scaling the densities to optimize the radial dimensions and average density levels (see Fig. 2). The program RobEM (http://cryoem.ucsd.edu/programDocs/runRobem.txt) was used to eliminate small islands of density, which were probably merely noise in the difference density map, for visualization.
TABLE 2.
Image data for cryoEM reconstruction
| Parameter | Value or identity |
|---|---|
| Specimen | MVMi VLPs with bound B7 Fab molecules |
| No. of micrographs | 14 |
| Defocus level range (μm) | 3.20-2.05 |
| No. of particles selected from micrographs | 9,682 |
| No. of particles used for reconstruction | 6,375 |
| Final resolution (Å) | 7.0a (6.8b) |
Resolution at a Fourier shell correlation coefficient of 0.5 between half data sets.
Resolution at a Fourier shell correlation coefficient of 0.3 between half data sets.
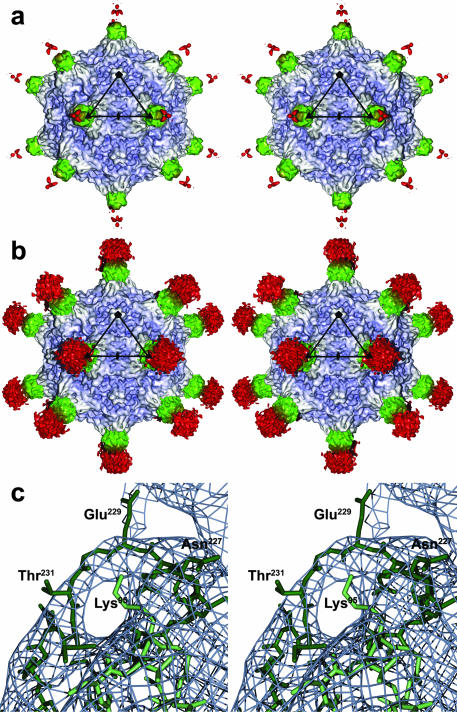
FIG. 2.
3D image reconstruction of MVMi in complex with B7 Fab molecules at 7-Å resolution. (a) Stereo view of a surface rendering of the complex, viewed down an icosahedral two-fold axis, showing the virus (blue-gray) and bound Fab. The variable and constant domains of the Fab fragment are colored green and red, respectively. The black triangle marks an icosahedral asymmetric unit. (b) Same as panel a, but the Fab density was rendered at a lower contour level. (c) A stereo diagram of a section of the cryoEM map shows density that can be attributed to loop 2 of MVM VP2 (dark green; residues 221 to 240) located on the shoulder of a three-fold spike. The presence of several resolved surface loops is an indicator of the quality of the cryoEM density map.
Fitting of the B7 homology model into the cryoEM density map.
The homology models of the B7 Fab variable domain (VL and VH) and the constant domain (CL and CH1) were independently fitted into the cryoEM difference density map by using the program EMfit, version SR5 (55) (Table 3). The fitting was constrained by icosahedral symmetry. This fitting procedure compares the observed cryoEM density with simultaneously positioned symmetry-related molecules. Due to the Fab binding close to the three-fold axes of symmetry, clashes between symmetry-related Fab molecules had to be ignored during the fitting procedure. The B7 variable domain was fitted first into the difference map by using main-chain and side chain atoms. A maximum distance restraint of 15 Å between Thr437 of MVMi VP2 and Ser91 of the light chain (CDR L3) and Asp203 of the heavy chain (CDR H3) of B7 Fab was used to maintain the general orientation of the Fab CDRs towards the viral surface. The Fab structure was completed by subsequent fitting of the Cα atoms of the constant domain while restraining the distance between the C and N termini of the variable and constant portions of the light and heavy chains, respectively, to less than 15 Å. The elbow angle of the resultant Fab structure was determined using the program RBOW (63). The contact region between the MVM three-fold spike (composed of the three symmetry-related VP2 molecules A, B, and C) and one Fab molecule was analyzed using the programs AREAIMOL (37) and CONTACTS of the CCP4 program suite (14) (see Fig. 3 and 4).
TABLE 3.
Statistics for fitting of the Fab atomic coordinates into the cryoEM difference density mapa
| Structural unit of Fab | Atom(s) fitted | sumf (%) | −den (%) | avgds (Å) |
|---|---|---|---|---|
| Variable domain (VL and VH) | All | 2.4 | 4.5 | 8.5 |
| Constant domain (CL and CH1) | Cα | 1.5 | 1.4 | 5.4 |
sumf is defined as the average density value for all atomic positions as normalized by the root-mean-square deviation from the mean of the map. −den is the percentage of atoms that are positioned in negative density. avgds is the average distance between restraining and target positions. For a more detailed definition of these terms, see the program EMfit (55).
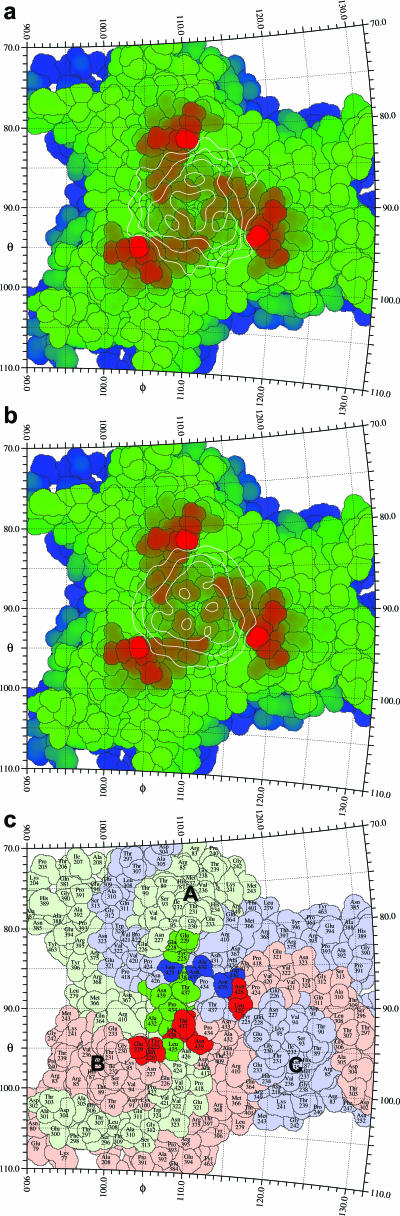
FIG. 3.
Stereographic road map projection of the MVM three-fold protrusion formed by surface loops of symmetry-related VP2 molecules. (a) Difference cryoEM density representing the variable domain of B7 radially projected onto the spike. VP2 amino acids are colored according to their distance from the viral center, with red amino acids being farthest away. (b) Same as panel a, but the density was calculated from the trimer of the fitted variable domain after assigning a relative occupancy of 0.33 to each atom because steric hindrance precludes the binding of more than one Fab molecule to the site. Note the similarity between the observed density in panel a and the calculated density of the fitted trimer in panel b. (c) A single B7 Fab molecule recognizes the three symmetry-related MVMi VP2 molecules forming the three-fold spike asymmetrically. The VP2 molecules A, B, and C are colored green, red, and blue, respectively. Amino acids that define the contact region of one bound Fab molecule are highlighted in darker hues of the same colors.
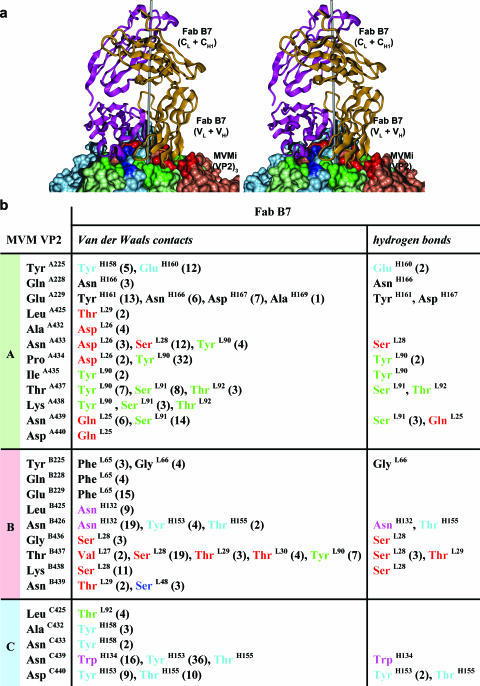
FIG. 4.
Interaction of one B7 Fab molecule with an MVMi VP2 trimeric spike. (a) Stereo view of the complex. The icosahedral three-fold axis (gray) is slightly tilted towards the viewer. The three VP2 molecules A, B, and C forming the protrusion at an icosahedral three-fold axis are colored green, blue, and red, respectively, as in Fig. 3c. The light and heavy chains of the Fab molecule are shown in gold and magenta. (b) Atomic contacts at the interface between three symmetry-related MVM VP2 molecules (A, B, and C) and one B7 Fab molecule. Contact residues of B7 Fab molecules that are part of the CDRs of the homology model are color coded: L1, red; L2, blue; L3, green; H1, magenta; and H2, cyan. The numbers in parentheses denote the numbers of contacts or putative bonds, if >1. The selection of van der Waals contacts was based on an interatomic distance of ≤4.0 Å. The prediction of hydrogen bonds was based on an interatomic distance of ≤3.5 Å and default geometry defined in the program CONTACTS of the CCP4 program suite (14). Residue numbers in A, B, and C molecules and light (L) and heavy (H) chains are indicated as superscripts.
Figure preparation.
Figures were created using the programs DINO: Visualizing Structural Biology (2002; http://www.dino3d.org), RIVEM (75), and POV-Ray (2004; Persistence of Vision Pty. Ltd. [http://www.povray.org]).
Accession numbers.
The consensus sequences of the coding regions for the heavy and light chains of B7 Fab have been deposited in GenBank under accession numbers EF506758 and EF506759. The cryoEM density map of the MVM-Fab complex has been deposited in the Electron Microscopy Data Bank under accession number 1326.
RESULTS AND DISCUSSION
Structural analysis of the Fab-VLP complex.
Complexes of purified MVMi VLPs with the Fab fragment of MAb B7 were analyzed by cryoEM. The 3D density map of MVMi VLPs in complex with the B7 Fab fragment has an estimated resolution of about 7 Å (Table 2; Fig. 1 and 2). The quality of the cryoEM reconstruction was confirmed by observing that the β-sheets in the jelly roll β-barrel and even some loops on the outer surface of the virus could be clearly recognized (Fig. 2c). The most significant difference in density between the complex and MVM, interpreted as the bound Fab molecule, was found close to the icosahedral three-fold axes, protruding from each of the spikes. The binding of the Fab molecule to the three-fold spikes is consistent with the location of MVMi escape mutations from the neutralizing B7 antibody, which had been mapped to the tips of the spikes (43). Due to steric interference close to the icosahedral three-fold axis, only one Fab molecule can bind to a three-fold spike at one time, resulting in a maximum of 20 bound Fab molecules per virus. Each of the three symmetry-related epitopes at the tip of any one spike has an equal chance of being recognized by an Fab molecule. Therefore, the cryoEM difference density represents the average of the three different Fab orientations, resulting in a radially distributed, bilobal appearance of the Fab electron density (Fig. 2). The radially inner and outer lobes presumably correspond to the variable and constant domains of B7 Fab, respectively. A similar density had been observed for an Fab molecule bound close to a five-fold symmetry axis of cucumber mosaic virus (8). The density related to the constant domain of Fab is about 0.4-fold lower than that related to the variable domain, suggesting flexibility of the elbow angle. The Fab density is almost as strong as the density of the MVM protein shell, where the three-fold-related Fab variable domains overlap, demonstrating about 90% occupancy of the spikes by B7 Fab. By using the fitting process described in Materials and Methods, an Fab molecule that had an elbow angle of about 170° was extracted (Fig. 4).
The contact region of one Fab molecule spans all three symmetry-related MVM VP2 molecules (A, B, and C) that form the spike (Fig. 3 and 4). The VP2 residues involved in the contact are 225, 228, 229, 425, and 426 and the loop of residues 432 to 440. These residues are completely conserved between MVMi and MVMp, which explains why the MAb B7 raised against MVMp (see Materials and Methods) binds and neutralizes MVMi. Some of these residues are employed in all three symmetry-related VP2 molecules, whereas others are used in only one or two VP2 molecules, resulting in an asymmetric epitope on a symmetric antigen. In other words, the antibody recognizes an epitope that is located at a symmetric vertex but that is formed by different sets of amino acid residues from each of the three symmetry-related VP subunits. The simultaneous recognition of three VP2 subunits explains the B7 specificity for intact capsids and its inability to bind individual VPs (41, 42, 47). The B7 epitope is at the location farthest away from the interfaces between VP trimers, which are intermediates in the MVM capsid assembly (52). However, B7 is not able to recognize VP trimers that accumulate intracellularly (52), suggesting that the assembly of trimers into a capsid involves conformational changes that propagate from the trimer edges to the vertex of the three-fold spike.
The B7 contact region seen in the cryoEM structure includes the MVM residues 433 to 439 previously identified by antibody escape mutant analysis as part of the B7 epitope (43). About 70% of the van der Waals contacts and putative hydrogen bonds are between loop residues 432 and 440 of MVM and the CDRs L1, L3, and H2 of B7 Fab. One B7 Fab molecule recognizes some of these residues in more than one of the three three-fold-related subunits at any one vertex (Fig. 4). This mode of binding would facilitate the escape of the virus from antibody recognition, as a single mutation could entail the simultaneous replacement of two or even three residues involved in the B7 epitope, leading to the loss of a substantial number of antigen-antibody interactions.
Dual mechanism of MVMi neutralization by B7.
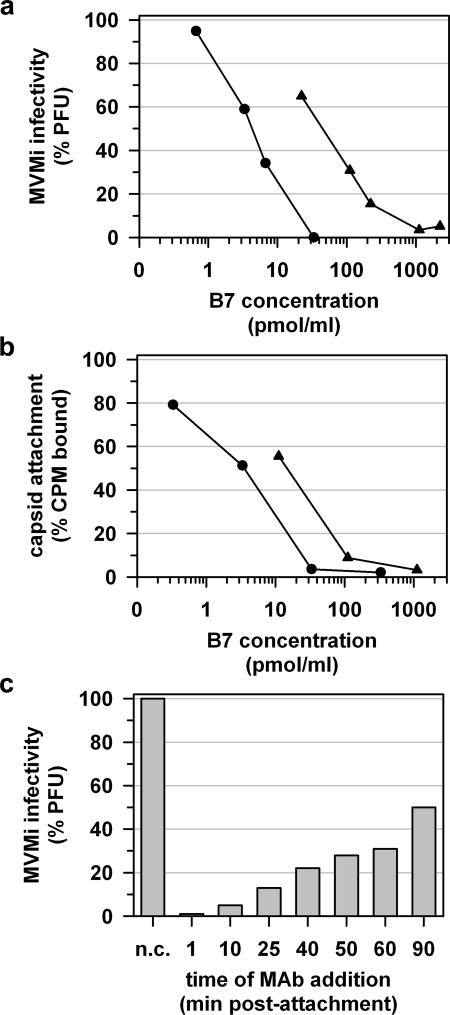
The orientation of bound B7 Fab, roughly perpendicular to the virus surface, makes it unlikely that the antibody would be able to bridge neighboring three-fold spikes. Indeed, as both B7 IgG and Fab fragments neutralize the MVMi plaque-forming capacity (Fig. 5a), antibody bivalency does not appear to be required for neutralization. The B7 antibody, as well as its Fab fragment, blocked the attachment of MVM capsids to permissive cells (Fig. 5b). This blockage is most likely the result of steric hindrance of virus-receptor contact by the antibody.
FIG. 5.
Neutralization of MVMi by MAb B7 and its Fab fragment. (a) MAb B7 and the B7 Fab fragment efficiently neutralize MVMi infection. Plaque formation in permissive cells is inhibited after the preincubation of MVMi virions with various amounts of antibody (•) or Fab (▴). The percentage of PFU is defined relative to the number of PFU of nonneutralized virus. (b) MAb B7 and the B7 Fab fragment interfere with virus-cell recognition. The number of 35S-labeled MVMi VLPs attached to cells decreases with increasing amounts of antibody (•) or Fab (▴). The percentage of counts per minute (CPM) is defined relative to the level of capsid attachment in the absence of MAb or the Fab fragment. (c) MAb B7 can neutralize MVMi infection postattachment. The reduction of MVMi plaque-forming capacity depends on the time of MAb B7 addition after the initiation of internalization. A result representative of two independent experiments is shown. The percentage of PFU is defined relative to the number of PFU of nonneutralized virus. n.c., negative control, no MAb added.
MVM infection can also be neutralized by MAb B7 added at different time points after virus attachment (Fig. 5c), indicating either that MAb can compete with receptor binding or that the antibody-spike interaction may block an essential step in the postbinding viral entry pathway. Similar to other parvoviruses, MVM enters the host cell via receptor-mediated endocytosis (72), although the cytoplasmic trafficking of the viral genome is a poorly understood process. Postattachment B7 binding to VP2 trimers may neutralize MVM by blocking pH-induced structural transitions of the virion (19), by hindering the exposure of the VP1 unique region and genome release (11, 15, 46, 53, 73), or by interfering with the nuclear invasion by the viral particle (62). Postattachment neutralizing mechanisms may enhance the B7 neutralization efficiency, if an antibody concentration that is sufficient for the inhibition of virus attachment cannot be achieved.
Acknowledgments
We are grateful to Yasmina Ruiz for help in the production of the VLPs and to Sharon Wilder, Sheryl Kelly, and Cheryl Towell for help in the preparation of the manuscript.
This work was supported by NIH grants AI-11219 to M.G.R. and AI-33468 to C.R.P. and M.G.R., “Comunidad de Madrid” grant S-SAL/0185/2006 to J.M.A., a grant from the Ministerio de Educación y Ciencia to M.G.M. (BIO2006-00793), and an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa. We are grateful for a Keck Foundation grant for the purchase of the CM300 field emission gun electron microscope used in these studies.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 2.Astell, C. R., E. M. Gardiner, and P. Tattersall. 1986. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J. Virol. 57:656-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 4.Ball-Goodrich, L. J., and P. Tattersall. 1992. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J. Virol. 66:3415-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbis, D. P., S. F. Chang, and C. R. Parrish. 1992. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology 191:301-308. [DOI] [PubMed] [Google Scholar]
- 6.Bloom, M. E., D. A. Martin, K. L. Oie, M. E. Huhtanen, F. Costello, J. B. Wolfinbarger, S. F. Hayes, and M. Agbandje-McKenna. 1997. Expression of Aleutian mink disease parvovirus capsid proteins in defined segments: localization of immunoreactive sites and neutralizing epitopes to specific regions. J. Virol. 71:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnard, G. D., E. K. Manders, D. A. Campbell, Jr., R. B. Herberman, and M. J. Collins, Jr. 1976. Immunosuppressive activity of a subline of the mouse EL-4 lymphoma. Evidence for minute virus of mice causing the inhibition. J. Exp. Med. 143:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman, V. D., E. S. Chase, A. W. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, C. S., T. Jensen, R. H. Meloen, W. Puijk, K. Sugamura, H. Sato, and W. J. Spaan. 1992. Localization of an immunodominant domain on baculovirus-produced parvovirus B19 capsids: correlation to a major surface region on the native virus particle. J. Virol. 66:6989-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownstein, D. G., A. L. Smith, R. O. Jacoby, E. A. Johnson, G. Hansen, and P. Tattersall. 1991. Pathogenesis of infection with a virulent allotropic variant of minute virus of mice and regulation by host genotype. Lab. Investig. 65:357-364. [PubMed] [Google Scholar]
- 11.Carreira, A., M. Menéndez, J. Reguera, J. M. Almendral, and M. G. Mateu. 2004. In vitro disassembly of a parvovirus capsid and effect on capsid stability of heterologous peptide insertions in surface loops. J. Biol. Chem. 279:6517-6525. [DOI] [PubMed] [Google Scholar]
- 12.Chang, S. F., J. Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinton, G. M., and M. Hayashi. 1976. The parvovirus MVM: a comparison of heavy and light particle infectivity and their density conversion in vitro. Virology 74:57-63. [DOI] [PubMed] [Google Scholar]
- 14.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 15.Cotmore, S. F., A. M. D'Abramo, Jr., C. M. Ticknor, and P. Tattersall. 1999. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology 254:169-181. [DOI] [PubMed] [Google Scholar]
- 16.Crawford, L. V. 1966. A minute virus of mice. Virology 29:605-612. [DOI] [PubMed] [Google Scholar]
- 17.D'Abramo, A. M., Jr., A. A. Ali, F. Wang, S. F. Cotmore, and P. Tattersall. 2005. Host range mutants of minute virus of mice with a single VP2 amino acid change require additional silent mutations that regulate NS2 accumulation. Virology 340:143-154. [DOI] [PubMed] [Google Scholar]
- 18.Engers, H. D., J. A. Louis, R. H. Zubler, and B. Hirt. 1981. Inhibition of T cell-mediated functions by MVM(i), a parvovirus closely related to minute virus of mice. J. Immunol. 127:2280-2285. [PubMed] [Google Scholar]
- 19.Farr, G. A., S. F. Cotmore, and P. Tattersall. 2006. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N terminus and the genome of minute virus of mice. J. Virol. 80:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galfrè, G., and C. Milstein. 1981. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 73:3-46. [DOI] [PubMed] [Google Scholar]
- 21.Gardiner, E. M., and P. Tattersall. 1988. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J. Virol. 62:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris, L. J., E. Skaletsky, and A. McPherson. 1998. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275:861-872. [DOI] [PubMed] [Google Scholar]
- 23.Hernando, E., A. L. Llamas-Saiz, C. Foces-Foces, R. McKenna, I. Portman, M. Agbandje-McKenna, and J. M. Almendral. 2000. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology 267:299-309. [DOI] [PubMed] [Google Scholar]
- 24.Hewat, E. A., and D. Blaas. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 15:1515-1523. [PMC free article] [PubMed] [Google Scholar]
- 25.Hewat, E. A., N. Verdaguer, I. Fita, W. Blakemore, S. Brookes, A. King, J. Newman, E. Domingo, M. G. Mateu, and D. I. Stuart. 1997. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 16:1492-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itah, R., J. Tal, and C. Davis. 2004. Host cell specificity of minute virus of mice in the developing mouse embryo. J. Virol. 78:9474-9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji, Y., D. C. Marinescu, W. Zhang, X. Zhang, X. Yan, and T. S. Baker. 2006. A model-based parallel origin and orientation refinement algorithm for cryoTEM and its application to the study of virus structures. J. Struct. Biol. 154:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajigaya, S., H. Fujii, A. Field, S. Anderson, S. Rosenfeld, L. J. Anderson, T. Shimada, and N. S. Young. 1991. Self-assembled B19 parvovirus capsids, produced in a baculovirus system, are antigenically and immunogenically similar to native virions. Proc. Natl. Acad. Sci. USA 88:4646-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufmann, B., U. Baxa, P. R. Chipman, M. G. Rossmann, S. Modrow, and R. Seckler. 2005. Parvovirus B19 does not bind to membrane-associated globoside in vitro. Virology 332:189-198. [DOI] [PubMed] [Google Scholar]
- 31.Kaufmann, B., G. E. Nybakken, P. R. Chipman, W. Zhang, M. S. Diamond, D. H. Fremont, R. J. Kuhn, and M. G. Rossmann. 2006. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. USA 103:12400-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kern, A., K. Schmidt, C. Leder, O. J. Müller, C. E. Wobus, K. Bettinger, C. W. der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimsey, P. B., H. D. Engers, B. Hirt, and C. V. Jongeneel. 1986. Pathogenicity of fibroblast- and lymphocyte-specific variants of minute virus of mice. J. Virol. 59:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klasse, P. J., and Q. J. Sattentau. 2002. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 83:2091-2108. [DOI] [PubMed] [Google Scholar]
- 35.Kontou, M., L. Govindasamy, H. J. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M. P. Rubio, R. McKenna, J. M. Almendral, and M. Agbandje-McKenna. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 79:10931-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langeveld, J. P., J. I. Casal, C. Vela, K. Dalsgaard, S. H. Smale, W. C. Puijk, and R. H. Meloen. 1993. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J. Virol. 67:765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, B., and F. M. Richards. 1971. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55:379-400. [DOI] [PubMed] [Google Scholar]
- 38.Le Gall-Reculé, G., V. Jestin, P. Chagnaud, P. Blanchard, and A. Jestin. 1996. Expression of muscovy duck parvovirus capsid proteins (VP2 and VP3) in a baculovirus expression system and demonstration of immunity induced by the recombinant proteins. J. Gen. Virol. 77:2159-2163. [DOI] [PubMed] [Google Scholar]
- 39.Livingston, R. S., D. G. Besselsen, E. K. Steffen, C. L. Besch-Williford, C. L. Franklin, and L. K. Riley. 2002. Serodiagnosis of mice minute virus and mouse parvovirus infections in mice by enzyme-linked immunosorbent assay with baculovirus-expressed recombinant VP2 proteins. Clin. Diagn. Lab. Immunol. 9:1025-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llamas-Saiz, A. L., M. Agbandje-McKenna, W. R. Wikoff, J. Bratton, P. Tattersall, and M. G. Rossmann. 1997. Structure determination of minute virus of mice. Acta Crystallogr. D 53:93-102. [DOI] [PubMed] [Google Scholar]
- 41.Lombardo, E., J. C. Ramírez, M. Agbandje-McKenna, and J. M. Almendral. 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lombardo, E., J. C. Ramírez, J. Garcia, and J. M. Almendral. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 76:7049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López-Bueno, A., M. G. Mateu, and J. M. Almendral. 2003. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J. Virol. 77:2701-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López-Bueno, A., M. P. Rubio, N. Bryant, R. McKenna, M. Agbandje-McKenna, and J. M. Almendral. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 80:1563-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López-Bueno, A., L. P. Villarreal, and J. M. Almendral. 2006. Parvovirus variation for disease: a difference with RNA viruses? Curr. Top. Microbiol. Immunol. 299:349-370. [DOI] [PubMed] [Google Scholar]
- 46.Mani, B., C. Baltzer, N. Valle, J. M. Almendral, C. Kempf, and C. Ros. 2006. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J. Virol. 80:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maroto, B., N. Valle, R. Saffrich, and J. M. Almendral. 2004. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J. Virol. 78:10685-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maxwell, I. H., A. L. Spitzer, F. Maxwell, and D. J. Pintel. 1995. The capsid determinant of fibrotropism for the MVMp strain of minute virus of mice functions via VP2 and not VP1. J. Virol. 69:5829-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, J. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, J. B. Wolfinbarger, and M. Agbandje-McKenna. 1999. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J. Virol. 73:6882-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMaster, G. K., P. Beard, H. D. Engers, and B. Hirt. 1981. Characterization of an immunosuppressive parvovirus related to the minute virus of mice. J. Virol. 38:317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paradiso, P. R. 1981. Infectious process of the parvovirus H-1: correlation of protein content, particle density, and viral infectivity. J. Virol. 39:800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riolobos, L., J. Reguera, M. G. Mateu, and J. M. Almendral. 2006. Nuclear transport of trimeric assembly intermediates exerts a morphogenetic control on the icosahedral parvovirus capsid. J. Mol. Biol. 357:1026-1038. [DOI] [PubMed] [Google Scholar]
- 53.Ros, C., C. Baltzer, B. Mani, and C. Kempf. 2006. Parvovirus uncoating in vitro reveals a mechanism of DNA release without capsid disassembly and striking differences in encapsidated DNA stability. Virology 345:137-147. [DOI] [PubMed] [Google Scholar]
- 54.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, and A. G. Mosser. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 55.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with x-ray crystallographic structures. J. Struct. Biol. 136:190-200. [DOI] [PubMed] [Google Scholar]
- 56.Rubio, M. P., A. López-Bueno, and J. M. Almendral. 2005. Virulent variants emerging in mice infected with the apathogenic prototype strain of the parvovirus minute virus of mice exhibit a capsid with low avidity for a primary receptor. J. Virol. 79:11280-11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saliki, J. T., B. Mizak, H. P. Flore, R. R. Gettig, J. P. Burand, L. E. Carmichael, H. A. Wood, and C. R. Parrish. 1992. Canine parvovirus empty capsids produced by expression in a baculovirus vector: use in analysis of viral properties and immunization of dogs. J. Gen. Virol. 73:369-374. [DOI] [PubMed] [Google Scholar]
- 58.Segovia, J. C., J. M. Gallego, J. A. Bueren, and J. M. Almendral. 1999. Severe leukopenia and dysregulated erythropoiesis in SCID mice persistently infected with the parvovirus minute virus of mice. J. Virol. 73:1774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segovia, J. C., A. Real, J. A. Bueren, and J. M. Almendral. 1991. In vitro myelosuppressive effects of the parvovirus minute virus of mice (MVMi) on hematopoietic stem and committed progenitor cells. Blood 77:980-988. [PubMed] [Google Scholar]
- 60.Simpson, A. A., B. Hébert, G. M. Sullivan, C. R. Parrish, Z. Zádori, P. Tijssen, and M. G. Rossmann. 2002. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 315:1189-1198. [DOI] [PubMed] [Google Scholar]
- 61.Smith, T. J. 2003. Structural studies on antibody-virus complexes. Adv. Protein Chem. 64:409-453. [DOI] [PubMed] [Google Scholar]
- 62.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 80:11040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanfield, R. L., A. Zemla, I. A. Wilson, and B. Rupp. 2006. Antibody elbow angles are influenced by their light chain class. J. Mol. Biol. 357:1566-1574. [DOI] [PubMed] [Google Scholar]
- 64.Stewart, P. L., C. Y. Chiu, S. Huang, T. Muir, Y. Zhao, B. Chait, P. Mathias, and G. R. Nemerow. 1997. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 16:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strassheim, M. L., A. Gruenberg, P. Veijalainen, J. Y. Sgro, and C. R. Parrish. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175-184. [DOI] [PubMed] [Google Scholar]
- 66.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tattersall, P., P. J. Cawte, A. J. Shatkin, and D. C. Ward. 1976. Three structural polypeptides coded for by minute virus of mice, a parvovirus. J. Virol. 20:273-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thouvenin, E., S. Laurent, M. F. Madelaine, D. Rasschaert, J. F. Vautherot, and E. A. Hewat. 1997. Bivalent binding of a neutralising antibody to a calicivirus involves the torsional flexibility of the antibody hinge. J. Mol. Biol. 270:238-246. [DOI] [PubMed] [Google Scholar]
- 69.Tresnan, D. B., L. Southard, W. Weichert, J. Y. Sgro, and C. R. Parrish. 1995. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology 211:123-132. [DOI] [PubMed] [Google Scholar]
- 70.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verdaguer, N., N. Sevilla, M. L. Valero, D. Stuart, E. Brocchi, D. Andreu, E. Giralt, E. Domingo, M. G. Mateu, and I. Fita. 1998. A similar pattern of interaction for different antibodies with a major antigenic site of foot-and-mouth disease virus: implications for intratypic antigenic variation. J. Virol. 72:739-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vihinen-Ranta, M., S. Suikkanen, and C. R. Parrish. 2004. Pathways of cell infection by parvoviruses and adeno-associated viruses. J. Virol. 78:6709-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wikoff, W. R., G. Wang, C. R. Parrish, R. H. Cheng, M. L. Strassheim, T. S. Baker, and M. G. Rossmann. 1994. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure 2:595-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao, C., and M. G. Rossmann. 2007. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoshimoto, K., S. Rosenfeld, N. Frickhofen, D. Kennedy, R. Hills, S. Kajigaya, and N. S. Young. 1991. A second neutralizing epitope of B19 parvovirus implicates the spike region in the immune response. J. Virol. 65:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]