Abstract
Based on a reverse genetics approach, we previously reported that bovine leukemia virus (BLV) mutants harboring deletions in the accessory R3 and G4 genes persist at very low proviral loads and are unable to induce leukemia or lymphoma in sheep, indicating that these R3 and G4 gene sequences are required for pathogenesis. We now show that lymphoma can occur, albeit infrequently (1 case of 20) and after extended periods of latency (7 years). Direct sequencing and reinfection experiments demonstrated that lymphomagenesis was not due to the reversion of the mutant to the wild type. Similar observations with another type of attenuated mutant impaired in the transmembrane protein (TM) YXXL signaling motifs were made. We conclude that the R3 and G4 genes and the TM YXXL motifs are not strictly required for pathogenesis but that their integrity contributes to disease frequency and latency.
Viral persistence results from a dynamic interplay between the host immune defense and the replicative capacity of the virus. Bovine leukemia virus (BLV) replication occurs via two main processes: the infection of new cellular targets and the mitosis of the host cell (recently reviewed in references 7 and 11). Although these two mechanisms can theoretically occur simultaneously, the former operates mainly in the early stages of infection, i.e., the seroconversion period. In contrast, viral spread by mitotic cell division accounts for most of the proviral loads during the asymptomatic and leukemic phases (20). Amazingly, the leukemic clone that generates the tumor originates from a cell that was infected soon after seroconversion. It seems, therefore, that the premalignant cell clone and/or its progeny persist throughout the long latency period. However, the molecular mechanisms that govern viral persistence and spread are still largely unknown.
In an attempt to define the viral determinants required for transformation, we first established a model system based on the infection of sheep with a cloned BLV provirus (36). Among a series of proviruses, the 344 strain recapitulated all aspects associated with a lymphoproliferative disease followed by the onset of leukemia, lymphoma, or lymphosarcoma. In order to discover the role of viral genes in the leukemogenesis process (which includes all events leading to leukemia, lymphoma, or lymphosarcoma), we used reverse genetics by introducing directed deletions, insertions, and point mutations into the 344 proviral clone (35). By this approach, we previously demonstrated that the R3 and G4 accessory genes are dispensable for infection but are required for the maintenance of high proviral loads (34). Furthermore, the G4 protein, but not R3, exhibits transforming potential in primary rat embryo fibroblasts when coexpressed with the Ha-ras oncogene (15). Indeed, the G4- and Ha-ras-double-positive cells form transformed foci in cell cultures and induce tumors in nude mice. Mechanistically, G4 interacts with farnesyl pyrophosphate synthetase, a protein involved in the mevalonate-squalene pathway (17). Finally, proviruses mutated in G4 were found to be nonpathogenic in 13 sheep over a period of 40 months postinoculation while the wild-type strain 344 induced leukemia or lymphoma in 19 of 23 infected animals (15).
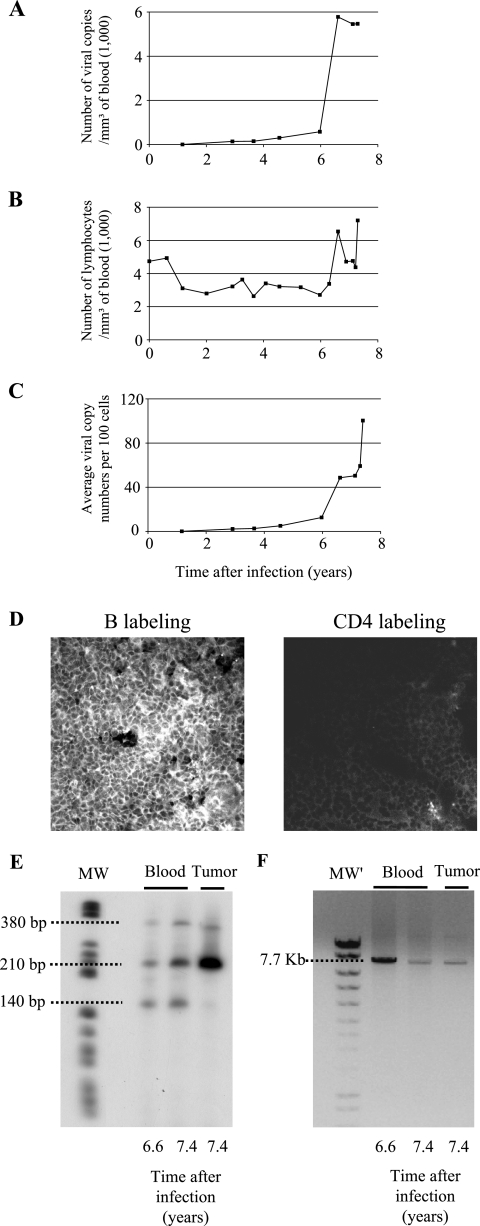
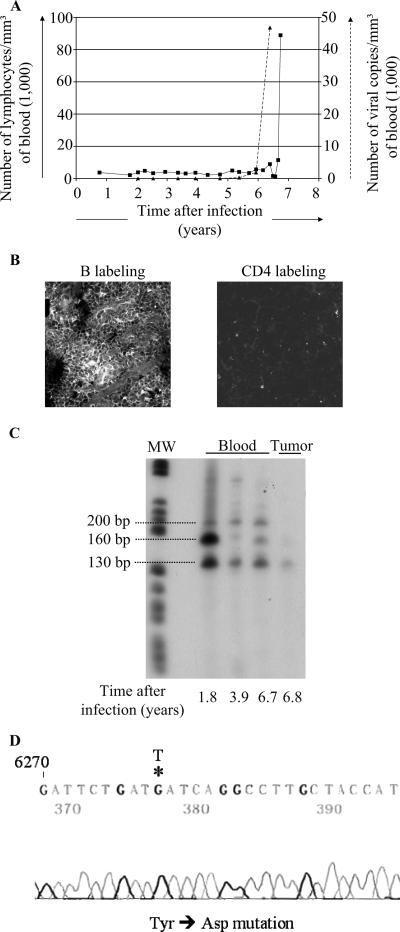
In Table 1, we now present the results of a 15-year survey of 53 sheep infected with either wild-type (n = 29) or mutant (n = 24) BLV proviruses. Some sheep died from unrelated causes (e.g., accidents, diarrhea, scabies, or old age) or were euthanized because the experiment was discontinued. The latency period preceding leukemogenesis in wild-type virus-infected sheep ranged between 13 and 80 months (mean, 36 months). In contrast, R3-G4 mutant proviruses were nonpathogenic in 19 animals but not in sheep 245. In this animal, the proviral loads, as determined by real-time PCR (18), increased sharply 6 years postinoculation, reaching almost 6,000 viral genome copies per mm3 of blood (Fig. 1A). Although the cell counts remained below the lymphocytic threshold arbitrarily set at 10,000 lymphocytes per mm3 (Fig. 1B), these levels corresponded to approximately one viral genome copy per cell (Fig. 1C). More importantly, multiple tumors appeared in several lymphoid tissues, including a mesenteric lymph node. A cryosection excised from this lymph node tumor and labeled with anti-CD21 showed that the tumor was of B-cell origin (Fig. 1D). Inverse PCR, performed as described in reference 20, demonstrated that three infected cell clones expanded in the blood 10 months before death and that one of them became predominant in the solid tumor (Fig. 1E). Finally, a complete proviral genome copy was inserted into the cellular genome, as shown by PCR (Fig. 1F) and Southern blot hybridization (data not shown).
TABLE 1.
Summary of results of infection of sheep with wild-type and mutant BLV proviruses
| Sheepa | Type of virus | Provirus strain | Date (mo-yr) of:
|
Cause of deathi | Period (mos) of latencyj | Mean duration (mos) of latencyj | |
|---|---|---|---|---|---|---|---|
| Seroconversion | Death | ||||||
| 210 | Wild type | 344b | 02-91 | 01-94 | Leukemia | 35 | 36 |
| 214 | Wild type | 344 | 10-91 | 01-93 | Leukemia | 16 | |
| 224 | Wild type | 344 | 09-91 | 03-94 | Leukemia | 31 | |
| 230 | Wild type | 344 | 07-91 | 09-92 | Leukemia | 15 | |
| 235 | Wild type | 344 | 12-91 | 10-97 | Leukemia | 71 | |
| 236 | Wild type | 344 | 11-91 | 09-95 | Leukemia | 47 | |
| 238 | Wild type | 344 (Denv + Dpol)c | 05-92 | 02-96 | Leukemia | 46 | |
| 241 | Wild type | 344 (Denv + Dpol) | 04-92 | 07-95 | Leukemia | 40 | |
| 244 | Wild type | 344 | 05-92 | 12-95 | Leukemia | 44 | |
| 252 | Wild type | 344 | 01-93 | 07-95 | Leukemia | 31 | |
| 260 | Wild type | 344 (IX)d | 10-92 | 12-95 | Leukemia | 39 | |
| 261 | Wild type | 344 (IX) | 10-92 | 04-96 | Leukemia | 43 | |
| 273 | Wild type | 344 | 11-93 | 09-95 | Leukemia | 23 | |
| 274 | Wild type | 344 | 11-93 | 10-95 | Leukemia | 24 | |
| 286 | Wild type | 344 | 05-95 | 06-96 | Leukemia | 13 | |
| 293 | Wild type | 344 (IX) | 07-96 | 04-01 | Leukemia | 58 | |
| 8 | Wild type | 344 (IX) | 06-95 | 01-02 | Leukemia | 80 | |
| 11 | Wild type | 344 (IX) | 06-95 | 11-96 | Leukemia | 18 | |
| 31 | Wild type | 344 (IX) | 07-95 | 11-96 | Leukemia | 17 | |
| 298 | Wild type | 344 (IX) | 10-95 | 06-99 | Leukemia | 45 | |
| 4229 | Wild type | 344 | 10-04 | 04-06 | Leukemia | 19 | |
| 109 | Wild type | 344 (IX) | 12-97 | 06-99 | Unrelated | (19) | (23) |
| 110 | Wild type | 344 (IX) | 09-98 | 06-99 | Unrelated | (10) | |
| 2672 | Wild type | 344 (IX) | 10-99 | 10-03 | Unrelated | (49) | |
| 1095 | Wild type | 344 (IX) | 09-00 | 09-03 | Unrelated | (37) | |
| 292 | Wild type | 344 (IX) | 06-96 | 08-99 | Unrelated | (27) | |
| 4536 | Wild type | 344 | 02-02 | 05-03 | Unrelated | (16) | |
| 2158 | Wild type | 344 | 10-03 | 05-05 | Unrelated | (20) | |
| 4188 | Wild type | 344 | 10-04 | 04-05 | Unrelated | (7) | |
| 237 | G4 mutant | IG4e | 04-93 | 08-99 | Unrelated | (77) | (48) |
| 240 | G4 mutant | IG4 | 06-93 | 08-99 | Unrelated | (75) | |
| 249 | G4 mutant | IG4 | 05-93 | 01-95 | Unrelated | (21) | |
| 271 | G4 mutant | IG4 | 11-93 | 04-95 | Unrelated | (18) | |
| 272 | G4 mutant | IG4 | 11-93 | 10-00 | Unrelated | (84) | |
| 1071 | G4 mutant | IG4 | 10-00 | 07-04 | Unrelated | (46) | |
| 1077 | G4 mutant | IG4 | 10-00 | 09-04 | Unrelated | (48) | |
| 4537 | G4 mutant | IG4 | 02-02 | 03-04 | Unrelated | (26) | |
| 4538 | G4 mutant | IG4 | 02-02 | 12-04 | Unrelated | (35) | |
| 245 | G4-R3 mutant | DXf | 11-93 | 04-01 | Leukemia | 90 | 90 |
| 246 | G4-R3 mutant | DX | 05-92 | 03-00 | Unrelated | (95) | (71) |
| 262 | G4-R3 mutant | DX | 11-93 | 07-96 | Unrelated | (33) | |
| 264 | G4-R3 mutant | DX | 11-92 | 09-02 | Unrelated | (119) | |
| 278 | G4-R3 mutant | DX | 05-95 | 06-99 | Unrelated | (50) | |
| 279 | G4-R3 mutant | DX | 05-95 | 06-99 | Unrelated | (50) | |
| 1086 | G4-R3 mutant | DX | 09-00 | 02-04 | Unrelated | (42) | |
| 1091 | G4-R3 mutant | DX | 09-00 | 02-04 | Unrelated | (42) | |
| 255 | G4-R3 mutant with RZ | DX (RZ)g | 07-92 | 06-99 | Unrelated | (84) | |
| 256 | G4-R3 mutant with RZ | DX (RZ) | 10-92 | 05-96 | Unrelated | (44) | |
| 257 | G4-R3 mutant with RZ | DX (RZ) | 07-92 | 01-03 | Unrelated | (151) | |
| 277 | TM mutant | 6073 Tyrh | 07-94 | 05-01 | Leukemia | 83 | 83 |
| 276 | TM mutant | 6073 Tyr | 06-94 | 10-95 | Unrelated | (17) | (39) |
| 284 | TM mutant | 6073 Tyr | 05-95 | 06-99 | Unrelated | (50) | |
| 285 | TM mutant | 6073 Tyr | 05-95 | 06-99 | Unrelated | (50) | |
Data for animals 245 and 277 are highlighted in bold.
Provirus 344 was cloned from lymphomatotic BLV-infected sheep 344.
Denv and Dpol are mutants with deletions of env and pol, respectively. Cotransfection with these mutants is equivalent to transfection with the wild-type provirus.
The IX strain contains a deletion in untranslated sequences (viral RNA coordinates 6614 to 6731) but behaves as the wild type.
The IG4 mutant contains a stop codon in the G4 open reading frame at position 6697.
The DX mutant has a deletion between nucleotides 6614 and 6997 which affects both R3 and G4 genes.
DX (RZ) is identical to DX but contains a Tax-targeted ribozyme sequence (RZ) inserted in place of the 6614-6997 deletion.
The transmembrane protein (TM) mutant has a single substitution of G instead of T at position 6073, creating a tyrosine-to-aspartic acid change in the gp30 protein.
Causes including leukemia, lymphoma, and lymphosarcoma are indicated as leukemia. For those animals surviving to the end of the experiment or dying from accidental disease (mainly diarrhea and scabies) or other causes, the cause is listed as unrelated.
Number of months separating seroconversion and death due to leukemia, lymphoma, and/or lymphosarcoma. When the numbers are in parentheses, the death was due to unrelated causes.
FIG. 1.
Induction of leukemia and lymphoma in sheep 245 infected with an attenuated strain of BLV harboring a deletion in the R3 and G4 accessory genes. Sheep 245 was infected with BLV provirus DX, which derives from the wild-type BLV strain 344 but contains a deletion in the R3 and G4 genes. At regular intervals postinfection, blood was collected by jugular venipuncture. (A) Proviral loads were quantified by real-time PCR and represented as numbers of viral genome copies per cubic millimeter of blood. (B) Leukocyte counts were determined by using a Coulter Counter, model ZN, and the relative proportions of lymphocytes were estimated by examination under a microscope after May-Grunwald and Giemsa staining. (C) Proviral loads reported relative to the number of peripheral blood cells. (D) Cryosections from the tumoral mesenteric lymph node were stained either with anti-CD21 (left panel) or with anti-CD4 (right panel) monoclonal antibodies in association with a fluorescein isothiocyanate conjugate and analyzed by confocal microscopy. B, B cell. (E) The profile of provirus integration sites was analyzed by inverse PCR of peripheral blood and tumor cells. The sizes of the fragments are indicated on the left. MW, molecular size markers (V; Roche). (F) DNA (250 ng) extracted from whole blood or tumor cells was subjected to nonquantitative long-PCR amplification of the integrated provirus using primers located in the 5′ and 3′ long terminal repeats (expected molecular size of the amplicon, 7.7 kb). MW′, molecular size markers (Smart Ladder; Eurogentec).
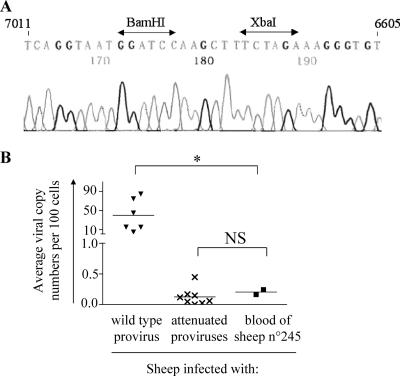
It thus appears that sheep 245 developed clinical signs characteristic of BLV-associated tumor induction. Although BLV does not naturally spread among sheep, it was possible that animal 245 may have been reinfected with a wild-type virus (for example, through iatrogenic transmission) and/or that a reversion abolishing the attenuated phenotype occurred. To exclude these possibilities, the proviral region encompassing the R3-G4 deletion was amplified by PCR and the amplicon was directly sequenced. Figure 2A demonstrates that the deletion located between the XbaI and BamHI restriction sites at viral coordinates 6614 and 6997 (23) was preserved. Furthermore, cloning and sequencing of individual PCR products (n = 10) led to the same conclusion (data not shown).
FIG. 2.
Lack of reversion of the R3-G4 deletion mutant and maintenance of the attenuated phenotype after injection of blood from sheep 245 into naïve recipient hosts. (A) Nucleotide sequence and corresponding sequence chromatogram of a PCR product from DNA from sheep 245 amplified using primers encompassing the R3-G4 deletion (coordinates 7011 to 6605 according to reference 23). (B) Blood from sheep 245 was injected into two BLV-free animals, 1086 and 1091 (▪). Their proviral loads (in numbers of genome copies per 100 cells) were quantified by real-time PCR at 7 months postinfection and compared to the levels reached in sheep infected with wild-type (▾) and attenuated (×) viruses (IG4, CRE, R3-G4, and 6073 Tyr mutants). Statistical relevance (NS, not statistically significant, and *, statistically significant) was calculated according to the two-tailed unpaired Student t test.
Although the R3-G4 deletion was still present in the proviral DNA of the tumor from sheep 245, other modifications in the viral genome may have occurred to reverse the attenuated phenotype. Therefore, peripheral blood from sheep 245 was used to infect two BLV-free animals (1086 and 1091) and the proviral loads were measured by real-time PCR. Figure 2B shows that the average numbers of viral genome copies in sheep 1086 and 1091 were in the range of those typically achieved by attenuated viruses (not statistically significant by the unpaired Student t test), far below the wild-type levels measured at similar periods after infection (7 months post inoculation; P < 0.05). We conclude that the virus infecting sheep 245 did not revert to the wild type and that its attenuated phenotype was preserved after transmission to new hosts.
Collectively, our data show that the R3-G4 deletion mutant may be pathogenic in sheep, albeit infrequently (1 of 20) and after a long latency period (e.g., 90 months). Interestingly, among all BLV mutants analyzed to date (5, 10, 15, 19, 26, 30-32, 34, 35), we found another example of leukemogenesis induced by an attenuated virus (Table 1). Indeed, one (277) of four sheep infected with BLV mutant 6073 Tyr developed leukemia (Fig. 3A) and lymphoma (Fig. 3B) concomitantly. Three clones were integrated in the tumor genome (Fig. 3C), and the T-to-G transversion located in the cytoplasmic domain of the viral gp30 transmembrane gene pertaining to this mutant was preserved (Fig. 3D).
FIG. 3.
Pathogenicity in sheep 277 of an attenuated BLV strain mutated in the transmembrane protein YXXL signaling motifs. Sheep 277 was infected with the attenuated BLV mutant 6073 Tyr carrying a T-to-G transversion located in the cytoplasmic domain of the viral gp30 transmembrane gene. Blood was collected at regular intervals postinfection. (A) Lymphocyte counts were determined by using a Coulter Counter, model ZN, and May-Grunwald and Giemsa staining, and proviral loads were quantified by real-time PCR. (B) Cryosections from a tumoral mesenteric lymph node stained with anti-CD21 (left panel) or with anti-CD4 (right panel) monoclonal antibodies and analyzed by confocal microscopy. B, B cell. (C) The profile of provirus integration sites was analyzed by inverse PCR of peripheral blood and tumor cells. The sizes of the fragments are indicated on the left. MW, molecular size markers (V; Roche). (D) Nucleotide sequence and corresponding sequence chromatogram (coordinates 6270 to 6299 according to reference 23) of a PCR product amplified from a tumor from sheep 277. The position of the T-to-G transversion is marked by an asterisk.
In summary, we show in this report that two types of mutations restricting BLV proviral loads do not completely abolish its pathogenic potential in sheep. A first direct conclusion is that the genes affected by the mutations (i.e., the R3, G4, and transmembrane genes) cannot be considered to be oncogenes per se but rather participate in viral pathogenicity, most likely by favoring efficient replication. In fact, we have previously used the reverse genetics approach in order to assess the role of viral proteins in the leukemogenesis process. We identified three categories of mutations: (i) those that completely abolish infectivity (10, 31, 33, 35), (ii) those that are apparently silent and do not affect replication (10, 19, 26, 33), and (iii) intermediates that do not completely abolish infectivity but reduce proviral loads (10, 15, 19, 31, 34). Sheep 245 and 277 described here were infected with recombinant viruses belonging to the third category (15). Since these two sheep eventually became leukemic and/or lymphomatotic, we have to reassess our previous interpretation postulating that the mutations abolished pathogenicity. We now propose that the R3-G4-transmembrane sequences are required for efficient maintenance and replication of the virus with the ultimate result of increasing the proviral loads.
It is known that, during leukemogenesis, events and conditions such as chromosomal instability that leads to aneuploidy, cellular gene mutations, deletions, and amplifications occur and accumulate while disease progresses (6, 25). Moreover, a viral protein like Tax also potentially induces defective DNA repair (21). These modifications of the cellular genome may favor leukemic cell growth by promoting cell proliferation or inactivating tumor suppressors. Even proviral sequences are frequently deleted in tumors, leading to defective viruses apparently unable to express any viral protein (16, 27). This process may allow escape from the host's immune response and promote the survival of the tumor clone. Cellular and viral genomic mutations are rare events thought to contribute to a multistep mechanism of leukemogenesis (28). In this context, we speculate that BLV replication during the asymptomatic phase abnormally increases the turnover of the virus-infected B-cell population and thereby increases the likelihood that genomic mutations will occur. The occurrence of these events would be restricted in sheep infected with attenuated mutants since the proviral loads are reduced due to impaired mitotic expansion or infectious replication. This simple model provides a straightforward rationale for the observed difference in pathogenic potential between attenuated and wild-type proviruses.
These data obtained from this BLV model system may be of interest concerning the related human T-cell lymphotropic virus type 1 (HTLV-1), responsible for adult T-cell leukemia and a neurodegenerative disease called tropical spastic paraparesis or HTLV-associated myelopathy (9, 39). Although rabbits, mice, rats, and monkeys are informative model systems, there is indeed no fully satisfactory experimental model for HTLV that includes concomitant viral infection and induction of disease. In rabbits, HTLV p12I and p13II/p30II accessory genes, the orthologs of R3 and G4 genes, are required to maintain high proviral loads, suggesting their implication in viral pathogenesis (3, 4). Whether the integrity of these genes contributes to disease progression in HTLV-1-infected subjects remains to be determined. However, p12I mutations in a significant proportion of tropical spastic paraparesis-HTLV-associated myelopathy and adult T-cell leukemia patients have been identified previously (8, 13).
The low but still significant pathogenic potential of BLV deletion mutants described in this report has major implications for the future development of vaccines based on attenuated viruses. A similar objection for rhesus macaques infected with simian immunodeficiency virus was raised previously, impairing human immunodeficiency virus vaccination trials (reviewed in references 24 and 29). Indeed, infection with a triply mutated simian immunodeficiency virus strain (carrying deletions in nef, vpr, and the 3′ long terminal repeat) leads to reduced viral loads and provides partial protection against challenge (37) but provokes AIDS progression in neonatal macaques (1, 38) as well as adult animals after prolonged observation (2, 12). To explain the divergent behavior of this vaccine strain (i.e., the induction of protective immunity in some animals and fatal immunodeficiency in others), the “threshold hypothesis” has been proposed (12, 29). In this model, an attenuated virus must replicate at a sufficiently high rate to trigger an immune response (the “vaccine threshold”). However, high viral loads exceeding the “disease threshold” cause disease. An effective vaccination thus occurs only if the viral burden falls into the discrete window between these two thresholds. Although BLV R3-G4 deletion mutant loads apparently fall within this threshold range and confer efficient protective immunity (14, 22), pathogenicity in sheep may occur after extended periods of time. Our present report thus highlights another important parameter in this model: the cumulative risk of disease onset that increases with time.
Acknowledgments
We thank the Sixth Research Framework Programme of the European Union (project INCA LSHC-CT-2005-018704), the Belgian Foundation against Cancer, the Bekales Foundation, and the Fonds National de la Recherche Scientifique (FNRS) for financial support. A.F. (research fellow), N.G. (“Télévie” fellow), M.B. (FRIA fellow), and R.K. and L.W. (research directors) are members of the FNRS.
We are grateful to Hervé Balon, François Debande, Rafaela Fandango, and Jean-Marie Londes for experimental assistance. We thank Yves Beckers and André Théwis for support in animal housing facilities.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Baba, T. W., Y. S. Jeong, D. Pennick, R. Bronson, M. F. Greene, and R. M. Ruprecht. 1995. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267:1820-1825. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T. W., V. Liska, A. H. Khimani, N. B. Ray, P. J. Dailey, D. Penninck, R. Bronson, M. F. Greene, H. M. McClure, L. N. Martin, and R. M. Ruprecht. 1999. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat. Med. 5:194-203. [DOI] [PubMed] [Google Scholar]
- 3.Bartoe, J. T., B. Albrecht, N. D. Collins, M. D. Robek, L. Ratner, P. L. Green, and M. D. Lairmore. 2000. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J. Virol. 74:1094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, N. D., G. C. Newbound, B. Albrecht, J. L. Beard, L. Ratner, and M. D. Lairmore. 1998. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood 91:4701-4707. [PubMed] [Google Scholar]
- 5.Debacq, C., M. T. Sanchez Alcaraz, F. Mortreux, P. Kerkhofs, R. Kettmann, and L. Willems. 2004. Reduced proviral loads during primo-infection of sheep by bovine leukemia virus attenuated mutants. Retrovirology 1:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dequiedt, F., R. Kettmann, A. Burny, and L. Willems. 1995. Mutations in the p53 tumor-suppressor gene are frequently associated with bovine leukemia virus-induced leukemogenesis in cattle but not in sheep. Virology 209:676-683. [DOI] [PubMed] [Google Scholar]
- 7.Florins, A., N. Gillet, B. Asquith, M. Boxus, C. Burteau, J. C. Twizere, P. Urbain, F. Vandermeers, C. Debacq, M. T. Sanchez-Alcaraz, I. Schwartz-Cornil, P. Kerkhofs, G. Jean, A. Thewis, J. Hay, F. Mortreux, E. Wattel, M. Reichert, A. Burny, R. Kettmann, C. Bangham, and L. Willems. 2007. Cell dynamics and immune response to BLV infection: a unifying model. Front. Biosci. 12:1520-1531. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa, Y., K. Usuku, S. Izumo, and M. Osame. 2004. Human T cell lymphotropic virus type I (HTLV-I) p12I is dispensable for HTLV-I transmission and maintenance of infection in vivo. AIDS Res. Hum. Retrovir. 20:1092-1099. [DOI] [PubMed] [Google Scholar]
- 9.Gallo, R. C. 2005. History of the discoveries of the first human retroviruses: HTLV-1 and HTLV-2. Oncogene 24:5926-5930. [DOI] [PubMed] [Google Scholar]
- 10.Gatot, J. S., I. Callebaut, J. P. Mornon, D. Portetelle, A. Burny, P. Kerkhofs, R. Kettmann, and L. Willems. 1998. Conservative mutations in the immunosuppressive region of the bovine leukemia virus transmembrane protein affect fusion but not infectivity in vivo. J. Biol. Chem. 273:12870-12880. [DOI] [PubMed] [Google Scholar]
- 11.Gillet, N., A. Florins, M. Boxus, C. Burteau, A. Nigro, F. Vandermeers, H. Balon, A. B. Bouzar, J. Defoiche, A. Burny, M. Reichert, R. Kettmann, and L. Willems. 2007. Mechanisms of leukemogenesis induced by bovine leukemia virus: prospects for novel anti-retroviral therapies in human. Retrovirology 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann-Lehmann, R., J. Vlasak, A. L. Williams, A. L. Chenine, H. M. McClure, D. C. Anderson, S. O'Neil, and R. M. Ruprecht. 2003. Live attenuated, nef-deleted SIV is pathogenic in most adult macaques after prolonged observation. AIDS 17:157-166. [DOI] [PubMed] [Google Scholar]
- 13.Iniguez, A. M., R. Gastaldello, S. Gallego, K. Otsuki, and A. C. Vicente. 2006. HTLV-1 p12I protein sequences from South America: truncated proteins and common genetic signatures. AIDS Res. Hum. Retrovir. 22:466-469. [DOI] [PubMed] [Google Scholar]
- 14.Kerkhofs, P., J. S. Gatot, K. Knapen, M. Mammerickx, A. Burny, D. Portetelle, L. Willems, and R. Kettmann. 2000. Long-term protection against bovine leukaemia virus replication in cattle and sheep. J. Gen. Virol. 81:957-963. [DOI] [PubMed] [Google Scholar]
- 15.Kerkhofs, P., H. Heremans, A. Burny, R. Kettmann, and L. Willems. 1998. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J. Virol. 72:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kettmann, R., J. Deschamps, Y. Cleuter, D. Couez, A. Burny, and G. Marbaix. 1982. Leukemogenesis by bovine leukemia virus: proviral DNA integration and lack of RNA expression of viral long terminal repeat and 3′ proximate cellular sequences. Proc. Natl. Acad. Sci. USA 79:2465-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefebvre, L., A. Vanderplasschen, V. Ciminale, H. Heremans, O. Dangoisse, J. C. Jauniaux, J. F. Toussaint, V. Zelnik, A. Burny, R. Kettmann, and L. Willems. 2002. Oncoviral bovine leukemia virus G4 and human T-cell leukemia virus type 1 p13(II) accessory proteins interact with farnesyl pyrophosphate synthetase. J. Virol. 76:1400-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lew, A. E., R. E. Bock, J. B. Molloy, C. M. Minchin, S. J. Robinson, and P. Steer. 2004. Sensitive and specific detection of proviral bovine leukemia virus by 5′ Taq nuclease PCR using a 3′ minor groove binder fluorogenic probe. J. Virol. Methods 115:167-175. [DOI] [PubMed] [Google Scholar]
- 19.Merezak, C., C. Pierreux, E. Adam, F. Lemaigre, G. G. Rousseau, C. Calomme, C. Van Lint, D. Christophe, P. Kerkhofs, A. Burny, R. Kettmann, and L. Willems. 2001. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: implications for viral latency. J. Virol. 75:6977-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moules, V., C. Pomier, D. Sibon, A. S. Gabet, M. Reichert, P. Kerkhofs, L. Willems, F. Mortreux, and E. Wattel. 2005. Fate of premalignant clones during the asymptomatic phase preceding lymphoid malignancy. Cancer Res. 65:1234-1243. [DOI] [PubMed] [Google Scholar]
- 21.Philpott, S. M., and G. C. Buehring. 1999. Defective DNA repair in cells with human T-cell leukemia/bovine leukemia viruses: role of tax gene. J. Natl. Cancer Inst. 91:933-942. [DOI] [PubMed] [Google Scholar]
- 22.Reichert, M., G. H. Cantor, L. Willems, and R. Kettmann. 2000. Protective effects of a live attenuated bovine leukaemia virus vaccine with deletion in the R3 and G4 genes. J. Gen. Virol. 81:965-969. [DOI] [PubMed] [Google Scholar]
- 23.Rice, N., and R. G. R. Stephens. 1987. Sequence analysis of the bovine leukemia virus genome, p. 115-144. In A. Burny and M. Mammerickx (ed.), Enzootic bovine leukosis and bovine leukemia virus. Nijhoff, The Hague, The Netherlands.
- 24.Ruprecht, R. M. 1999. Live attenuated AIDS viruses as vaccines: promise or peril? Immunol. Rev. 170:135-149. [DOI] [PubMed] [Google Scholar]
- 25.Schnurr, M. W., R. F. Carter, I. D. Dube, V. E. Valli, and R. M. Jacobs. 1994. Nonrandom chromosomal abnormalities in bovine lymphoma. Leuk. Res. 18:91-99. [DOI] [PubMed] [Google Scholar]
- 26.Twizere, J. C., P. Kerkhofs, A. Burny, D. Portetelle, R. Kettmann, and L. Willems. 2000. Discordance between bovine leukemia virus tax immortalization in vitro and oncogenicity in vivo. J. Virol. 74:9895-9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van den Broeke, A., Y. Cleuter, G. Chen, D. Portetelle, M. Mammerickx, D. Zagury, M. Fouchard, L. Coulombel, R. Kettmann, and A. Burny. 1988. Even transcriptionally competent proviruses are silent in bovine leukemia virus-induced sheep tumor cells. Proc. Natl. Acad. Sci. USA 85:9263-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinberg, R. A. 1989. Oncogenes, antioncogenes, and the molecular bases of multistep carcinogenesis. Cancer Res. 49:3713-3721. [PubMed] [Google Scholar]
- 29.Whitney, J. B., and R. M. Ruprecht. 2004. Live attenuated HIV vaccines: pitfalls and prospects. Curr. Opin. Infect. Dis. 17:17-26. [DOI] [PubMed] [Google Scholar]
- 30.Willems, L., A. Burny, D. Collete, O. Dangoisse, F. Dequiedt, J. S. Gatot, P. Kerkhofs, L. Lefebvre, C. Merezak, T. Peremans, D. Portetelle, J. C. Twizere, and R. Kettmann. 2000. Genetic determinants of bovine leukemia virus pathogenesis. AIDS Res. Hum. Retrovir. 16:1787-1795. [DOI] [PubMed] [Google Scholar]
- 31.Willems, L., J. S. Gatot, M. Mammerickx, D. Portetelle, A. Burny, P. Kerkhofs, and R. Kettmann. 1995. The YXXL signalling motifs of the bovine leukemia virus transmembrane protein are required for in vivo infection and maintenance of high viral loads. J. Virol. 69:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willems, L., C. Grimonpont, P. Kerkhofs, C. Capiau, D. Gheysen, K. Conrath, R. Roussef, R. Mamoun, D. Portetelle, A. Burny, E. Adam, L. Lefebvre, J. C. Twizere, H. Heremans, and R. Kettmann. 1998. Phosphorylation of bovine leukemia virus Tax protein is required for in vitro transformation but not for transactivation. Oncogene 16:2165-2176. [DOI] [PubMed] [Google Scholar]
- 33.Willems, L., P. Kerkhofs, L. Attenelle, A. Burny, D. Portetelle, and R. Kettmann. 1997. The major homology region of bovine leukaemia virus p24gag is required for virus infectivity in vivo. J. Gen. Virol. 78:637-640. [DOI] [PubMed] [Google Scholar]
- 34.Willems, L., P. Kerkhofs, F. Dequiedt, D. Portetelle, M. Mammerickx, A. Burny, and R. Kettmann. 1994. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc. Natl. Acad. Sci. USA 91:11532-11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willems, L., R. Kettmann, F. Dequiedt, D. Portetelle, V. Voneche, I. Cornil, P. Kerkhofs, A. Burny, and M. Mammerickx. 1993. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 67:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willems, L., D. Portetelle, P. Kerkhofs, G. Chen, A. Burny, M. Mammerickx, and R. Kettmann. 1992. In vivo transfection of bovine leukemia provirus into sheep. Virology 189:775-777. [DOI] [PubMed] [Google Scholar]
- 37.Wyand, M. S., K. H. Manson, M. Garcia-Moll, D. Montefiori, and R. C. Desrosiers. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 70:3724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyand, M. S., K. H. Manson, A. A. Lackner, and R. C. Desrosiers. 1997. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat. Med. 3:32-36. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, M. 2005. Discovery of HTLV-1, the first human retrovirus, its unique regulatory mechanisms, and insights into pathogenesis. Oncogene 24:5931-5937. [DOI] [PubMed] [Google Scholar]