Abstract
The PPPY motif in the matrix (MA) domain of human T-cell leukemia virus type 1 (HTLV-1) Gag associates with WWP1, a member of the HECT domain containing family of E3 ubiquitin ligases. Mutation of the PPPY motif arrests particle assembly at an early stage and abolishes ubiquitination of MA. Similar effects are seen when Gag is expressed in the presence of a truncated form of WWP1 that lacks the catalytically active HECT domain (C2WW). To understand the role of ubiquitination in budding, we mutated the four lysines in MA to arginines and identified lysine 74 as the unique site of ubiquitination. Virus-like particles produced by the K74R mutant did not contain ubiquitinated MA and showed a fourfold reduction in the release of infectious particles. Furthermore, the K74R mutation rendered assembly hypersensitive to C2WW inhibition; K74R Gag budding was inhibited at significantly lower levels of expression of C2WW compared with wild-type Gag. This finding indicates that the interaction between Gag and WWP1 is required for functions other than Gag ubiquitination. Additionally, we show that the PPPY− mutant Gag exerts a strong dominant-negative effect on the budding of wild-type Gag, further supporting the importance of recruitment of WWP1 to achieve particle assembly.
Retroviruses recruit the multivesicular body (MVB) biogenesis machinery to accomplish particle release (for reviews, see references 7, 34, and 56). Peptide motifs found in retroviral Gag proteins serve as docking sites for components of the MVB pathway in order to divert the machinery to the site of particle assembly (43). MVBs are the penultimate endosomal compartment of the degradative pathway that delivers activated cell surface receptors to the lysosome. The signal for the recruitment of the receptor into the lysosomal degradation pathway is a low-level ubiquitination—usually one or two ubiquitin moieties are added at one to four lysine residues following phosphorylation of the receptor (16, 20). Several E3-type ubiquitin ligases have been identified which perform this function, among them WWP1 and other members of the Nedd4 family (29). After internalization, the ubiquitinated receptor is recognized by the HRS/STAM complex (45), which hands it off to the ESCRT I complex, thus shunting it into the degradative rather than the recycling compartment of the early endosome (2). The ubiquitin modification serves as a tag on the receptor along the pathway consisting of the ESCRT II and III complexes. Finally, ubiquitin is removed from the receptor by a deubiquitinase and the vesicle buds into the MVB with the help of the VPS4 ATPase complex (1, 24).
The region in retroviral Gag harboring the MVB machinery interaction motifs was designated the late domain (LD) (40), as it controls the late step of virus budding in the infectious cycle. Three motifs have been identified in retroviruses so far; they are PPXY (61), PT/SAP (12, 21), and YPXL/LXXLF (44, 53) and connect to Nedd4 family E3 ubiquitin ligases (4, 17, 19, 28, 54, 64), the TSG101 subunit of ESCRT I (11, 32, 59), and ALIX (31, 53), respectively. Most viruses have more than one motif; for example PTAP and YPXL are found in p6 of the human immunodeficiency virus type 1 (HIV-1) Gag, while Mason-Pfizer Monkey virus (MPMV) (63), murine leukemia virus (MLV) (65), and HTLV-1 (4, 19, 48, 60) all have PPPY and PS/TAP motifs. The sequence PPPYVEPTAP is found just before the proteolytic cleavage site between matrix (MA) and capsid (CA) domains in HTLV-1 Gag. Previous studies by us and others have shown that the integrity of the PPPY motif is more important for HTLV-1 release than that of the PTAP motif (4, 19, 26, 48, 60). A striking aspect of the HTLV-1 PPPY− phenotype is that virus budding is arrested at an early stage. In contrast to the tethered, immature particles reported for PPPY− mutants of Rous sarcoma virus (RSV) (41) and MLV (65), HTLV-1 PPPY− Gag accumulates under the plasma membrane, as if contraction into spherical particles is compromised. It is interesting to note in this context that PPPY− mutants of MPMV can still form procapsids but are unable to cause the plasma membrane to envelope them, suggesting that part of the defect is in promoting membrane curvature (13).
WWP1 is a member of the Nedd4 family of HECT-E3 ubiquitin ligases and was recently shown by Martin-Serrano et al. to have the highest affinity, along with the closest family members WWP2 and Itch/AIP4, for the PPPY motifs in the late domains of RSV and MLV (30). WWP1 contains three major domains; an amino-terminal C2 domain, consisting of a Ca2+-dependent lipid binding domain; four WW domains in the middle of the protein; and the C-terminal HECT (homologous to E6-AP carboxy terminus) domain. The HECT domain contains ubiquitin ligase activity with cysteine 890 at the active site (23, 47). WW domains recognize PPXY motifs and share two tryptophan residues spaced between 20 and 25 amino acids apart with a core of several aromatic residues. Some Nedd4 constructs containing only the C2 and WW domains strongly inhibit MLV and HTLV-1 budding (30). Consistent with the greater role of the PPPY motif than the PTAP motif in HTLV-1 release, we found that dominant-negative (DN) WWP1 (C2WW) was a more potent inhibitor of virus release than DN TSG101 (19) or TSG101 small interfering RNA (Heidecker et al., unpublished).
Most Gag proteins have been shown to be ubiquitinated, mainly in the vicinity of the late domain motifs (36-38, 51, 52). However, the role of ubiquitination in retroviral budding is debated; several findings argue for a functional contribution of the ubiquitin modification to virus release, whereas others suggest that it is merely a bystander effect (41, 52, 58). It has been shown that ubiquitination of HIV-1 Gag increases its affinity for TSG101 (11) and that inhibiting ubiquitination by sequestering ubiquitin in the proteasome pathway interferes with particle release (39). However, appending ubiquitin to the end of Gag in the RSV system did not enhance particle release in the presence of a DN E3-ubiquitin-ligase LD1 compared to wild-type (WT) Gag (57). Furthermore, mutation of the lysine residues in the p6 domain of HIV-1 Gag or the p12 domain of MLV Gag had little effect in virus infectivity assays (36). However, Spidel et al. showed that substituting the five lysine residues in the C-terminal half of the RSV MA with arginines resulted in a drastic impairment of budding; this defect could be largely ablated by restoring the lysine at any of the five positions. Also, recent work by Gottwein et al. demonstrated that many lysines in the HIV-1 Gag protein could be ubiquitinated and that a high proportion of them need to be changed to manifest an effect on overall ubiquitination levels and on particle release (14). A likely explanation for these seemingly contradictory results is that the importance of ubiquitination varies among viruses and may also depend on the cellular conditions.
We have shown that up to 40% of MA in HTLV-1 particles is ubiquitinated. Ubiquitinated forms of MA (p19) are detected on Western blots as bands migrating at 27 kDa and 35 kDa, indicating that monoubiquitination and diubiquitination occur (19). The finding that the truncated WWP1 protein, which lacks the catalytically-active HECT domain (C2WW), inhibits budding suggests that ubiquitination of HTLV-1 Gag plays an important role in particle release (4, 19, 48, 60).
In this study, we show that the substrate for ubiquitination is the lysine at position 74 in HTLV-1 Gag. Changing the lysine residue to arginine (K74R mutation) abolished ubiquitination of MA, which made it possible to determine the role of ubiquitination in HTLV-1 assembly and release. In contrast to mutation of the PPPY motif, which abolished release of infectious virus, the K74R mutant retained about 20% of WT budding activity. In addition, budding of the K74R mutant was hypersensitive to inhibition by C2WW. We also addressed whether the budding defect of the PPPY− mutant could be rescued by coexpression of WT Gag and found that the mutant phenotype was dominant.
MATERIALS AND METHODS
Plasmids.
The HTLV-1 provirus expression plasmid pCMVHT-1 has been described previously (9, 18, 50). pCMVHT-1ΔX has a defective env gene due to the deletion of an XhoI fragment. pCMVHT-1-M and pCMVHT-1ΔX-M carry the pol gene of the HTLV-1 virus present in T-cell line MT2 (33). The part of the HTLV-1 gag gene encoding the MA and CA domains was subcloned into pCMVpA-RRE, which contains the cytomegalovirus (CMV) immediate-early promoter and the HIV Rev response element (10). The MA-CA expression plasmids were cotransfected with the HIV-1 Rev expression plasmid, pRS-HRev, to ensure export of the truncated Gag mRNA. LD mutants affecting the PPPY and PTAP motifs, PPPG (P3G) and AAAP (A3P), respectively, were described previously (18). Mutations were generated by overlapping PCR and transferred into the appropriate expression plasmids on restriction fragments, which were sequenced in their entirety. WWP1 expression plasmids were generated in the retroviral expression vector pUCHRatt, which contains the HIV sequences of pHR recloned in pUC19 with the Gateway attA cassette inserted after the CMV promoter (Invitrogen, Carlsbad, CA) (9, 25, 35). The plasmids pCMVΔ8.2, pCMV-VSVg, pHTC-GFPluc, and pCMV-β Gal have been described previously (9, 35). Plasmid DNAs were isolated using QIAGEN (Hilden, Germany) columns and protocols.
Cell culture, transfections, virus preparation, and infections.
Human 293T cells and HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. Transfections were done essentially as previously described (18). Six to 18 μg of plasmid DNA was transfected into 5 × 106 293T cells using calcium phosphate precipitation in N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES)-buffered saline. Some experiments were done on a smaller scale with 293T or HeLa cells using Fugene transfection reagent (Roche). Supernatants were harvested 40 h posttransfection and filtered through 0.45-μm filters. For immunoblot analysis, viruses were pelleted through a 20% glycerol cushion at 100,000 × g for 90 min for large-scale transfections or pelleted for 2 h at 22,000 × g in a microcentrifuge in the cold. Cells were washed with phosphate-buffered saline (PBS) and lysed on ice in radioimmunoprecipitation assay buffer (100 mM NaCl, 10 mM Tris [pH 7.4], 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% Triton X-100) with protease inhibitor cocktail (Sigma) for 15 min and then kept on ice. Lysates were cleared at 20,000 × g for 15 min. Single-cycle cell-free infection assays were previously described (9). Luciferase assays were performed using the Luciferase Assay System from Promega.
Western blot analysis and ELISA.
Denaturing protein gel electrophoresis was carried out on 4 to 12% NUPAGE gels in morpholineethanesulfonic acid (MES) or morpholinepropanesulfonic acid (MOPS) buffer according to the manufacturer's directions (Invitrogen, Carlsbad, CA). Gels were transferred to Immobilon membranes (Millipore, Bedford, MA) and blocked in Tris-buffered saline with 3% dried milk. Blots were probed in the same solution containing anti-HTLV-1 p19(MA) mouse monoclonal antibody (Zeptometrix, Buffalo, NY), affinity-purified rabbit anti-HTLV-1 MA peptide (SRPAPPPPSSPTHDPPDSDP) antisera, or mouse anti-ubiquitin antibody (Santa Cruz Biotechnologies, Inc). Washing and exposure to the appropriate horseradish-peroxidase-linked secondary antibody (Cell Signaling) were done in Tris-buffered saline-0.2% Tween. Blots were developed with ChemigloWest (AlphaInnotech, San Leandro, CA) and visualized on an AlphaInnotech Imager. Alternatively, IRDye-linked secondary antibodies were used and blots were analyzed by infrared imaging (Li-Cor). Enzyme-linked immunosorbent assay (ELISA) determinations were carried out on filtered tissue culture supernatants and cell extracts using anti-HTLV-1 p19 ELISA kits from Zeptometrix according to the manufacturer's protocol.
Immunofluorescent microscopy.
For immunofluorescent microscopy analysis, HeLa cells were seeded on glass coverslips. Following transfection and growth, cells were fixed in 3.5% paraformaldehyde (Sigma) in PBS for 15 min. After washing with PBS, cells were permeabilized and blocked with 0.25% saponin (Sigma) in PBS with 3% normal goat serum. Samples were incubated with antibodies and washed in the same solution. Samples were mounted in Prolong (Molecular Probes, Eugene, OR) and inspected on a Zeiss deconvoluting microscope.
TEM.
Detailed procedures for standard thin-section transmission electron microscopy (TEM) were previously described (55), with the following modifications. Cultured cells were scraped, centrifuged at 150 × g, and fixed in 2% glutaraldehyde in cacodylate buffer (0.1 M, pH. 7.4) overnight at 4°C followed by 1% osmium fixation for 1 h at room temperature. Cell pellets were dehydrated in graded ethanol (e.g., 35%, 50%, 70%, 95%, and 100%) and propylene oxide (100%). Cell pellets were infiltrated overnight in a 1:1 mixture of propylene oxide and epoxy resin, embedded in pure resin, and cured at 55°C for 48 h. The cured block was thin sectioned, mounted on naked 200-mesh copper grids, and stained in uranyl acetate and lead citrate. The digital image was obtained with an electron microscope equipped with digital camera system.
RESULTS
Ubiquitination of HTLV-1 Gag occurs on a single lysine in MA.
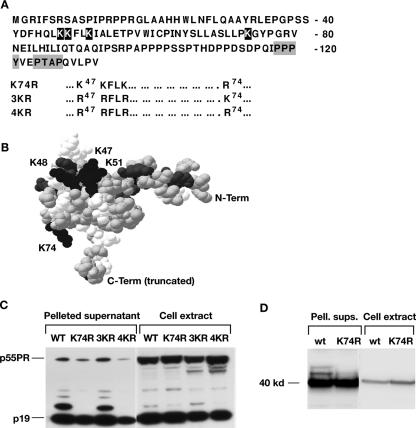
We previously showed that HTLV-1 MA is ubiquitinated and that the fraction of MA present in ubiquitinated forms ranged from 10 to 40% between experiments (19). To test whether ubiquitination of Gag is necessary for particle release, we first identified sites of ubiquitination in MA. There are four lysines in HTLV-1 MA: one at position 74 and three clustered at positions 47, 48, and 51 (Fig. 1A and B). All four positions are conserved in HTLV-3, but in HTLV-2 arginines are present at positions 47 and 48 (5, 62). A modeled structure of HTLV-1 MA was generated by Swiss-Model (42, 49) (Fig. 1B) based on the nuclear magnetic resonance structure of HTLV-2 MA (6). The clustered lysines together with other basic residues form a positively charged surface, which is thought to interact with negatively charged heads of phospholipids in the membrane. This patch is located on one side of the globular head of MA, while K74 is located on the other side of the molecule.
FIG. 1.
K74 is the site of HTLV-1 Gag ubiquitination. (A) Lysine residues in the HTLV-1 MA domain are indicated in white print, and the LD motifs, PPPY and PTAP, are boxed. The amino acid changes in the mutants K74R, 3KR and 4KR are shown underneath the MA sequence. (B) The structure of HTLV-1 MA was modeled by Swiss-Model based on the nuclear magnetic resonance structure of HTLV-2 MA (6). The lysine residues are labeled and shown in black; the other basic residues are shown in gray. The sequence was truncated at residue 100 for the purpose of modeling, as the C terminus is unstructured. (C) Immunoblot analysis of cell extracts and pelleted supernatants from 293T cultures transfected with pCMVHT-1ΔX, pCMVHT-1Δ-K74R, pCMVHT-1ΔX-3KR, and pCMVHT-1ΔX-4KR. Blots were probed with mouse monoclonal anti-HTLV-1 p19(MA) antibody. (D) Immunoblot analysis of cell extracts and pelleted supernatants (Pell. sups.) from 293T cells transfected with pCMV-MACA-RRE and pCMV-MACA-K74R-RRE. Blots were treated as described above.
We mutated the codons for all four lysines (mutant 4KR), the three clustered lysines (3KR), and lysine 74 (K74R) to arginine codons in the context of the provirus expression plasmid pCMVHT-1ΔX (9). Ubiquitination was examined by immunoblotting cell extracts and pelleted supernatants from transfected cells. The samples from the K74R and the 4KR mutants did not show any of the bands indicative of ubiquitinated MA, whereas the pattern of the 3KR mutant was the same as for the WT (Fig. 1C). This finding indicates that either the K74 residue is the only target for mono- and diubiquitination in MA or secondary ubiquitination in MA depends on initial modification at K74. In either case, ubiquitination is specific for the conserved lysine at position 74 in MA.
We next asked whether there are additional substrate sites for ubiquitination in Gag. Although we have not observed ubiquitination of capsid (CA) or nucleocapsid (NC) proteins on immunoblots of either virus-like particles (VLPs) or cellular extracts (data not shown), it is possible that it occurs when the primary target is missing, as E3 ubiquitin ligases are not very discriminating in their substrate site selection. We limited our search for alternate substrate sites to CA as the MA-CA portion of Gag is sufficient to form VLPs efficiently (46), indicating that ubiquitination only within this part of Gag is relevant to budding. We generated pCMV-MACA and pCMV-MACA-K74R expression plasmids and performed immunoblot analysis of pelleted supernatants and cellular extracts of 293T cells transfected with these plasmids (Fig. 1D). The pelleted supernatants from WT MA-CA samples showed the additional bands indicative of ubiquitination, visible above the band of the MA-CA protein migrating with the predicted mobility of 43 kDa. These bands were absent in the MA-CA-K74R samples. In addition, the MA-CA-K74R protein was less competent at promoting VLP release than WT MA-CA; WT MA-CA and MA-CA-K74R released 77% ± 3% and 55% ± 2% of the total anti-MA-reactive material into the supernatant, respectively (average of three independent experiments). These results indicate that the K74 residue is the only ubiquitination site in MA-CA and that other ubiquitination events in Gag, should they occur, are not relevant for budding.
The K74R mutation reduces HTLV-1 particle release.
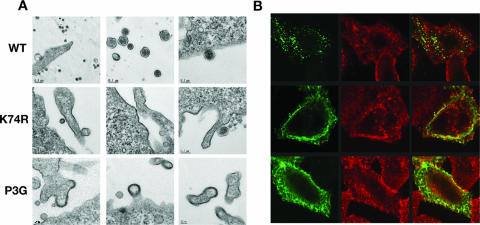
We next compared the phenotype of the K74R mutant to that of the PPPY− mutant (P3G) by TEM and immunofluorescence microscopy. We performed TEM on 293T cells transfected with WT, K74R, and P3G versions of pCMVHT-1ΔX. Virus-like particles with mature HTLV-1 morphology were readily detected in cells expressing WT pCMVHT-1ΔX (Fig. 2A, top panels). In contrast, very few HTLV-1 particles were observed in K74R samples and no tethered viral particles, commonly observed for LD mutants, were detected. Instead, we observed a thickening of the membranes comparable to those seen in the P3G samples, which we and others had previously reported for PPPY− mutants (Fig. 2A, middle and bottom panels).
FIG. 2.
The K74R and PPPY− mutants have similar phenotypes. (A) Representative TEM images of 293T cells transfected with pCMVHT-1ΔX (WT [top row]), pCMVHT-1Δ-K74R (middle row), or pCMVHT-1ΔX-P3G (bottom row). (B) Confocal images of HeLa cells expressing pCMVHT-1ΔX, pCMVHT-1Δ-K74R, or pCMVHT-1ΔX-P3G. Cells were stained with monoclonal anti-p19(MA)/anti-mouse Alexa488 (green), and wheat germ agglutinin-tetramethyl rhodamine isothiocyanate (TRITC) (red), which stains plasma membranes and the trans-Golgi compartment.
Figure 2B shows typical examples of HeLa cells transfected with WT, K74R, and P3G versions of pCMVHT-1ΔX stained with anti-HTLV-1 MA monoclonal antibody with membranes visualized with rhodamine-labeled wheat germ agglutinin. WT Gag/MA was seen as discrete cytoplasmic puncta with very little staining in the plasma membrane. In contrast, K74R and P3G Gag/MA accumulated in the plasma membrane, staining in large sheets. Taken together, these results suggest that ubiquitination of K74 contributes to the efficient morphogenesis and/or release of viral particles.
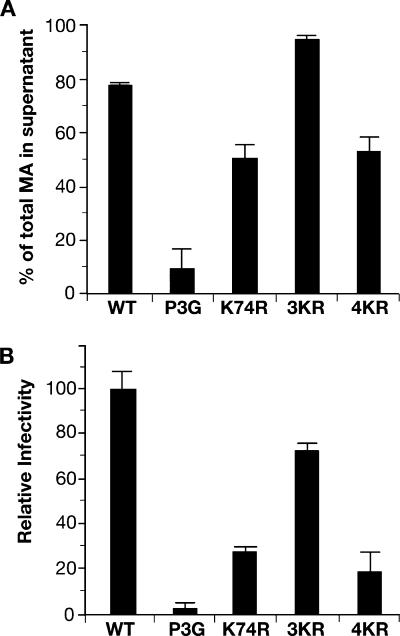
To quantify the effects of the K74R mutation on particle release, we performed HTLV-1 p19(MA) ELISA analyses on culture supernatants and cell extracts of 293T cells transfected with the WT and mutant HTLV-1 expression plasmids. The results of three independent experiments are summarized in Fig. 3A and show that the K74R mutant consistently released MA-reactive material about 40% less efficiently than the WT. Although this is a significant reduction, it is a less severe defect than that seen with the P3G mutant (80 to 90% reduction). Similar results were reported by LeBlanc et al., who tested the effect of changing basic residues in MA to leucines (27).
FIG. 3.
The K74R mutation decreases, but does not abolish, virus release. (A) The amounts of MA-reactive material in cell extracts and supernatants were determined by HTLV-1 p19(MA) ELISA. The percentage of the total MA released into the supernatants is shown for 293T cells transfected with pCMVHT-1ΔX (WT), pCMVHT-1ΔX-P3G, pCMVHT-1Δ-K74R, pCMVHT-1ΔX-3KR, or pCMVHT-1ΔX-4KR. The graph summarizes three independent experiments. (B) VLPs produced after transfection of 293T cells with the indicated HTLV-1 expression vectors were examined in single-cycle replication assays. Infectivity is represented relative to total MA protein in the culture, and the value for pCMVHT-1ΔX was set at 100%; error bars indicate standard deviations.
We next analyzed the effects of the K74R, 3KR, and 4KR mutations in single-cycle infection assays using an HTLV-1-based transfer vector with a luciferase gene (9). The infectivities of the K74R and 4KR particles were reduced to 25 and 20%, respectively, compared to the WT (Fig. 3B). Again, the consequence of the K74R mutation was less severe than that seen with P3G, which reduced infectivity to levels that were not significantly above background. A small decrease in infectivity was measured for the 3KR mutant. The values obtained for release of mutant VLPs based on infectivity were lower than the values determined by ELISA, because some Gag is always released from cells in the form of vesicles that do not resemble either mature or immature virus particles (Fig. 2B). This is especially noticeable at high expression levels. For this reason, we think that the infectivity data are a more reliable indicator of virus assembly and release in this context.
In summary, the K74R mutation abolished Gag ubiquitination and diminished release of infectious VLPs to 20% of WT levels, indicating that Gag ubiquitination contributes to the efficiency of virus budding. By comparison, mutation of the PPPY motif, which abolishes recruitment of WWP1 and Gag ubiquitination, decreased the release of infectious VLPs to barely detectable levels. The difference between the phenotypes of K74R and P3G mutants suggests that WWP1 is required for events in addition to Gag ubiquitination.
Inhibition of WT and K74R particle release by mutant WWP1.
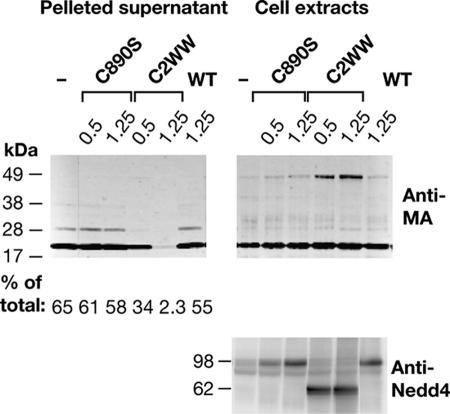
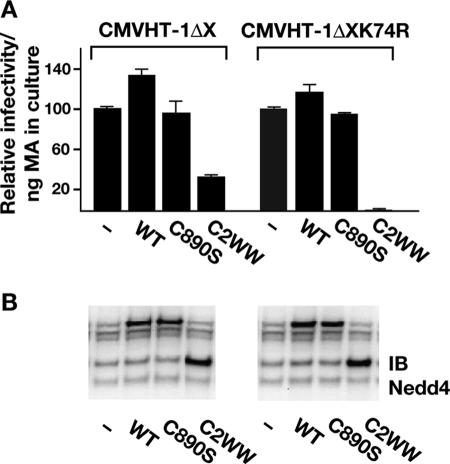
If ubiquitination of Gag is important for virus release, preventing this step by overexpression of a catalytically inactive ubiquitin ligase should be inhibitory. We compared the effect of a WWP1 mutant with a cysteine-to-serine substitution in the active site at position 890 (C890S) to the DN effect of the HECT domain deletion mutant C2WW. Surprisingly, threefold and sixfold overexpression of the C890S mutant, relative to the endogenous protein, did not inhibit VLP release significantly and did not result in a detectable change in MA ubiquitination (Fig. 4 A and B). In contrast, similar levels of expression of C2WW prevented ubiquitination and, at the higher level, almost abolished VLP release.
FIG. 4.
Effects of WWP1 mutants C890S and C2WW on MA ubiquitination and VLP release. 293T cells were transfected with 100 ng of pCMVHT-1ΔX and the indicated amounts of pUCHR empty vector (−), pUCHRWWP1 (WT), pUCHRWWP1-C890S, or pUCHRWWP1-C2WW. Cells and cell culture supernatants (concentrated by ultracentrifugation through a 20% glycerol cushion) were lysed in radioimmunoprecipitation assay buffer, and equal amounts of extracts were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblots were probed with monoclonal anti-HTLV-1 p19(MA) (upper panel) and rabbit anti-Nedd4 antibodies (lower panel), followed by IRDye680-labeled antimouse and IRDye 800-labeled antirabbit antibodies. Molecular size standards (kDa) are shown on the left-hand side. The numbers under the supernatant part of the anti-MA blot indicate the fraction of the total MA (cell extract plus supernatant) released into the supernatant, derived by quantitative analysis of immunoblots.
Next, we tested whether assembly and release of the K74R mutant were affected by ectopic expression of WT and mutated forms of WWP1. If ubiquitination were the major function of the E3 ubiquitin ligase interaction with Gag, one would expect that DN WWP1 had little effect on the release of K74R VLPs. However, if the Gag-WWP1 complex serves as an assembly platform for the recruitment of other proteins, the prediction would be that DN WWP1 inhibits K74R particle release to the same extent seen with WT Gag. We measured the amount of p19 ELISA, and as shown in Fig. 5A, ectopic expression of WWP1 increased the release of WT and K74R VLPs by 30 and 18%, respectively. The catalytically inactive version of WWP1, C890S, caused no change in WT and K74R VLP release. As in the previous experiment, the truncated form of WWP1, C2WW, reduced the release of WT HTLV-1 VLPs by 70%. Surprisingly, release of K74R VLPs was highly sensitive to the DN effect of C2WW; the amount of infectious K74R in the supernatant dropped 25-fold with the addition of C2WW. This result does not fit the predictions stated above; rather it suggests that the domain of Gag around the K74 residue interacts with the HECT domain and stabilizes the Gag-WWP1 interaction. This means that the interaction of Gag with full-length WWP1 is more stable than with the C2WW protein, and as a consequence, more C2WW is needed to compete with the endogenous WT protein. In contrast, the K74R mutant Gag only interacts via the WW domain and thus less C2WW protein is sufficient to compete out WT WWP1. Taken together, the results in this section indicate that the interaction between the HECT domain of WWP1 and the K74 region of Gag is more important to the release of infectious particles than Gag ubiquitination.
FIG. 5.
Budding of the K74R mutant is hypersensitive to inhibition by C2WW. (A) Release of infectious VLPs was assessed in HTLV-1 single-cycle replication assays with either 100 ng pCMVHT-1ΔX or pCMVHTΔX-1-K74R packaging plasmids. 293T cells were transfected with viral vectors in combination with 700 ng pUCHR empty vector (−), pUCHRWWP1 (WT), pUCHRWWP1-C890S, or pUCHRWWP1-C2WW as indicated. Infectivities of CMVHT-1ΔX and CMVHT-1ΔX K74R are represented relative to the pUCHR empty vector controls, which are set at 100% and standardized for the amount of MA expressed in the culture, as determined by HTLV-1 p19(MA) ELISA (data not shown). The experiment was performed in duplicate, and the mean values and standard errors are shown. The experiment was performed two more times with similar results. (B) Immunoblot (IB) analysis of one set of transfected 293T cell extracts from panel A probed with rabbit anti-Nedd4 antibody.
The P3G mutation has a DN effect on VLP release and infectivity.
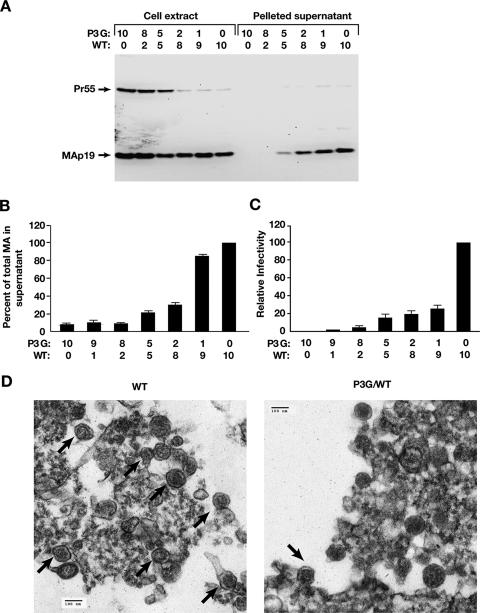
We showed previously that the P3G mutation in the late domain of HTLV-1 causes a severe defect in virus budding. If this were a consequence of the inability of P3G to recruit WWP1 to ubiquitinate a fraction of Gag at the site of virus assembly and release, one would expect that a small amount of WT Gag could restore budding. If, on the other hand, the WWP1-Gag complex is a necessary structural component for particle assembly, it seems likely that even small amounts of P3G Gag could have profound effects on the proper formation of three-dimensional particles. We performed cotransfection experiments with pCMVHT-1ΔX and pCMVHT-1ΔX-P3G plasmids, mixed at different ratios to give a total of 10 μg of DNA. Immunoblotting of cell extracts and pelleted supernatants with anti-MA antibody revealed that WT Gag was not able to rescue assembly and release of P3G Gag (Fig. 6A). Instead, mixtures containing as little as 20% P3G Gag expression construct inhibited HTLV-1 budding, and at equal amounts of P3G and WT Gag, a significant drop in release of VLPs was evident. Quantitative data from ELISA measurements of MA-reactive material present in culture supernatants and cell extracts showed that P3G Gag had a strong DN effect on budding, reducing release fivefold when equal amounts of both Gag proteins were present (Fig. 6B). When P3G Gag accounted for 80 or 90% of the total Gag, VLP release was indistinguishable from that of P3G Gag alone. The DN effect of P3G Gag was even more pronounced in infectivity assays, inhibiting VLP release by 80% when only 1 in 10 Gag molecules carried the P3G mutation (Fig. 6C). Control experiments showed the infectivity of pCMVHTΔX was not diminished when pCMV-β Gal was used instead of pCMVHTΔX-P3G.
FIG. 6.
The P3G Gag mutant is a DN inhibitor of HTLV-1 assembly and release. (A) Immunoblot analysis of virus particle release. 293T cells were transfected with the indicated amounts (μg) of pCMVHT-1ΔX (WT) and pCMVHT-1ΔX-P3G plasmids. Forty-eight hours after transfection, cell extracts and pelleted supernatants were prepared and equivalent amounts of each sample were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblots were probed with rabbit anti-HTLV-1 p19(MA) and developed with horseradish peroxidase-conjugated secondary antibodies and visualized on an AlphaInnotech Imager. Under the conditions used in this experiment, the total amounts of MA protein produced and released in cell cultures transfected with 10 μg of pCMVHTΔX or with 1 μg of pCMVHTΔX and 9 μg carrier plasmid (pCMV-β gal) were the same. (B) Quantitative analysis of VLP release (expressed as the fraction of the total MA present in the supernatant) in cultures transfected with the indicated amounts (μg) of pCMVHT-1ΔX and pCMVHT-1ΔX-P3G plasmids. Transfections were performed as described above, and the amount of MA in supernatants and cell extracts was determined by HTLV-1 p19(MA) ELISA. Release is expressed relative to that of pCMVHT-1ΔX (WT), which is set at 100%. The graph represents the average of two experiments, each done in duplicate; error bars indicate standard deviations. (C) Single-cycle infectivity assays of HTLV-1 viral vectors with the indicated mixtures of pCMVHT-1ΔX (WT) and pCMVHT-1ΔX-P3G. The graph summarizes two experiments, each done in triplicate. Infectivity is expressed relative to that obtained with 10 μg of pCMVHT-1ΔX (WT), which was set at 100%. (D) TEM analysis of material pelleted from supernatants of 293T cells transfected with 10 μg of pCMVHT-1ΔX (WT [left]) or a mixture of 5 μg of the two plasmids (P3G/WT [right]). Thirty-six hours after transfection, supernatants were filtered and particles were pelleted by ultracentrifugation through a 20% glycerol cushion. Pelleted material was fixed in glutaraldehyde and processed for TEM. Arrows indicate particles with HTLV-1 morphology.
We performed TEM analysis of material pelleted through glycerol cushions from supernatants of 293T cultures transfected with mixture of the two plasmids at a ratio of 1 to 1 (Fig. 6D) and compared it to the same material from cells transfected with pCMVHTΔX. Numerous virus particles with HTLV-1 morphology (Fig. 6D, left panel) can be seen in pelleted supernatants from cells transfected with pCMVHT-1ΔX amid other vesicles and amorphous material. About 10-fold-fewer particles were observed in the pelleted supernatants of cells with the mixture of pCMVHT-1ΔX and pCMVHT-1ΔX-P3G (Fig. 6D, right panel). We also analyzed the supernatants of cells transfected with pCMVHT-1ΔX-P3G and as expected found no viral particles (data not shown).
In summary, transfection experiments performed with combinations of plasmids expressing WT and P3G Gag showed that P3G Gag has a DN effect on virus release, especially when measured by VLP infectivity. These findings suggest that the stoichiometry of the interaction of Gag with WWP1 has to be near equimolar or that specific positions in the Gag multimer have to be able to associate with WWP1 for virus assembly and release to occur.
DISCUSSION
We and others have previously shown that budding of HTLV-1 was absolutely dependent on the binding of the PPPY motif to a Nedd4 E3-ubiquitin ligase family member (3, 4, 19, 60). We identified the family member WWP1 as a binding partner and showed that WWP1 lacking the enzymatically active HECT domain exerted a strong DN effect on virus release (3, 4, 19, 60). The role of Gag ubiquitination in the release of retrovirus particles is not well understood, in part because no specific lysine could be pinpointed as the target for the relevant ubiquitination (14, 15, 51). In the work presented here, we identified a single lysine in MA, K74, as the substrate for ubiquitination of HTLV-1 Gag. This result was unexpected, as ubiquitination in other retroviruses can occur on several lysine residues in Gag and mutation of individual targets has little effect on the overall ubiquitination status (14, 51). However, based on the structure of HTLV-1 MA, it seems likely that the other lysines in MA are inaccessible to the ubiquitin ligase, because they are on the other side of the molecule and probably are involved in interactions with the membrane.
The ubiquitination of a single lysine residue in MA made it possible to examine the roles of the Gag-WWP1 interaction and the resulting ubiquitination in the assembly and release of HTLV-1 particles. The K74R mutant released infectious particles with 25% the efficiency of WT virus, in contrast to the PPPY− mutant, which was noninfectious and released very few VLPs. Surprisingly, the K74R mutant was hypersensitive to the DN effect of the HECT domain-deleted form of WWP1. If ubiquitination itself were the main reason for the interaction between Gag and WWP1, one would expect that removing the ubiquitination substrate sites in Gag should have severe effects on virus budding. Given the absolute requirement for the recruitment of a HECT ubiquitin ligase to the budding complex, these results indicate that while important, ubiquitination at K74 is not as essential for virus assembly and release as the physical interaction with WWP1. The interaction of WWP1 with Gag may have several functions that are not mutually exclusive.
In addition to ubiquitination of Gag, WWP1 may be required to ubiquitinate other factors that are brought to the budding HTLV-1 particle. Many of the interactions in the MVB pathway are held together by multiple binding events, each of which has a relatively high dissociation constant. It is thought that this is necessary to facilitate the progression from one step to the next. In particular, interactions of ubiquitin binding motifs to ubiquitin are known to be weak and are often stabilized in a multimeric complex (22).
It is still unclear how the HTLV-1 Gag-WWP1 complex connects to downstream components of the MVB machinery. In fact, the mechanisms by which PPXY motifs in general connect to the MVB pathway have not been defined. The fact that a DN mutant of Vps4 inhibits release of virus particles, indicates that HTLV-1 (G. Heidecker, data not shown), like all other retroviruses tested to date, utilizes this last step of MVB biogenesis. The HECT domain of WWP1 is about 45 kDa in size and, based on its function, has to interact with at least three other proteins: ubiquitin, which covalently but transiently binds to C890; E2 ubiquitin conjugase, which delivers the activated ubiquitin; and the acceptor lysine in the substrate protein. It is generally assumed that the C2 and WW domains are mainly responsible for substrate specificity. Martin-Serrano et al. recently presented evidence that the HECT domain of WWP1 is able to connect to the late endosomal compartment and could thus play a role in the recruitment of MVB components to the site of assembly for PPPY-dependent viruses (30). This is consistent with our observation that overexpression of the enzymatically inactive C890S mutant of WWP1 did not result in a significant reduction of release or infectivity of WT HTLV-1 particles. Martin-Serrano et al. (30) presented evidence that WWP1 was a better effector for PPPY motifs from MLV, RSV, and Ebola virus than Nedd4.1 and that the DN effects of WWP1 C890S were small compared to those of HECT-truncated WWP1. It is possible that a stronger DN effect can be achieved with C890S than we observed, depending on relative expression levels of HTLV-1 and ubiquitin ligase constructs. Nevertheless, the DN effect of C2WW was significantly stronger than that of C890S and resulted in a 25-fold drop in the release of infectious VLPs. These findings support the hypothesis that it is the recruitment of the HECT domain structure rather than its activity that is important for HTLV-1 budding.
For HTLV-1 PPPY− mutants, morphogenesis is arrested at an early stage prior to the rounding up of nascent particles. This suggests that the recruitment of the Nedd4 ligase affects the structure of Gag and facilitates the assembly of HTLV-1 particles. In contrast, RSV and MLV, which also have PPPY as their dominant LD motif, form tethered, immature virus particles when the motif is mutated or a DN version of the ubiquitin ligase is present (8, 11). The hypothesis that the interaction between the HTLV-1 MA domain and WWP1 is important for the structure of the Gag multimer is supported by the finding that PPPY− Gag exerts a strong DN effect on virus assembly and release. Similar studies have been performed with RSV and MPMV. Contradictory results have been reported in the RSV system: one study showed that budding of mutants could be rescued when coexpressed with WT Gag protein (61), while the other reported a DN phenotype for PPPY− constructs (51), similar to that seen in the MPMV system (13). In neither of the latter two cases were the DN effects as pronounced as in the HTLV-1 system. The strength of the DN effect suggests that either every Gag molecule binds to a WWP1 molecule or specific, spatially defined Gag molecules in oligomers interact with WWP1 at some point in the assembly process.
Our finding that eliminating the WWP1 ubiquitin ligase substrate site in Gag makes budding hypersensitive to the DN effect of C2WW supports the idea that the interaction of K74 with the HECT domain is important for HTLV-1 budding. It is likely that the driving force for Gag-WWP1 interaction is the binding of PPPY motif by the WW domains of WWP1, which positions the HECT domain to interact with K74. The latter interaction may induce changes in the conformation of the complex necessary for virion morphogenesis and release. In addition, the HECT domain may recruit other cellular proteins that connect to the MVB pathway.
Acknowledgments
We thank Jennifer Brown for graphics.
This work was supported by the Intramural Research Program of the National Institutes of Health and the National Cancer Institute, Center for Cancer Research. This publication has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bache, K. G., A. Brech, A. Mehlum, and H. Stenmark. 2003. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blot, V., F. Perugi, B. Gay, M. C. Prevost, L. Briant, F. Tangy, H. Abriel, O. Staub, M. C. Dokhelar, and C. Pique. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117:2357-2367. [DOI] [PubMed] [Google Scholar]
- 4.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. Methods 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calattini, S., S. A. Chevalier, R. Duprez, P. Afonso, A. Froment, A. Gessain, and R. Mahieux. 2006. Human T-cell lymphotropic virus type 3: complete nucleotide sequence and characterization of the human Tax3 protein. J. Virol. 80:9876-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, A. M., M. A. Massiah, B. G. Turner, W. I. Sundquist, and M. F. Summers. 1996. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J. Mol. Biol. 264:1117-1131. [DOI] [PubMed] [Google Scholar]
- 7.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 8.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Soc. Exp. Biol. Med. 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derse, D., S. A. Hill, P. A. Lloyd, H.-k. Chung, and B. A. Morse. 2001. Examining human T-lymphotropic virus type 1 infection and replication by cell-free infection with recombinant virus vectors. J. Virol. 75:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derse, D., S. A. Hill, G. Princler, P. Lloyd, and G. Heidecker. 2007. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. USA 104:2915-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 12.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottwein, E., J. Bodem, B. Müller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottwein, E., S. Jäger, A. Habermann, and H.-G. Krausslich. 2006. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 Gag cause a late budding defect. J. Virol. 80:6267-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, E., and H.-G. Kräusslich. 2005. Analysis of human immunodeficiency virus type 1 Gag ubiquitination. J. Virol. 79:9134-9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haglund, K., S. Sigismund, S. Polo, I. Szymkiewicz, P. P. Di Fiore, and I. Dikic. 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5:461-466. [DOI] [PubMed] [Google Scholar]
- 17.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidecker, G., S. Hill, P. A. Lloyd, and D. Derse. 2002. A novel protease processing site in the transframe protein of human T-cell leukemia virus type 1 PR76gag-pro defines the N terminus of RT. J. Virol. 76:13101-13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidecker, G., P. A. Lloyd, K. Fox, K. Nagashima, and D. Derse. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicke, L. 2002. Protein regulation by monoubiquitin. Nat. Struct. Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 21.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurley, J. H., S. Lee, and G. Prag. 2006. Ubiquitin-binding domains. Biochem. J. 399:361-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingham, R. J., G. Gish, and T. Pawson. 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23:1972-1984. [DOI] [PubMed] [Google Scholar]
- 24.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 25.Landy, A. 1989. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 58:913-949. [DOI] [PubMed] [Google Scholar]
- 26.Le Blanc, I., M. C. Prévost, M.-C. Dokhélar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Blanc, I., A. R. Rosenberg, and M.-C. Dokhélar. 1999. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73:1860-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marmor, M. D., and Y. Yarden. 2004. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23:2057-2070. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, M. S., E. T. Bodine, S. Hill, G. Princler, P. Lloyd, H. Mitsuya, M. Matsuoka, and D. Derse. 2007. Phenotypic and genotypic comparison of human T-cell leukemia virus type 1 reverse transcriptase from infected T-cell lines and patient samples. J. Virol. 81:4422-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 35.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 37.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ott, D. E., L. V. Coren, R. C. Sowder, J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 43.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 44.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raiborg, C., K. G. Bache, A. Mehlum, and H. Stenmark. 2001. Function of Hrs in endocytic trafficking and signalling. Biochem. Soc. Trans. 29:472-475. [DOI] [PubMed] [Google Scholar]
- 46.Rayne, F., F. Bouamr, J. Lalanne, and R. Z. Mamoun. 2001. The NH2-terminal domain of the human T-cell leukemia virus type 1 capsid protein is involved in particle formation. J. Virol. 75:5277-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membrane Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 48.Sakurai, A., J. Yasuda, H. Takano, Y. Tanaka, M. Hatakeyama, and H. Shida. 2004. Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect. 6:150-156. [DOI] [PubMed] [Google Scholar]
- 49.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuh, M., S. A. Hill, and D. Derse. 1999. Defective and wild-type human T-cell leukemia virus type I proviruses: characterization of gene products and trans-interactions between proviruses. Virology 262:442-451. [DOI] [PubMed] [Google Scholar]
- 51.Spidel, J. L., R. C. Craven, C. B. Wilson, A. Patnaik, H. Wang, L. M. Mansky, and J. W. Wills. 2004. Lysines close to the Rous sarcoma virus late domain critical for budding. J. Virol. 78:10606-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 54.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 55.Tobin, G. J., K. Nagashima, and M. A. Gonda. 1996. Immunologic and ultrastructural characterization of HIV pseudovirions containing Gag and Env precursor proteins engineered in insect cells. Methods 10:208-218. [DOI] [PubMed] [Google Scholar]
- 56.Urbe, S. 2005. Ubiquitin and endocytic protein sorting. Essays Biochem. 41:81-98. [DOI] [PubMed] [Google Scholar]
- 57.Vana, M. L., Y. Tang, A. Chen, G. Medina, C. Carter, and J. Leis. 2004. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J. Virol. 78:13943-13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 60.Wang, H., N. J. Machesky, and L. M. Mansky. 2004. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J. Virol. 78:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolfe, N. D., W. Heneine, J. K. Carr, A. D. Garcia, V. Shanmugam, U. Tamoufe, J. N. Torimiro, A. T. Prosser, M. Lebreton, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. M. Switzer. 2005. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc. Natl. Acad. Sci. USA 102:7994-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]