Abstract
Our functional mapping study of murine gammaherpesvirus 68 (MHV-68, or γHV-68) revealed that a mutant harboring a transposon at the ORF49 locus (ORF49null) evidenced a highly attenuated in vitro growth. ORF49 resides adjacent to and in an opposite direction from RTA, the primary switch of the gammaherpesvirus life cycle. A FLAG-tagged ORF49 protein was able to transcomplement ORF49null, and a revertant of ORF49null restored its attenuated growth to a level comparable to that of the wild type. The FLAG-tagged ORF49 protein promoted the ability of RTA to activate downstream target promoters and enhanced virus replication from the ORF50null virus in the presence of RTA. Furthermore, ORF49 enhanced wild-type virus replication by increasing the RTA transcript levels. Our data indicate that ORF49 may perform an important function in MHV-68 replication in cooperation with RTA.
Gammaherpesviruses, including Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), herpesvirus saimiri, and murine gammaherpesvirus 68 (MHV-68, or γHV-68), are important pathogens as they are associated with various malignancies (19). MHV-68 has been used as an animal model system for human gammaherpesviruses, based on its ability to establish both lytic and latent infections in vivo as well as in vitro (6, 16, 23, 25).
Although latent infection is critical for gammaherpesvirus-associated malignancies, reactivation from latency and lytic replication have been suggested to be important in the persistence and pathogenesis of gammaherpesviruses (13). An immediate-early protein, RTA (BRLF1 or ORF50), represents a sensitive switch system in the gammaherpesviruses (2-4, 15, 24, 27, 29, 30). ZTA (BZLF1 or ZEBRA) and RTA in EBV (gamma-1 herpesvirus) perform essential functions in the disruption of viral latency, thereby activating downstream genes in a cooperative fashion (2-4, 7, 17, 30). However, there is no functional ZTA homologue in gamma-2 herpesviruses and RTA alone is sufficient and necessary for the initiation of lytic replication (12, 24, 27-29).
Our functional mapping study using a genome-wide mutant library of MHV-68 revealed that a mutant harboring a transposon at the ORF49 locus (ORF49null) manifested attenuated growth, thereby suggesting the important functions of ORF49 in virus replication in vitro (22). As ORF49 is located adjacent to and in a reverse orientation relative to ORF50, the major coding region of RTA, it is conceivable that ORF49 may function in cooperation with RTA, the master switch gene of the virus life cycle.
ORF49 is thought to be a positional homologue of the BRRF1 gene (Na) of EBV, which is located in an opposite orientation between the immediate-early genes ZTA and RTA and expressed under the control of ZTA (20). A recent study demonstrated that Na is a transcription factor that activates the ZTA IE promoter through its effects on c-Jun and suggested that Na enhances the RTA-mediated induction of lytic EBV infection in certain cell lines (9). In this report, we determined the function of ORF49 in the replication of MHV-68 by using a genetic approach.
MATERIALS AND METHODS
Cells, viruses, and plaque assays.
BHK21 (baby hamster kidney fibroblast cell line), 293T, and Vero (green monkey kidney cell line) cells were cultured in complete Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and supplemented with penicillin and streptomycin (10 units/ml) (HyClone). MHV-68 virus was originally obtained from the American Type Culture Collection (VR1465). The ORF49null and the ORF50null mutant viruses were generated by in vitro Mu transposition with an infectious bacterial artificial chromosome (BAC) clone of MHV-68 (pMHV-68) and purified STM transposons as described previously (22). A marker rescue mutant of ORF49null (ORF49nullMR) was generated by a two-step allelic exchange method as described by Smith and Enquist (21). The donor strain was Escherichia coli GS111 carrying the shuttle plasmid pGS284/49WT, and the recipient strain was GS500 (recA+) harboring the BAC DNA of ORF49null (pORF49null). The rescued site was confirmed by DNA sequencing, and the genome integrity of pORF49nullMR was examined by restriction enzyme digestion and Southern blot analysis as described previously (29). Subsequently, either pORF49null or pORF49nullMR was transfected into BHK21 cells by using Lipofectamine Plus (Invitrogen). The titers of the produced viruses were determined by plaque assays, using a monolayer of Vero cells overlaid with 1% methylcellulose. After 5 days of infection, the cells were fixed and stained with 2% crystal violet in 20% ethanol. Plaques were then counted to determine the titers. The representative pictures of distinct plaque morphologies were taken using an SZX9 stereomicroscope (Olympus).
Molecular cloning.
MHV-68 ORF49 (nucleotides [nt] 66741 to 67643) was cloned into a vector, pCMV-FLAG2 or pEGFP-C1, using the EcoRI and BamHI sites. The inserted ORF49 DNA fragment was amplified via PCR from the MHV-68 genome (pMHV-68), using the primers ORF49F (5′-aga gaattc AACAGAAAACATG GGGGATG-3′) and ORF49R (5′-cgc ggatcc CTACGAGGGAATTT CTGC-3′). In all sequences, the lowercase letters represent restriction enzyme sites and additional nucleotides for cloning the PCR product, and the uppercase letters represent viral or cellular sequences.
To construct the shuttle plasmid for the generation of pORF49nullMR, a 720-bp fragment (nt 66961 to 67680) was PCR amplified from the MHV-68 genome with the primers ORF49AF (5′-acgc gtcgac TCCACAATCGCTTCTAACTC-3′) and ORF49BR (5′-cctt gcatgc TGCTTCGATCCGTGCCAGTCCTATAAGAAC-3′) and cloned into pGS284 by using the SalI and SphI sites. The 720-bp fragment contains the transposon insertion site (nt 67320) at the center, with approximately 360 bp of the genomic sequence on each side. Each construct was DNA sequenced to confirm that there were no additional mutations, deletions, or insertions.
Cell transfection.
Plasmid DNAs were prepared with the standard method by using a plasmid maxi or midi kit according to the manufacturer's recommendations (QIAGEN). For BHK-21 cell transfection, approximately 6 × 105 cells were transferred to six-well culture plates 1 day prior to the experiment. Plasmid DNA or BAC DNA was transfected into cells by the use of Lipofectamine Plus (Invitrogen). The transfection into Vero cells was performed by the use of Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen). Cells were usually assayed at 2 days posttransfection, unless otherwise indicated. For the luciferase reporter assays, 3.5 × 105 293T cells were transfected with 50 to 250 ng of FLAG-ORF49 and 50 ng of a reporter plasmid into 12-well plates in the presence or absence of the RTA-expressing plasmid (0.2 ng) by using a calcium phosphate transfection method. Each transfection for reporter assays was performed in triplicate and contained 40 ng of the β-galactosidase expression plasmid as an internal control. At 24 h posttransfection, cells were washed with 1× phosphate-buffered saline (PBS) and subjected to reporter assays.
Viral DNA isolation and analysis by quantitative real-time PCR.
Cell extracts were resuspended with lysis buffer (40 mM Tris [pH 7.7], 20 mM EDTA [pH 8.0], 0.2 M NaCl, and 1% sodium dodecyl sulfate) and incubated with proteinase K (0.5 mg/ml) overnight at 56°C. Genomic DNAs, including viral DNAs, were isolated by a standard method of phenol chloroform extraction and ethanol precipitation. Isolated viral DNAs were then treated with DpnI to remove input plasmid DNAs of the virus genome. The whole genomic DNA (50 ng) was used for real-time PCR. Real-time PCR was performed using SYBR green PCR and specific primers in a 20-μl reaction mixture for M1 (M1F, 5′-CCTGGCCATGGTTACATACTC-3′; M1R, 5′-GGAACATAATCCATAAGCAGGGT-3′) (18). Real-time PCR was run at 50°C for 2 min and then 45 cycles at 95°C for 10 s, 58°C for 15 s, and 72°C for 20 s, followed by melting curve analysis. Real-time PCRs were performed in duplicate on an iCycler (Bio-Rad), and the results were analyzed according to the manufacturer's instructions.
Antibodies, Western blotting, and fluorescence and indirect-immunofluorescence assays.
Cell extracts were analyzed with the following primary antibodies: rabbit polyclonal antibody to ORF26 (1:500), ORF45 (1:400), M9 (1:500), or MHV-68 (1:2,000) (10) and mouse monoclonal antibodies to FLAG tag (1:5,000), β-actin (1:500), or α-tubulin (1:1,000). Anti-rabbit or anti-mouse immunoglobulin G (IgG) conjugated with horseradish peroxidase (Santa Cruz Biotechnology) was used as a secondary antibody. The proteins in the Western blots were detected by chemiluminescence detection (ECL Plus system; Amersham Pharmacia Biotech), and the signals were detected by using an image analyzer, Chemi-DOC XRS (Bio-Rad) or ProXPRESS 2D (Perkin Elmer). The same membranes were reprobed with antibody against actin or tubulin as a loading control.
For fluorescence and indirect-immunofluorescence assays, transfected cells were washed with 1× PBS and then fixed with absolute methanol at −20°C for 10 min and permeabilized in ice-cold 1× PBS for 5 min. The slides were then incubated with monoclonal anti-FLAG at 37°C for 1 h and incubated with rhodamine-conjugated goat anti-mouse IgG (Jackson) at 37°C for 1 h. To stain the cell nucleus, mounting solution containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Inc.) was used. Slides were examined, and representative images were photographed on a confocal laser scanning microscope (LSM 5 Exciter) with LSM analysis software (Carl-Zeiss).
Promoter reporter assays.
The luciferase reporter assay system (Promega) was used to measure promoter activity. Transfected 293T cells in 12-well plates were washed with 1× PBS and incubated with 200 μl of 1× reporter lysis buffer provided by the manufacturer. Lysates were frozen, thawed once, and centrifuged at top speed in a microcentrifuge for 5 min. Lysates were analyzed for luciferase activity by using a 20/20n luminometer (Turner BioSystem) and for β-galactosidase activity with β-galactosidase substrate buffer by using a Victor3 plate reader (Perkin Elmer). The β-galactosidase activates were used as an internal control.
Quantitative RT-PCR and RT-PCR.
Total RNA was extracted from transfected cells by using TriReagent (Molecular Research Center) according to the manufacturer's instructions. The mRNA was subsequently reverse transcribed into cDNA by using SuperScript III RNase H− reverse transcriptase (Invitrogen). A set of gene-specific primers for ORF50 (50F, 5′-GGCCGCAGACATTTAATGAC-3′; 50R, 5′-GCCTCAACTTCTCTGGATATGCC-3′) was used for reverse transcription (RT)-quantitative PCR. The PCR product amplified by β-actin primers (actinF, 5′-GTATCCTGACCCTGAAGTACC-3′; actinR, 5′-TGAAGGTCTCAAACATGATCT-3′) was used as an internal control. The primers used for RT-PCR were SH-50 (5′-GATTCCCCTTCAGCCGATAAG-3′ as the forward primer and 5′-CAGACATTGTAGAAGTTCACCT-3′ as the reverse primer) for the spliced RTA transcript, ORF57SP (5′-ACCAAATGATGGAAGGACTAC-3′ as the forward primer and 5′-GCAGAGGAGAGTTGTGGAC-3′ as the reverse primer) for the spliced ORF57 transcript, ORF29 (5′-TTCTCATTGGCA TCTTTGAGG-3′ [R1] as the forward primer and 5′-GGAAAATGGGGTGATCCTGT-3′ [L4] as the reverse primer) for ORF29, and SH-73 (5′-GATGAGGGAAGTGTTGGTGATG-3′ as the forward primer and 5′-CTCGTGAGTAGCGCCGACTAG-3′ as the reverse primer) for the spliced transcript of ORF73 (1).
RESULTS AND DISCUSSION
Highly attenuated in vitro growth of MHV-68 ORF49null virus.
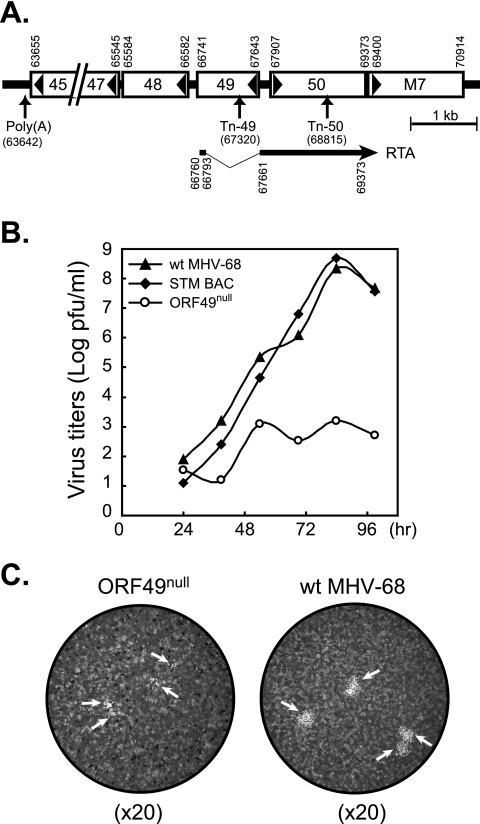
The ORF49null virus was one of the six mutants that exhibited attenuated in vitro growth among a genome-wide random mutant library of MHV-68 (22). This mutant library was generated using in vitro Mu transposition, with an infectious BAC clone of MHV-68 as a target template. Replication-competent mutants were reconstituted by transfecting their BAC DNAs into permissive cells (22). The transposon-inserted site of ORF49null (nt 67320; Tn49) is indicated in Fig. 1A. To confirm the attenuated phenotype of the ORF49null virus, the ORF49null virus was subjected to multiple-step growth curve analysis. Wild-type MHV-68 virus and an STM BAC mutant harboring a transposon at the sequence of the BAC plasmid were also tested as a control. While the wild type and the STM BAC virus were able to grow over 108 PFU/ml, ORF49null never reached higher than 103 to 104 PFU/ml, indicating significant impairment of virus growth in vitro (Fig. 1B). It was also noted that the average plaque size of the ORF49null virus was smaller than that of the wild type (Fig. 1C), thereby suggesting the important functions of ORF49 in virus replication in vitro.
FIG. 1.
Attenuated in vitro growth of the ORF49null virus. (A) Diagram of the genomic locus harboring ORF49. Open reading frames are shown in open boxes; the directions of the open reading frames are indicated by the arrowheads within the boxes. Numbers indicate nucleotide positions in the MHV-68 genome (26). The positions of the Mu transposon inserted into ORF49null and ORF50null are shown as Tn49 and Tn50 (22). An encoded portion of spliced RTA transcripts is shown with an arrow (29). (B) Analysis of in vitro multiple-step growth curves of ORF49null, STM BAC, and wild-type (wt) MHV-68 in fibroblasts. The viruses were infected into BHK21 cells at multiplicities of infection of 0.05, and total cell lysates were harvested at different time points. STM BAC has a transposon insertion at the sequence of the BAC plasmid, serving as a control. The growth properties of individual STM mutants were compared with those of STM BAC and wild-type MHV-68 by using plaque assays. (C) Plaque morphology of the ORF49null and the wild-type virus. Representative pictures were taken from a well of a plaque assay performed on Vero cells with similar titers of each virus by using a stereo microscope (×20 magnification).
Characterization of the MHV-68 ORF49 protein.
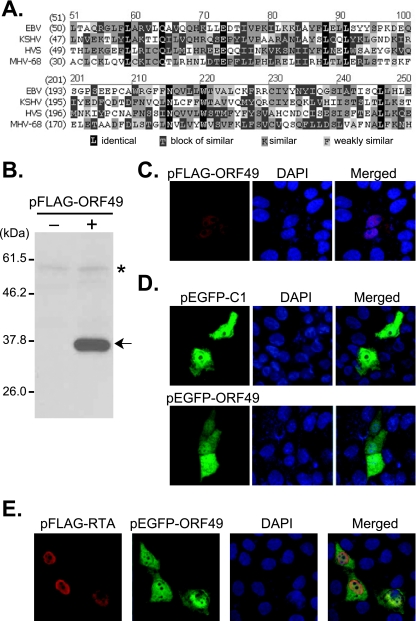
MHV-68 ORF49 (nt 66741 to 67643) has been predicted to be translated in a leftward direction and to share its poly(A) site with ORF45, -46, -47, and -48 (Fig. 1A) (26). The genomic sequence of ORF49 overlaps in reverse orientation with the first exon sequence of RTA (amino acids 1 to 12) (29). Upon analysis by Vector NTI (Invitrogen), the MHV-68 ORF49 protein sequence indicates limited identities (13% to 17%) to its homologues in other gammaherpesviruses (Fig. 2A). In order to determine the functions of the ORF49 protein, the MHV-68 ORF49 genome sequence was cloned into a vector, pCMV-FLAG2 or pEGFP-C1. When pFLAG-ORF49 was transiently transfected into BHK21 cells, the FLAG-tagged ORF49 protein was expressed as a recombinant protein of the predicted size (∼35 kDa) (Fig. 2B). The enhanced green fluorescent protein (EGFP)-fused ORF49 protein was also expressed as a recombinant protein of a similar size in addition to EGFP alone (data not shown).
FIG. 2.
MHV-68 ORF49 encodes a putative homologue of the EBV BRRF1 gene product and is located in both the cytoplasm and the nucleus. (A) Comparison of predicted amino acid sequences of MHV-68 ORF49 with those of homologues in other members of the gammaherpesvirus subfamily. Shown is a comparison of MHV-68 ORF49 (amino acids 30 to 79 and 170 to 219) with a relatively greater homology. (B) Expression of FLAG-tagged ORF49. Vero cell lysates transfected with either vector alone or pFLAG-ORF49 (200 ng) were separated on 12% gel. Monoclonal anti-FLAG antibody was used for Western analysis. The asterisk indicates nonspecific bands used as a loading control. (C, D) Subcellular localization of the ORF49 protein. The Vero cells were transfected with pFLAG-ORF49 (C), pEGFP-C1 (D, upper), or pEGFP-ORF49 (D, lower) and then fixed at 48 h posttransfection. FLAG-tagged ORF49 was analyzed by indirect-immunofluorescence assays with anti-FLAG monoclonal antibody, followed by incubation with rhodamine-conjugated goat anti-mouse IgG (red). The GFP signals (green) from EGFP and EGFP fusion protein were visualized after fixation. (E) Subcellular localization of FLAG-tagged RTA and EGFP-fused ORF49 in cotransfected cells. Vero cells were cotransfected with FLAG-tagged RTA and EGFP-fused ORF49 and subjected to indirect-immunofluorescence assays as described for panels C and D. The nucleus was stained with DAPI (blue). All fluorescence images were obtained with a confocal laser scanning microscope.
To examine the subcellular localization of the ORF49 protein, Vero cells were transfected with pFLAG-ORF49 and subjected to indirect-immunofluorescence assays with anti-FLAG antibody using a confocal laser scanning microscope. At 48 h posttransfection, FLAG-tagged ORF49 was detected both in the cytoplasm and in the nucleus (Fig. 2C). To verify this result, either pEGFP-ORF49 or pEGFP-C1 was transfected, and the results showed that ORF49 was evenly distributed throughout the cytoplasm and the nucleus, the pattern of which is similar to that of the pEGFP vector control (Fig. 2D).
Ectopic expression of the FLAG-ORF49 protein transcomplemented the attenuated growth of the ORF49null virus.
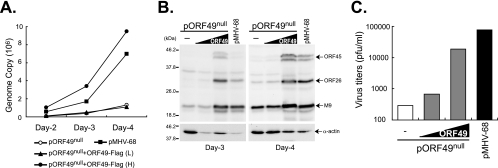
In order to determine whether the FLAG-ORF49 protein is capable of rescuing the attenuated phenotype of ORF49null, the BAC DNA of ORF49null (pORF49null) was transfected into BHK21 cells along with increasing doses of pFLAG-ORF49. The BAC DNA harboring the entire genome of MHV-68 (pMHV-68) was also transfected as a control.
At the indicated time points, DNA was extracted from the cells and digested with DpnI to remove the transfected BAC DNA. Real-time PCR using primers against MHV-68 M1 was performed (18). While pORF49null transfection alone evidenced highly attenuated virus replication, cotransfection of pFLAG-ORF49 significantly increased the viral genome copy number of pORF49null to a level comparable to that of the wild type (Fig. 3A). Western analyses using polyclonal antibodies against ORF45 (viral tegument protein), ORF26 (capsid protein), and M9 proteins (ORF65, small capsid protein) (10) also showed increased viral gene expressions from pORF49null in pFLAG-ORF49 cotransfected cells in comparison to results with pFLAG-CMV (Fig. 3B).
FIG. 3.
Recombinant ORF49 protein of MHV-68 transcomplemented the attenuated growth of ORF49null. BHK21 cells were cotransfected with no pFLAG-ORF49 or 100 ng (L) or 950 ng (H) of pFLAG-ORF49 in the presence of the BAC DNA of the ORF49null virus (50 ng). MHV-68-cloned BAC DNA (pMHV-68; 50 ng) was also transfected as a control. The transfected cells were harvested at the indicated time points. (A) Enhanced viral DNA replication of the ORF49null virus by FLAG-ORF49. Total genomic DNAs were extracted from the cells and digested with DpnI to remove the transfected BAC DNA. Real-time PCR using primers against MHV-68 M1 was performed to quantitate viral DNA, as described previously (18). (B) Increased viral gene expressions of the ORF49null virus. Total cellular lysates were prepared, separated on 12% gel, and subjected to Western analysis using polyclonal antibodies against ORF45 (tegument protein), ORF26 (capsid protein), and M9 (small capsid protein). α-Actin was used as a loading control. (C) FLAG-ORF49 promoted virion production of the ORF49null virus. Cultured supernatants were harvested at 4 days posttransfection and subjected to plaque assays. Similar results were obtained from at least three independent experiments, and shown is a representative result.
Moreover, cotransfected pFLAG-ORF49 increased infectious virion production in a dose-dependent manner, whereas the titers of infectious virions from pORF49null were quite low, less than ∼300 PFU/ml (Fig. 3C). Consistent with our previous results, the average plaque size of the ORF49null virus was smaller than that of the wild type and was not changed upon transcomplementation by ORF49 protein (data not shown). Therefore, our results indicated that the attenuated growth phenotype of the ORF49null virus was due to disrupted expression of the MHV-68 ORF49 protein and that our cloned pFLAG-ORF49 protein expressed a functional ORF49 protein.
Generation of an ORF49null marker rescue virus.
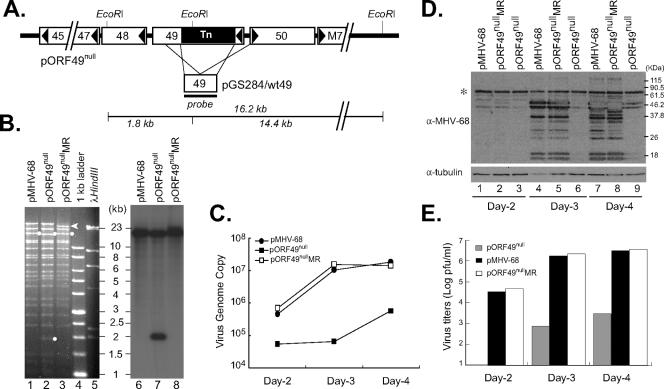
To further confirm the results from transcomplementation of ORF49null, we generated a marker rescue mutant of the ORF49null virus by using a conventional two-step allelic exchange method with a wild-type ORF49 shuttle vector as described in Materials and Methods. A marker rescue mutant (ORF49nullMR) was analyzed by EcoRI digestion and Southern blotting and showed the expected DNA fragments except a top band indicated with an arrowhead (Fig. 4A and B). The largest DNA fragment of ORF49nullMR bearing terminal repeats migrated faster than that of either the wild type or ORF49null. The cells were transfected with pORF49null and pORF49nullMR as well as pMHV-68. As demonstrated in viral DNA replication, viral gene expression, and infectious-virion production, ORF49nullMR restored the attenuated phenotype of ORF49null to a level similar to that of the wild type (Fig. 4C to E). The multiple-step growth curves of ORF49nullMR were also similar to those of the wild type (data not shown). Taken together, these results reinforce our hypothesis that disrupted expression of the MHV-68 ORF49 protein results in severe impairment of virus growth in vitro.
FIG. 4.
Generation of a marker rescue mutant of ORF49null. (A) Schematic diagram of construction of a marker rescue mutant of ORF49null. Shown are the genomic organization of ORF49 with a transposon insertion and neighboring genes (pORF49null) (top), a homologous region used to construct marker rescue mutant of ORF49null (middle), and DNA fragments from EcoRI digestion of ORF49null (bottom). The sizes of the diagrams are proportional to their actual genomic sizes. (B) Southern analysis of ORF49null and ORF49nullMR. The BAC DNAs of pMHV-68, pORF49null, and pORF49nullMR were digested with EcoRI and analyzed with a 32P-labeled Southern probe specific to the ORF49-containing fragment as indicated in panel A. (C to E) BHK21 cells were transfected with the BAC DNAs (50 ng) of ORF49null and ORF49nullMR. MHV-68-cloned BAC DNA (pMHV-68; 50 ng) was also transfected as a control. The transfected cells were harvested at the indicated time points and analyzed for viral DNA replication (C), viral gene expressions (D), and virion production (E) as described in the legend to Fig. 3. Western analysis was performed with rabbit serum against MHV-68-infected rabbit cell lysates (α-MHV-68) and with α-tubulin as a control. The asterisk indicates nonspecific bands.
ORF49 functionally cooperates with RTA in virus replication and transactivation.
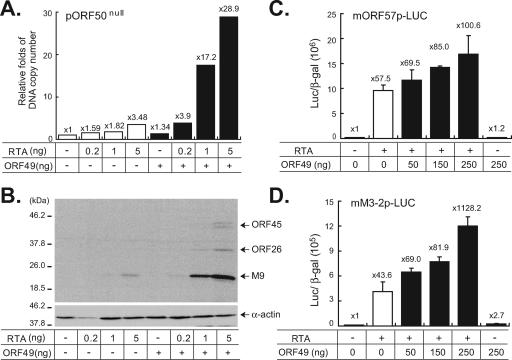
In order to determine whether ORF49 can functionally cooperate with RTA to regulate virus replication, we utilized ORF50null (nt 68815; Tn50) harboring a transposon insertion at ORF50, which would not replicate without the RTA protein provided in trans (Fig. 1A). The BAC DNA of ORF50null was transfected into BHK21 cells with various amounts of pFLAG-RTA in the absence or presence of ORF49 (Fig. 5). Coexpression with ORF49 further promoted viral DNA replication and gene expression from ORF50null (Fig. 5A and B), thereby suggesting that ORF49 functions in cooperation with RTA to enhance viral gene expression and DNA replication.
FIG. 5.
ORF49 facilitates RTA functions. BHK21 cells were cotransfected either with vector alone or with pFLAG-ORF49 (800 ng) in the presence of BAC DNA of ORF50null (pORF50null; 200 ng) and various amounts of an RTA expression plasmid (0, 0.2, 1, and 5 ng). The cells were then harvested at 4 days posttransfection. (A) FLAG-ORF49 increased the viral DNA replication of the ORF50null virus. Total genomic DNAs were extracted from the transfected cells, digested with DpnI, and subjected to quantitative real-time PCR. The viral DNA copy numbers were calculated relative to that of the pORF50null transfected samples. (B) FLAG-ORF49 promoted viral gene expression of the ORF50null virus. Total cell lysates from the transfected cells were prepared, separated on 12% gel, and subjected to Western analysis using polyclonal antibodies against ORF45, ORF26, and M9. α-Actin was used as a loading control. (C, D) FLAG-ORF49 enhanced RTA-mediated transactivation. A luciferase reporter construct harboring approximately 600-bp upstream sequences of ORF57 or M3 was cotransfected into 293 cells with increasing doses of ORF49 (0, 50, 150, and 250 ng) in the absence or presence of RTA (10 ng). A β-galactosidase expression plasmid (40 ng) was also transfected as an internal control. Shown are luciferase activities (n-fold), normalized to the corresponding β-galactosidase activities, relative to those of the reporter alone. Results are presented as the means for triplicates with their standard deviations.
Considering that RTA functions as a potent transcription activator and that the EBV Na protein has been shown to encode a transcription factor (9, 20, 24, 29), the further increase of virus replication in ORF50null by ORF49 may be attributable to the further activation of downstream target genes by ORF49. In order to evaluate this possibility, we utilized two lytic promoter constructs, ORF57 (early) and M3 (early-late) of MHV-68 (11, 14). RTA alone activated both promoters, and cotransfection with increasing amounts of ORF49 further enhanced RTA-mediated transactivation in a dose-dependent manner (Fig. 5C and D). However, unlike the EBV Na protein, ORF49 itself failed to activate these promoters, suggesting that ORF49 participates in RTA-mediated transactivation on the lytic promoters, rather than directly activating the expressions of these genes.
ORF49 promotes virus replication of MHV-68 by increasing RTA expression.
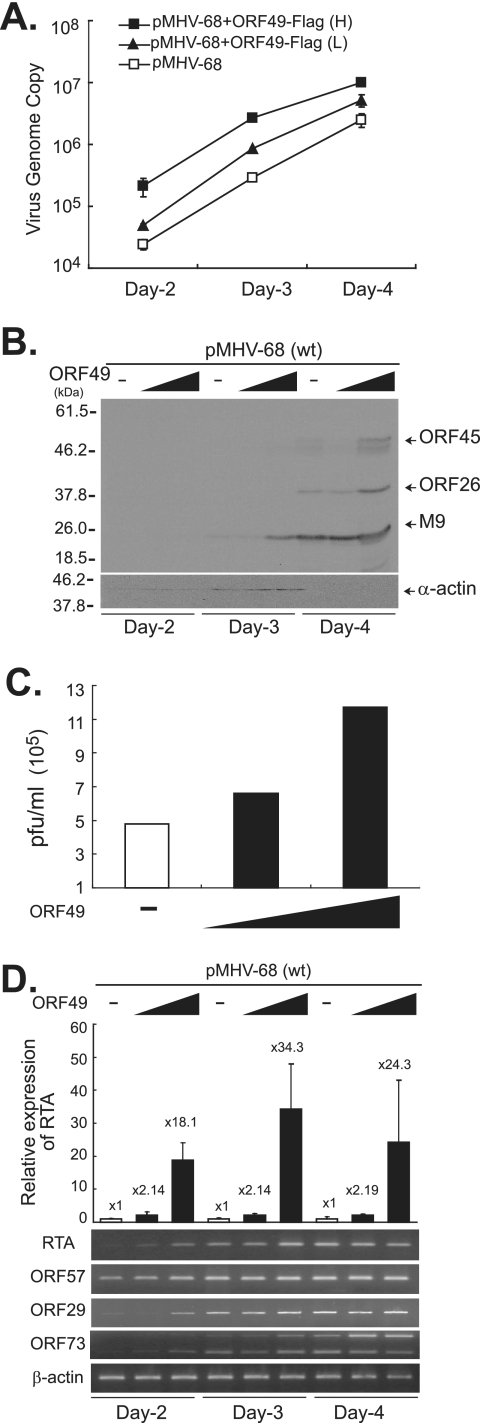
In our previous experiments using ORF50null, we employed suboptimal doses of RTA (<5 ng) to emulate the early phases of virus replication. We then attempted to determine whether ORF49 may function in a similar way when enough RTA is expressed during virus replication. BHK21 cells were transfected with pMHV-68 in the absence or presence of pFLAG-ORF49. The results of Western analyses, real-time PCR, and plaque assays were all consistent with the notion that ORF49 overexpression further promoted viral gene expression, DNA replication, and the production of infectious virions from the wild-type virus (Fig. 6A to C).
FIG. 6.
ORF49 promotes the replication of the MHV-68 virus via an increase in RTA transcription. No pFLAG-ORF49 or 100 ng (L) or 950 ng (H) of pFLAG-ORF49 was cotransfected into BHK21 cells with pMHV-68 (10 ng). The cells were harvested at 2, 3, and 4 days posttransfection. (A) FLAG-ORF49 increased viral DNA replication of the wild type (wt) in a dose-dependent manner. Total genomic DNAs were extracted and subjected to DpnI digestion prior to quantitative real-time PCR. (B) FLAG-ORF49 enhanced viral gene expression with faster kinetics. Total cellular lysates were prepared, separated on 12% gel, and subjected to Western analysis using anti-ORF45, anti-ORF26, and anti-M9 antibodies. α-Actin was utilized as a loading control. (C) Total virion production from the wild type was increased by FLAG-ORF49. Similar results were obtained from at least three independent experiments, and shown is a representative plaque assay result. (D) FLAG-ORF49 increased the levels of transcripts of RTA (immediate early), ORF57 (early), ORF29 (late), and ORF73 (latent) from wild-type MHV-68. Quantitative real-time RT-PCRs using ORF50-specific primers were conducted, and the relative ORF50 expression levels (n-fold) were calculated based on β-actin gene expression. Results are presented as the means for triplicates with their standard deviations. Another set of primers specific for the spliced transcript of RTA was utilized for RT-PCRs, and a representative gel picture is shown. The levels of ORF57, ORF29, and ORF73 transcripts were also measured via RT-PCR, and representative results are shown. β-Actin gene expression was utilized as a control.
Since RTA is known to autoregulate its own expression (2, 5, 17), this may be due to the functional cooperation of ORF49 with RTA on RTA's own promoter. Employing cDNAs converted from the total RNAs of these transfected cells, we conducted quantitative real-time RT-PCR in order to measure transcripts from ORF50 (immediate early). The β-actin gene was employed as a control. Compared with wild-type MHV-68 alone, coexpression with ORF49 resulted in a significant increase in the expression of the RTA transcripts in a dose-dependent manner at all tested times, suggesting that ORF49 functionally cooperated with RTA of MHV-68 on RTA's own promoter (Fig. 6D). The induction (n-fold) peaked at 3 days posttransfection. Using another set of primers (SH-50) that cross the splicing junction of the RTA transcript, we obtained similar results (Fig. 6D). Transcription of downstream genes, such as the ORF57 (early), ORF29 (late), and ORF73 (latent) genes, was also expressed at higher levels, with faster kinetics in ORF49 cotransfected cells, in a dose-dependent manner, than in cells transfected with pMHV-68 alone. These results indicate that ORF49 may facilitate viral replication via promotion of the function for activation of downstream target genes and for autoactivation of the RTA promoter, which in turn increases the RTA expression.
To further examine whether this functional cooperation of ORF49 with RTA is, at least in part, due to colocalization of these proteins, we cotransfected pFLAG-RTA and pEGFP-ORF49 into Vero cells and examined their subcellular localizations by using a confocal laser scanning microscope. At 48 h posttransfection, the nuclear localization of the FLAG-tagged RTA protein was overlapped with that of ORF49 (Fig. 2E). However, coexpression of RTA did not alter the subcellular localization of ORF49, as the ORF49 protein was also found in the cytoplasm. Whether or not the nuclear colocalization of RTA and ORF49 refers to direct physical interactions of these two proteins is currently under investigation.
In this report, we have investigated the functional role of the ORF49 protein in viral replication. We utilized two separate mutants of ORF49 and ORF50 to delineate the phenotypes and functional cooperation of these two genes, whereas the previous study on EBV BRRF1 utilized an RTA and BRRF1 double-knockout mutant virus (2, 5, 17). The results from transcomplementation of ORF49null by the FLAG-tagged ORF49 protein and from the ORF49null marker rescue mutant strongly support the hypothesis that the highly attenuated growth of the ORF49null virus was attributed to the disrupted expression of ORF49. MHV-68 ORF49 was capable of further enhancing lytic promoters that had been already activated by RTA and promoting a limited virus replication of ORF50null, thereby suggesting that ORF49 facilitates the function of RTA at the protein level.
While EBV Na alone activated the ZTA IE promoter, MHV-68 ORF49 alone did not directly activate the lytic promoters. A recent study on KSHV ORF49 reported that it cooperatively activated several KSHV lytic promoters with RTA but was not able to activate them by itself (8). Interestingly, there was no cooperation of KSHV ORF49 with RTA to activate the RTA promoter (8). In contrast, MHV-68 ORF49 was able to further increase the transcription of RTA as well as other genes, which may collectively contribute to enhanced virus replication. Therefore, our results consistently indicated that ORF49 facilitates the function and/or expression of RTA to promote the replication of the MHV-68 virus.
Acknowledgments
This work was supported by the Korea University Grant and the Korea Research Foundation Grant funded by the Korean government (MOEHRD) (R04-2004-000-10048-0) to M.J.S.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Allen, R. D., III, S. Dickerson, and S. H. Speck. 2006. Identification of spliced gammaherpesvirus 68 LANA and v-cyclin transcripts and analysis of their expression in vivo during latent infection. J. Virol. 80:2055-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 6.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González, C. M., E. L. Wong, B. S. Bowser, G. K. Hong, S. Kenney, and B. Damania. 2006. Identification and characterization of the Orf49 protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 80:3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong, G. K., H. J. Delecluse, H. Gruffat, T. E. Morrison, W. H. Feng, A. Sergeant, and S. C. Kenney. 2004. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. J. Virol. 78:4983-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia, Q., T. T. Wu, H. I. Liao, V. Chernishof, and R. Sun. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J. Virol. 78:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, S., I. V. Pavlova, H. W. Virgin IV, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin, D. F., B. D. Kuppermann, R. A. Wolitz, A. G. Palestine, H. Li, C. A. Robinson, et al. 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. N. Engl. J. Med. 340:1063-1070. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 77:10488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mellinghoff, I., M. Daibata, R. E. Humphreys, C. Mulder, K. Takada, and T. Sairenji. 1991. Early events in Epstein-Barr virus genome expression after activation: regulation by second messengers of B cell activation. Virology 185:922-928. [DOI] [PubMed] [Google Scholar]
- 16.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Philos. Trans. R. Soc. Lond. B 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ragoczy, T., and G. Miller. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75:5240-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rickabaugh, T. M., H. J. Brown, T. T. Wu, M. J. Song, S. Hwang, H. Deng, K. Mitsouras, and R. Sun. 2005. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 RTA reactivates murine gammaherpesvirus 68 from latency. J. Virol. 79:3217-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr Virus, p. 2575-2628. In D. M. Knipe and P. M. Howley (ed.), Fields Virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 20.Segouffin-Cariou, C., G. Farjot, A. Sergeant, and H. Gruffat. 2000. Characterization of the epstein-barr virus BRRF1 gene, located between early genes BZLF1 and BRLF1. J. Gen. Virol. 81:1791-1799. [DOI] [PubMed] [Google Scholar]
- 21.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song, M. J., S. Hwang, W. H. Wong, T. T. Wu, S. Lee, H. I. Liao, and R. Sun. 2005. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc. Natl. Acad. Sci. USA 102:3805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speck, S. H., and H. W. Virgin. 1999. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr. Opin. Microbiol. 2:403-409. [DOI] [PubMed] [Google Scholar]
- 24.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 26.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]