Abstract
The ORF57 protein expressed by Kaposi's sarcoma-associated herpesvirus (KSHV) during lytic replication is essential for KSHV virion production. ORF57 enhances gene expression by increasing accumulation of target gene mRNAs. ORF57 interacts with the cellular export factor REF and with RNA, suggesting that it may provide target mRNAs with access to REF, which mediates nuclear RNA export by binding to TAP/NXF1. A mutational analysis of ORF57 was performed to study the role of REF binding, RNA interaction, and multimerization in ORF57 function. ORF57 was shown to directly bind RNA. The ability to bind REF did not correlate with ORF57 function in enhancing mRNA accumulation. ORF57 enhanced the nuclear levels of mRNA and PAN, a nuclear KSHV RNA, and the activity of various ORF57 mutants on the levels of mRNA paralleled their ability to enhance nuclear PAN accumulation, suggesting that ORF57 may also act on messenger RNAs by export-independent effects on RNA stability. Finally, an ORF57 mutant lacking a region homologous to a nucleolar localization signal in herpesvirus saimiri was constructed. This mutant retained function, demonstrating that, unlike the ORF57 homolog in herpesvirus saimiri, nucleolar trafficking is not required for ORF57 function in enhancing mRNA accumulation.
Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 belongs to a family of proteins that facilitate herpesviral gene expression during lytic viral replication (for reviews, see references 34 and 37). KSHV ORF57, similar to Epstein-Barr virus (EBV) SM (BMLF1, Mta, EB2) and herpes simplex virus (HSV) ICP27, is essential for virion production and enhances expression of both early and late KSHV lytic cycle genes (12, 14, 33, 35). The majority of herpesvirus genes expressed during the lytic cycle of virus replication does not contain introns. The lack of introns may pose an intrinsic obstacle to gene expression as splicing is intimately linked to nuclear export of cellular mRNAs (6, 27). Proteins comprising the mRNA export machinery (TREX complex) are deposited on mRNA molecules consequent to splicing and facilitate nuclear export by interaction with TAP/NXF1, a central mediator of export, via the nuclear pore. Cellular and viral mRNAs that are not spliced are nevertheless exported by a variety of mechanisms. Both cellular histone RNAs and intronless viral mRNAs contain discrete constitutive transport elements capable of interacting directly with components of the cellular export apparatus (16, 20). Other unspliced mRNAs are exported with various degrees of efficiency despite having no identified transport elements. For some of the poorly exported intronless mRNAs, the TREX component REF, and/or TAP protein, appear to be limiting, since intranuclear injection of REF or TAP in Xenopus oocyte nuclei enhances export (31). In contrast, other intronless RNAs are exported efficiently, and exogenous REF or TAP do not significantly enhance export. Such RNAs are presumed, by virtue of their length or sequence characteristics, to be inherently more capable of directly recruiting TAP or TAP adaptor proteins.
The finding that ORF57 and several of its homologs in other herpesviruses interact with REF has led to the suggestion that they act to recruit REF to intronless viral mRNAs, thereby facilitating export (23, 39). Direct evidence for such a model is provided by microinjection experiments in which HSV ICP27 enhanced export of HSV mRNAs and formed complexes with both REF and TAP (19). However, several lines of evidence suggest that the mechanism of these proteins involves more than serving as a bridge to REF interaction. Although REF is recruited to sites of HSV transcription by ICP27, ICP27 mutants that fail to bind REF are nevertheless exported to the cytoplasm (4). ICP27 has also been shown to interact directly with TAP/NXF1, and this interaction was required for ICP27 export (4). The EBV SM protein interacts with REF and enhances cytoplasmic accumulation of mRNA but also increases the nuclear accumulation of mRNA (11, 15, 28, 32). Finally, it has been suggested that ORF57 may act synergistically with the immediate-early KSHV transcriptional activator ORF50 to enhance transcription of ORF50-responsive promoters, suggesting nuclear functions in addition to promoting nuclear export (18, 22).
In the present study ORF57 was specifically mutated to identify regions involved in REF binding, multimerization, and RNA binding and to determine the requirement of these activities in enhancing RNA accumulation. The possibility of export-independent functions of ORF57 was also investigated by examining the effects of ORF57 on PAN (nut-1, T1.1), a KSHV RNA that is exclusively nuclear (10, 36).
MATERIALS AND METHODS
Plasmids and ORF57 mutant construction.
The ORF57 cDNA from BCBL1 KSHV cloned in the mammalian expression vector pCDNA3 (Invitrogen) has been previously described (13). Site-specific ORF57 mutants were generated by PCR, cloned in pCDNA3 (Invitrogen), and sequenced. FLAG and influenza virus hemagglutinin (HA)-tagged versions of each mutant gene were constructed by cloning into pCDNA3 after insertion of an epitope tag sequence generated by PCR. The FLAG-tagged REF expression vector was kindly provided by Gideon Dreyfuss (17). The PAN expression vector was constructed by PCR amplification of PAN sequence from nucleotides 28690 to 30255 of the KSHV genome and cloning in pCDNA3. The KSHV gB expression vector (29) was kindly provided by R. Longnecker. The ORF59 expression vector consisted of the ORF59 sequence cloned in pCDNA3 (14).
Transfections, reporter assays and cell lines.
HeLa cells were maintained in Dulbecco modified Eagle medium and 10% fetal calf serum. Transfections were performed with Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. Chloramphenicol acetyltransferase (CAT) assays were performed by transfecting HeLa cells with CAT reporter plasmids and ORF57 expression plasmids, harvesting cell lysates 48 h after transfection, and measuring CAT activity as previously described (32).
Immunofluorescence microscopy and immunoprecipitation.
Adherent HeLa cells were washed and fixed with 100% ice-cold methanol 36 h after transfection and stained with rabbit anti-ORF57 antibody and goat anti-rabbit immunoglobulin G (IgG) conjugated with Alexa Fluor 594 (Invitrogen). Stained cells were visualized by fluorescence microscopy. Nucleoli were stained with anti-fibrillarin monoclonal antibody D77 (1) (kindly provided by J. Aris) and goat anti-mouse IgG conjugated with fluorescein isothiocyanate.
Immunoprecipitations were performed with lysates of HeLa cells harvested 48 h after transfection. Cells were washed and lysed in 250 mM NaCl, 50 mM Tris (pH 7.2), 0.5% NP-40, and protease inhibitor cocktail (Sigma) by incubation at 4°C for 15 min. Clarified lysates were precleared with normal rabbit IgG (Bethyl) and protein A-agarose beads, followed by immunoprecipitation with either anti-HA monoclonal antibody (Covance), anti-FLAG polyclonal antibody (Sigma), or normal rabbit IgG. Immunoprecipitations were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with anti-FLAG or anti-HA antibody.
RNA isolation and analysis.
RNA was isolated from cells with RNA-Bee (Teltest) and RNeasy columns (QIAGEN) as previously described (32). Northern blotting was performed as previously described (32). Blots were hybridized with 32P-labeled, gene-specific DNA probes generated by random oligonucleotide primed Klenow DNA polymerase. Quantitation was performed by direct radiometry with a Packard InstantImager and normalized to GAPDH mRNA levels. U6-specific RNA probes were generated by using T7 RNA polymerase. Cytoplasmic and nuclear RNAs were prepared by lysing cells in 100 mM NaCl, 50 mM Tris (pH 8.0), 5 mM MgCl2, 1 mM dithiothreitol (DTT), 0.5% NP-40 on ice, separating nuclei by centrifugation, and isolating RNA from each fraction as described above.
Quantitative reverse-transcription PCR (qRT-PCR).
For each sample to be analyzed, 1 mg of RNA was reverse-transcribed with Superscript II reverse transcriptase (Invitrogen), and either gene-specific primers or oligo(dT) primers. The gene-specific primers were as follows: gB 5′, ACACTACTTCATCACCCGCAACGA; gB 3′, TCGCGAGTCGTTTCTCTGCACT; GAPDH 5′, AGGGTCATCATCTCTGCCCCCTC; GAPDH 3′, TGTGGTCATGAGTCCTTCCACGAT; ORF59 5′, AAAGGCAGTGGAGACGTTAG; and ORF59 3′, GAGGTGAGGTTGTCCCCGTA.
qPCR was performed with iQ SYBR green Supermix (Bio-Rad) according to the manufacturer's protocol using a MyiQ iCycler (Bio-Rad). No template controls and no RT controls were included in each analysis. Each sample was analyzed in triplicate and normalized to GAPDH RNA.
RNA cross-linking assay.
Cos7 cells were transfected with ORF57 or ORF57 mutant expression vector DNA using Lipofectamine Plus (Invitrogen). At 48 h after transfection, cells were washed and harvested by scraping and centrifugation. Cell pellets were lysed by incubation at 4°C for 15 min in 100 μl of 20 mM HEPES (pH 7.9), 10 mM NaCl, 10% glycerol, 3 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.4 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Sigma), with frequent mixing. The lysed cell suspension was centrifuged at 4°C for 5 min at 700 × g. Supernatant was transferred to a fresh tube, and one-third volume of high-salt buffer (20 mM HEPES [pH 7.9], 400 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.4 mM phenylmethylsulfonyl fluoride, 1 mM DTT, and protease inhibitor cocktail) was added. Aliquots of extract were snap-frozen at −80°C. Radiolabeled ORF59 mRNA was synthesized with [32P]UTP and T7 RNA polymerase. To allow ORF57-RNA complex formation to occur, 2 × 106 cpm of radiolabeled RNA was incubated for 30 min at 30°C with 8 μl of cell extract, 2 μl of 20 mM magnesium acetate, 2 μl of 10 mM ATP, 2 μl of 200 mM K glutamate, 2 μl of 50 mM creatine phosphate, 1 μl of tRNA (1 μg/μl), and 1 μl of RNasin in a total volume of 20 μl. RNA-protein mixtures were then cross-linked by UV irradiation on ice with a Stratalinker (Stratagene) for a total of 0.6 J. RNA was hydrolyzed by incubation with 100 μg of RNase A/ml for 1 h at 37°C. ORF57 protein was then immunoprecipitated from the incubation mixtures as described above and analyzed by SDS-PAGE and autoradiography of dried gels.
RESULTS
Targeted mutation of ORF57.
Several regions of the ORF57 gene were targeted for mutational analysis (Fig. 1). Although the herpesvirus proteins homologous to KSHV ORF57 are generally similar, there is a significant degree of sequence divergence, and the regions analyzed in the present study do not contain highly conserved residues. These regions were chosen based on clues that they might be important in function. There are two RGG motifs in the ORF57 amino acid sequence. RGG motifs have been shown to be important for specific binding of FMRP RNA (30), and an RGG-containing region of HSV ICP27 is essential for binding RNA in vivo (25). In addition, an arginine-rich motif (RPRRRPRDRL; amino acids [aa] 118 to 127) was identified by using a prediction model for nuclear localization signals (NLSs) (8). An extended stretch of basic amino acids (aa 117 to 148) is also present, which includes the first RGG (aa 134 to 136) and the putative NLS (aa 118 to 127). A potential leucine zipper is present from aa 339 to 360. ORF57 is known to multimerize (24), and this region might therefore be important in homo- or heteromultimerization. It has been hypothesized that ORF57 function is dependent on complex formation with the cellular export factor REF, and a region from aa 181 to 215 was previously implicated as a REF-binding domain in vitro (23). However, the role of this region in REF interaction and ORF57 function in the context of the entire molecule has not been evaluated.
FIG. 1.
Diagram of ORF57 protein. Regions that were targeted for mutation or deletion are shown. aa 118 to 127 contain the 10-aa motif RPRRRPRDRL identified as a potential NLS. The amino acid numbers of RGG1 and RGG2 motifs are shown in parentheses. An extended basic region (EBR) spans aa 117 to 148 and includes both the potential NLS and RGG1. A region from aa 172 to 210 reported to contain a REF-binding domain (RBD) is also shown. The four leucines comprising a potential leucine zipper and their amino acid locations are depicted. The first two leucines were mutated to prolines individually or in combination, and each of the other motifs was deleted (see the text). The designation of each mutant gene construction is shown on the right.
The following mutant ORF57 genes were constructed to investigate the role of these regions in RNA binding, homomultimerization, REF binding, and enhancing RNA accumulation. Each RGG motif was specifically deleted (ΔRGG1 and ΔRGG2). The potential NLS (aa 118 to 127) was deleted (ΔNLS). The extended basic region encompassing the NLS and RGG1 was also deleted (ΔEBR). The putative REF-interaction domain (aa 181 to 215) was also specifically deleted (ΔRBD). The first and second leucines (L339 and L346) in the potential leucine zipper were individually mutated to proline since proline is expected to have the most disruptive effect on the alpha-helix of a leucine zipper (7). A third mutant in which both leucines were substituted with proline was also constructed. These three mutants were designated L1, L2, and L1,2, respectively. (Fig. 1).
Specific ORF57 amino acids are critical for transactivating function.
ORF57 increases the expression of chloramphenicol acetyltransferase (CAT) at the posttranscriptional level in reporter assays (13). We used a reporter assay in which ORF57 or empty vector pCDNA3 was transfected into HeLa cells with a CAT reporter plasmid. As shown in Fig. 2A, deletion of the second RGG (ΔRGG2), the putative REF-binding domain (ΔRBD), or mutation of L346 (L2), all led to a significant loss of activity. The RGG2 and L2 mutations abolished activity, and the ΔRBD mutation led to loss of ∼70% of activity. Interestingly, deletion of either the putative NLS, RGG1, or the basic region encompassing the above two sites (ΔEBR) did not affect activity. Mutation of L339 (L1) decreased activity by ca. 50%. These results therefore indicated that the second RGG motif, the potential leucine zipper, and REF-binding domain were all important for activity but that the first RGG1 and basic region were not critical.
FIG. 2.
(A) Effect of ORF57 mutations on function in enhancing CAT expression. HeLa cells were transfected with each mutant or wt ORF57, and CAT assays were performed to compare the activity of each mutant to wt ORF57. The fold activation is shown relative to the CAT activity in the presence of empty vector (lane C). (B) Effect of ORF57 mutations on ORF59 mRNA accumulation. The enhancement levels of ORF59 gene expression by wt ORF57 and each mutant were compared. RNA from cells transfected with target ORF59 mRNA expression vector and each mutant was analyzed by Northern blotting with an ORF59 probe (top). The blot was stripped and reprobed with a human GAPDH probe as a loading control (bottom). (C) RNA from cells transfected as in panel B above were analyzed by qRT-PCR. All values were normalized to cellular GAPDH RNA levels. (D) Immunoblotting of lysates from cells transfected with each mutant or wt ORF57 was performed with anti-ORF57 antibody.
In order to confirm the results of the reporter assays, the effect of each mutation was also assessed by comparing the ability of each mutant to enhance the expression of the KSHV ORF59 mRNA. ORF59 mRNA does not accumulate efficiently in the absence of ORF57 expression and therefore serves as a suitable assay for ORF57 function (14, 21). HeLa cells were transfected with an ORF59 expression vector and either ORF57 plasmid or empty vector, and the levels of ORF 59 mRNA were measured by Northern blotting. The relative amounts of each RNA was also measured by qRT-PCR. As shown in Fig. 2B and C, the effects of ORF57 mutants on ORF59 mRNA were similar to those obtained with CAT reporter experiments but revealed some differences in relative activity. ΔRGG2, L2, L1,2, and ΔRBD were essentially inactive, whereas the L1 and Δ NLS mutants were slightly reduced compared to wild-type (wt) ORF57 in enhancing accumulation of ORF59 mRNA. The RGG1 mutant was fully active. These results suggested that aa 118 to 127, since they are not essential, do not serve as a nonredundant NLS. Whereas RGG1 or L339 (L1) were not essential for function, RGG2 and L346 (L2) were required for function. These results are summarized in Table 1. In order to ensure that loss of function was not due to instability of any of the mutant proteins, cells were transfected with mutant and wt ORF57 expression vectors, and lysates were examined by immunoblotting (Fig. 2D).
TABLE 1.
Relative activity of wt ORF57 and ORF57 mutants in gene activation, dimerization, REF binding, and cross-linking to RNA in vitroa
| ORF57 structure | Relative activityb
|
|||
|---|---|---|---|---|
| Activation function | Multimerization | REF binding | RNA cross-linking | |
| wt | ++++ | +++ | +++ | +++ |
| ΔNLS | +++ | +++ | ND | ND |
| ΔRGG1 | ++++ | +++ | +++ | +++ |
| ΔRBD | - | + | + | - |
| L1 | +++ | ++ | + | - |
| L2 | - | ++ | ++ | - |
| L1,2 | - | + | ++ | - |
| ΔRGG2 | - | +++ | - | - |
The ability of ORF57 and various site-specific mutants to enhance ORF59 target mRNA accumulation, homomultimerize, or bind to REF in vivo, and physically bind to labeled RNA in vitro is summarized (see the text and Fig. 2 and 4 to 7 for details).
++++, Very strong activity; +++, strong activity; ++, moderate activity; +, some activity; −, no activity. ND, not determined.
Nuclear localization is intact in ORF57 mutants.
ORF57, like its homologs in other herpesviruses, such as HSV ICP27, and EBV SM, localizes to the nucleus in a diffuse speckled pattern. When visualized by immunofluorescence with anti-ORF57 antibody, although there is fainter staining of the nucleoli than in the nucleoplasm (Fig. 3, wt A), in ca. 10 to 20% of the cells ORF57 is concentrated in the center of nucleoli (Fig. 3 wt B). In order to determine whether the mutants which had lost activity had done so as a consequence of altered intracellular location, we examined all of the mutant proteins by immunofluorescence microscopy of transfected cells. It was recently reported that nucleolar localization of the herpesvirus saimiri (HVS) ORF57 homolog was essential for nuclear mRNA export and gene activation (3). Although the HVS and KSHV ORF57 sequences are not colinear in the region involved, one of the two nucleolar localization signals identified in HVS (KRPR) is also present in KSHV ORF57. The ΔNLS mutant that we constructed as a potential NLS mutant effectively disrupts this KRPR motif, deleting the last 3 aa and removing several additional basic amino acids. We were therefore particularly interested in the intranuclear localization of this mutant. Immunofluorescence micrographs of each mutant are shown in Fig. 3. Nuclear localization of all of the mutants was similar to that of wt ORF57, with a diffuse, speckled nuclear pattern, and relative sparing of the nucleoli (Fig. 3). These results demonstrate that the loss of activity in the nonfunctional mutants is not due to a defect in nuclear localization. The ΔNLS and the ΔEBR mutants, in which the motif shown to be a nucleolar localization signal in HVS is disrupted, showed a more complete exclusion from nucleoli than wt ORF57 or the other mutants, and no cells were seen in which nucleolar concentration occurred, as with wt ORF57 or the other mutants (Fig. 3). In order to verify that the areas from which ΔNLS was excluded were nucleoli, cells were stained with anti-fibrillarin antibody which stains the central portion of nucleoli (1) simultaneously with anti-ORF57 antibody. As can be seen in Fig. 3, ΔNLS ORF57 is excluded from nucleoli, demonstrating that this domain is involved in nucleolar localization.
FIG. 3.
Nuclear and nucleolar localization of wt KSHV ORF57 and mutants by immunofluorescence microscopy. HeLa cells were transfected with each mutant ORF57 or wt ORF57 plasmid and fixed 36 h after transfection. Slides were stained with anti-ORF57 specific antibody. The first panel (wt A) shows the appearance of the majority of wt ORF57 transfected cells, with fainter staining of the nucleoli than the surrounding nucleoplasm. The second panel shows the appearance of a minority of the cells in which nucleolar concentration was observed (wt B). The last two panels on the right show the ΔNLS mutant double stained with ORF57 antibody (ORF57) or anti-fibrillarin antibody to identify nucleoli (nucl).
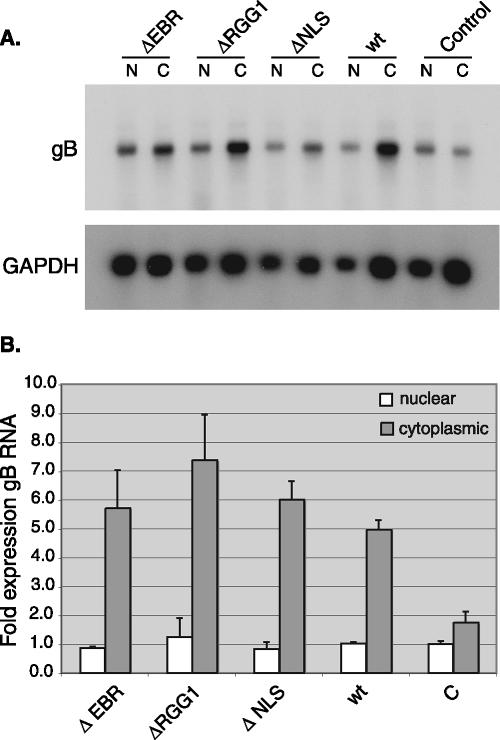
Since it had been recently reported that trafficking of HVS ORF57 was required for the nuclear export of intronless target mRNA (3), the fully retained activity of ΔNLS in enhancing ORF59 mRNA accumulation suggested either that the KSHV ORF57 effect on ORF59 mRNA is not dependent on enhanced nuclear export or that nucleolar trafficking is not essential for KSHV ORF57 function. We therefore examined the function of ΔNLS further. Since HVS gB was used as the target gene in the previous study with HVS ORF57, we studied the effect of ΔNLS and ΔEBR on the nuclear and cytoplasmic levels of KSHV gB. HeLa cells were transfected with a KSHV gB expression vector and either wt ORF57, ΔNLS, or ΔEBR, and cytoplasmic and nuclear fractions of RNA were prepared and analyzed by Northern blotting and by qRT-PCR. As shown in Fig. 4, in the presence of wt or mutant ORF57, cytoplasmic gB RNA levels increase, resulting in an increase in the ratio of cytoplasmic to nuclear RNA. These results demonstrate that both wt ORF57 and the two nucleolar localization mutants enhance cytoplasmic accumulation of gB mRNA and lead to an increased ratio of cytoplasmic to nuclear gB mRNA. These results therefore demonstrate that, whereas KSHV ORF57 increases the cytoplasmic accumulation of gB mRNA, impairing nucleolar localization does not affect ORF57 function significantly in this regard.
FIG. 4.
Effect of nucleolar localization mutants on cytoplasmic accumulation of KSHV gB mRNA. (A) RNA was prepared from nuclear and cytoplasmic fractions of cells transfected with gB expression vector and either wt ORF57 or mutant ORF57 plasmids. Equal amounts of each RNA were analyzed by Northern blotting and hybridized with radiolabeled gB probe. (B) Cells were transfected with wt ORF57 or mutant ORF57 plasmids and a gB expression vector as for panel A above. RNA from nuclear and cytoplasmic fractions was measured by qRT-PCR.
Interaction of ORF57 mutant proteins with target RNA.
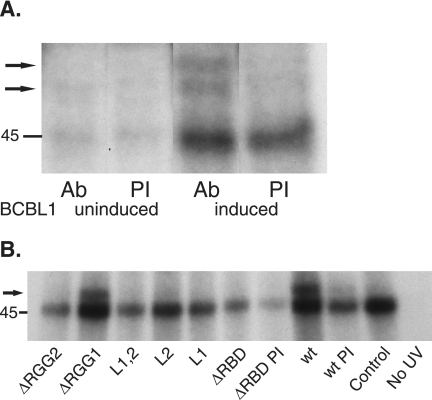
It is likely that ORF57 acts by stabilizing RNA transcripts in the nucleus, as well as enhancing their nuclear export (13). It has been previously reported that ORF57 could be immunoprecipitated in complexes that contain mRNA and bind radiolabeled RNA in vitro, but the interaction was dependent on the addition of nuclear extract (21). Thus, ORF57 may interact with RNA by direct binding or by forming complexes with cellular RNA-binding proteins. In order to examine whether ORF57 directly contacts target RNA molecules, we used a radioactive label transfer assay. In vitro-transcribed and radioactively labeled ORF59 mRNA was incubated with extracts from KSHV-positive primary effusion lymphoma cells (BCBL1) which had been chemically induced with 12-O-tetradecanoylphorbol-13-acetate (TPA) to permit KSHV lytic replicative cycle and ORF57 expression. After an incubation period to allow for complex formation, the RNA-protein mixtures were UV-irradiated to cross-link protein and RNA molecules that were in direct contact. The irradiated samples were then treated with RNase A to hydrolyze the labeled RNA to completion. ORF57 protein was immunoprecipitated and analyzed by SDS-PAGE and autoradiography. As seen in Fig. 5A, ORF57 is labeled by cross-linking to labeled ORF59 mRNA nucleotides, strongly suggesting that ORF57 directly interacts with mRNA in vitro. In BCBL1 cells, a doublet was detected, most likely due to the presence of multiple phosphorylated forms of ORF57 in BCBL1 cells, as previously described (24).
FIG. 5.
Radiolabeled uridine residue transfer from RNA in direct contact with ORF57 protein. (A) BCBL1 cells were induced to permit lytic replication by treatment with TPA. Cell lysate was prepared from mock-treated (uninduced) or TPA-treated (induced) cells and incubated with radiolabeled, in vitro-transcribed ORF59 mRNA. Protein-RNA cross-linking was performed by UV irradiation, followed by hydrolysis of RNA with RNase A. Immunoprecipitation was performed with anti-ORF57 antibody (Ab) or preimmune serum (PI), and RNA-labeled protein was detected by SDS-PAGE and autoradiography. The location of a 45-kDa marker is shown on the left, and ORF57-specific bands are denoted by arrows. (B) Lysates containing each mutant or wt ORF57 protein were prepared from transfected cells. Incubation and label transfer were performed as in panel A above. Also shown is a wt ORF57 lysate that was incubated with radiolabeled RNA but not cross-linked by UV and treated in parallel (No UV). Immunoprecipitation performed with preimmune serum as a control is also shown (PI).
In order to determine whether one or more of the inactive mutants we had constructed were defective in RNA binding, a similar assay was performed with wt and each mutant ORF57 protein. In vitro-transcribed and radioactively labeled ORF59 mRNA was incubated with extracts from cells transfected with each ORF57 mutant, and cross-linking assays were performed as described above. The results, shown in Fig. 5B, indicate that the inactive mutants L2 and ΔRGG2 were deficient in RNA binding compared to wt ORF57 or the fully functional RGG1 mutant. Because the ΔRBD mutant is smaller in size than wt ORF57, we cannot exclude the possibility that ΔRBD is labeled but cannot be detected because of the presence of a background band at ca. 45 kDa, which is present in all samples.
These data indicate that physical contact with RNA is dependent on several discrete residues in ORF57 and that multiple mutations which abrogate function also affect RNA binding. We should emphasize that although individual mutations affected RNA binding, this does not necessarily imply that each mutated region is an RNA-binding domain. Rather, the observed effects may be due to changes in conformation caused by a physically remote mutation or due to several regions contributing to the formation of an RNA binding motif. The L1 mutant, which was active in enhancing ORF59 mRNA accumulation, was also studied. Surprisingly, the L1 mutant was not labeled in the ORF59 mRNA binding assay. The most likely interpretation of this finding is that while the interaction of ORF57 with RNA is affected by mutation of leucine 339, this alteration does not eliminate function.
ORF57 multimerization and REF binding.
ORF57 has previously been shown to homodimerize (2). It is not known whether multimerization of ORF57 is essential for function. Since several of the mutants were inactive, we wanted to determine whether any were affected in the ability to multimerize. Multimerization was assessed by cotransfecting plasmids that expressed each mutant in a FLAG or HA epitope-tagged form. All of the epitope-tagged mutant and wt ORF57 proteins were reactive with the appropriate antibody (data not shown). Immunoprecipitation was then carried out with anti-FLAG or anti-HA antibody, and immunoblotting was performed with anti-HA antibody (Fig. 6). Multimerization was evident with the wt ORF57, RGG1, L1, L2, and RGG2 mutants. Although multimerization was somewhat decreased with the leucine mutants, it was still clearly detectable. These data indicate that the potential leucine zipper in ORF57 is not required for homomultimerization. Multimerization was most impaired in the ΔRBD mutant but still detectable over control. These data therefore demonstrate that the functional defect in L2 and RGG2 mutants is not due to an inability to multimerize and, further, that although multimerization is decreased by mutation of L1, the L1 mutant nevertheless retains function, suggesting that homomultimerization is not critical for activity (Table 1).
FIG. 6.
Multimerization of wt and mutant ORF57 proteins. Homomultimerization of each mutant and wt ORF57 was assessed by transfecting cells with both a FLAG-tagged and an HA-tagged version of each construct. Lysates were immunoprecipitated with either anti-HA (H), anti-FLAG (F), or control IgG (C). Lysates were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody. Ten percent of each input lysate was run on each gel (I). The identity of each mutant is shown above the corresponding panel. Markers (45 and 66 kDa) are indicated on the left. The second, lower band seen in the anti-HA immunoprecipitations is the immunoglobulin heavy chain. This band is observed only in the anti-HA immunoprecipitations since the immunoblot is performed with conjugated anti-mouse immunoglobulin secondary antibody, and only the anti-HA antibody is of murine origin.
It has been suggested that ORF57 and its homologs in other herpesviruses act to facilitate target RNA export by simultaneously binding the RNA and cellular REF protein, which mediates the export of spliced cellular mRNAs (5, 15, 23). Furthermore, it has been reported that aa 172 to 210 of ORF57 contain a REF interaction domain (23). However, it has also been reported that ORF57 function in enhancing cytoplasmic ORF57 accumulation is not inhibited by the depletion of REF with small interfering RNA (21). In order to determine whether the ability to bind REF was specifically impaired in any of the mutants and whether the ability to bind REF correlated with function, we tested REF binding by each ORF57 mutant. Coimmunoprecipitation of lysates from cells transfected with FLAG-tagged REF and each HA-tagged ORF57 mutant was performed and analyzed as described above (Fig. 7). Of the inactive mutants, L2 and L1,2 retained ca. 50% of wt REF binding activity, whereas ΔRGG2 and ΔRBD lost almost 90% of REF binding activity. The functional ΔRGG1 mutant retained full REF-binding function, whereas L1 retained ca. 30% of wt binding. Thus, the degree of REF binding did not correlate with function (see Table 1.). Furthermore, mutation of the RGG2 motif decreased REF binding almost as severely as deletion of the entire region from aa 172 to 210 which had been proposed to contain the REF interaction domain. Thus, it is clear that there may be more than one discrete REF interaction domain. Finally, although REF interaction may involve aa 172 to 210, this region is not solely a REF interaction domain since deletion of this region in the context of the entire protein led to the loss of multiple functions (Table 1).
FIG. 7.
REF complex formation by wt and mutant ORF57 proteins. The ability of each mutant or wt ORF57 to bind REF was assessed by transfecting cells with both a FLAG-tagged REF expression plasmid and an HA-tagged ORF57 expression plasmid. Immunoprecipitation was performed with anti-FLAG (F), anti-HA antibody (H), or control IgG (C), and immunoblotting was performed with anti-HA monoclonal antibody. Ten percent of each input lysate was also run on each gel (I). The identity of each mutant is shown to the right of the corresponding panel. Markers (45 and 66 kDa) are indicated on the left. The second, lower band seen in the anti-HA immunoprecipitations is immunoglobulin heavy chain.
Effects of ORF57 on nuclear RNA accumulation.
The lack of a direct correlation between REF binding and function among the various mutants suggested that gene activation by ORF57 may not be strictly related to an export function. In addition, we have previously reported increases in both nuclear and cytoplasmic levels of reporter gene mRNAs mediated by ORF57 (13). Other researchers have also suggested that REF is not essential for ORF57 enhanced RNA export (21). We therefore examined the effect of the ORF57 mutant proteins on a nuclear KSHV RNA. The polyadenylated nuclear RNA (PAN, T1.1) is a highly abundant noncoding transcript expressed during the KSHV lytic cycle replication that is exclusively nuclear (36) and has been shown to be upregulated by ORF57 (14, 18). Each mutant was cotransfected with a PAN expression vector, and the levels of PAN RNA were measured by Northern blotting (Fig. 8A). Significantly, the effect of each mutant on nuclear PAN RNA was similar to its effects on ORF59 mRNA. The ΔRGG1 and L1 mutants were fully active in enhancing PAN accumulation, whereas the nonfunctional mutants were inactive. These data suggested that ORF57 may act by enhancing nuclear accumulation of target messenger RNAs as well. PAN RNA has recently been demonstrated to undergo decay in the nucleus by deadenylation and 3′ degradation (9). We have previously shown by nuclear run-on assays that ORF57 does not increase the transcript initiation rate from the cytomegalovirus (CMV) promoter used in these experiments (13). Therefore, it is likely that ORF57 increases nuclear stability of target mRNAs, although other effects on export, elongation, or transcriptional termination are also possible (See Discussion).
FIG. 8.
Effect of ORF57 on nuclear RNA accumulation. (A) The effects of wt ORF57 and each mutant on PAN gene expression were compared. RNAs from cells transfected with PAN expression vector and each mutant were analyzed by Northern blotting with a PAN-specific probe. The blot was stripped and reprobed with a human GAPDH probe as a loading control (below). (B) The effect of wt ORF57 on the nuclear and cytoplasmic accumulation of ORF59 mRNA was measured. Cells were separated into nuclear and cytoplasmic fractions, and RNA from each fraction was analyzed by Northern blotting with ORF59-specific probe. The amounts of cytoplasmic (lanes C) and nuclear (lanes N) ORF59 mRNA in the presence (ORF57) and absence (control) of ORF57 are shown. (C) Nuclear and cytoplasmic ORF59 RNAs in the presence or absence of ORF57 were quantitated by qRT-PCR. The results from two independent experiments are shown. Note the difference in the axes between the nuclear and cytoplasmic measurements.
The effect of ORF57 on PAN RNA was distinct from its effect on gB mRNA, where ORF57 led to a dramatic increase in the cytoplasmic-to-nuclear ratio of mRNA, as well as the total levels of gB mRNA. These findings are consistent with an effect on mRNA export but could also involve increases in nuclear and cytoplasmic stability in the presence of ORF57. To ask whether the ORF57 could be shown to increase nuclear accumulation of mRNA, we compared the effect of ORF57 on the nuclear and cytoplasmic accumulation of ORF59 mRNA (Fig. 8C). Similar to gB mRNA, steady-state accumulation of ORF59 RNA in the cytoplasm was greatly increased by ORF57. However, the total nuclear levels of ORF59 mRNA were also increased by ORF57. These findings demonstrate that ORF57 is capable of enhancing accumulation of mRNA in the nucleus regardless of its effects on nuclear export.
DISCUSSION
The mutational analysis of ORF57 function revealed several novel aspects of KSHV ORF57 function. First, ORF57 was demonstrated to be an RNA-binding protein, making direct physical contact with RNA residues. Mutation of three regions of ORF57—the leucine heptad domain, the second RGG motif, and aa 172 to 210—affected RNA binding. Interestingly, however, the specific RNA interaction observed in the labeling assay was nonessential for ORF57 transactivation function. A similar finding was made in the case of the human CMV UL69 homolog of ORF57, where deletion of an RNA recognition motif did not affect nucleocytoplasmic shuttling or the ability of the protein to facilitate nuclear RNA export (38). It should be noted, however, that although the inability of the L1 mutant to be labeled by cross-linked RNA suggests that direct RNA contact is abolished, it is possible that the uridine RNA residues that make direct contact with ORF57 are few in number and that the L1 mutant may still interact directly with RNA, albeit in an altered conformation that does not permit uridine label transfer. Although none of the mutant proteins exhibited gross changes in stability, nuclear localization, or solubility, we cannot exclude conformational changes that could potentially affect interaction with RNA. Finally, although we consider this less likely, we also cannot formally rule out the possibility that a cellular protein the same size as ORF57 serves as a bridge between ORF57 and RNA molecules.
KSHV ORF57, like its homologs in HSV (26) and HVS (3), appears to localize at least partially to the nucleolus. We have shown here that a basic region of 10 aa, which was previously shown to act as one of three functionally redundant NLSs (21), may also serve as a nucleolar localization signal. Deletion of this region disrupts a KRPR motif and removes several additional contiguous arginine residues and causes nucleolar exclusion of KSHV ORF57. Although this effect on nucleolar localization is similar to that observed with HVS ORF57 upon mutation or deletion of corresponding KRPR residues (3), the KSHV ORF57 mutant is not significantly affected in its ability to enhance target mRNA accumulation in the cytoplasm. Therefore, while nucleolar trafficking appears to be a common characteristic of KSHV ORF57 protein homologs, it does not appear to be essential for KSHV ORF57 function.
The effect of KSHV ORF57 on the nuclear and cytoplasmic accumulation of three different target RNAs—gB, ORF59, and PAN—was examined. The first two are KSHV lytic cycle mRNAs, and PAN is a nuclear polyadenylated RNA of unknown function. ORF57 increased the cytoplasmic accumulation of both mRNAs, although they were constitutively exported from the nucleus at different efficiencies. The increase in the cytoplasmic/nuclear ratio of these mRNAs in the presence of ORF57 suggests that it enhances the nuclear export of both gB and ORF59 mRNAs. It should be noted, however, that a direct role of ORF57 in export can only be inferred from steady-state cytoplasmic and nuclear RNA levels, and effects on cytoplasmic RNA stability cannot be ruled out. Importantly, separation of cytoplasmic and nuclear RNAs demonstrated that ORF57 increases nuclear accumulation of ORF59 mRNAs. It had previously been shown that ORF57 increases cellular levels of PAN, and we confirmed that PAN RNA levels are increased. Significantly, the activity of the various mutant ORF57 proteins on ORF59 mRNA paralleled their activity on PAN RNA. Since all of these target RNAs were expressed from a CMV promoter, whose activity has been shown to be unaffected by ORF57 (13, 18), these data indicate that ORF57 enhances nuclear RNA stability, or possibly, transcript elongation. Consistent with these findings, even a mutant protein (L1), which was significantly impaired in its ability to bind REF, nevertheless retained activity in enhancing RNA accumulation. Thus, while an interaction with REF may be important for ORF57 effects on nuclear RNA export, ORF57 should not be considered to be merely an adaptor protein that recruits REF or other cellular export proteins to viral intronless mRNAs. In summary, KSHV ORF57 binds RNA, enhances nuclear accumulation of coding and noncoding RNAs, and enhances cytoplasmic accumulation of mRNAs regardless of their ability to be constitutively exported. The mechanism of ORF57-enhanced cytoplasmic RNA accumulation remains to be fully characterized and may involve interactions with cellular export proteins, as well as effects on cytoplasmic RNA.
Acknowledgments
This study was supported by Public Health Service grants CA081133 and CA119905 from the National Cancer Institute (to S.S.).
We thank Richard Longnecker, Gideon Dreyfuss, and John Aris for plasmids and antibodies used in this study.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Aris, J. P., and G. Blobel. 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol. 107:17-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bello, L. J., A. J. Davison, M. A. Glenn, A. Whitehouse, N. Rethmeier, T. F. Schulz, and J. Barklie Clements. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207-3215. [DOI] [PubMed] [Google Scholar]
- 3.Boyne, J. R., and A. Whitehouse. 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA 103:15190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, I. H., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 79:3949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, H., K. Dufu, C. S. Lee, J. L. Hsu, A. Dias, and R. Reed. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127:1389-1400. [DOI] [PubMed] [Google Scholar]
- 7.Chou, P. Y., and G. D. Fasman. 1973. Structural and functional role of leucine residues in proteins. J. Mol. Biol. 74:263-281. [DOI] [PubMed] [Google Scholar]
- 8.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad, N. K., S. Mili, E. L. Marshall, M. D. Shu, and J. A. Steitz. 2006. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell 24:943-953. [DOI] [PubMed] [Google Scholar]
- 10.Conrad, N. K., and J. A. Steitz. 2005. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 24:1831-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruffat, H., J. Batisse, D. Pich, B. Neuhierl, E. Manet, W. Hammerschmidt, and A. Sergeant. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta, A. K., V. Ruvolo, C. Patterson, and S. Swaminathan. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 74:1038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, Z., and S. Swaminathan. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 16.Huang, Y., K. M. Wimler, and G. G. Carmichael. 1999. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 18:1642-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataoka, N., and G. Dreyfuss. 2004. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J. Biol. Chem. 279:7009-7013. [DOI] [PubMed] [Google Scholar]
- 18.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, X., and J. E. Mertz. 1995. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 9:1766-1780. [DOI] [PubMed] [Google Scholar]
- 21.Majerciak, V., K. Yamanegi, S. H. Nie, and Z. M. Zheng. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365-28378. [DOI] [PubMed] [Google Scholar]
- 22.Malik, P., D. J. Blackbourn, M. F. Cheng, G. S. Hayward, and J. B. Clements. 2004. Functional co-operation between the Kaposi's sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 85:2155-2166. [DOI] [PubMed] [Google Scholar]
- 23.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001-33011. [DOI] [PubMed] [Google Scholar]
- 24.Malik, P., and J. B. Clements. 2004. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 32:5553-5569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Mears, E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mears, W. E., V. Lam, and S. A. Rice. 1995. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 69:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514-1518. [DOI] [PubMed] [Google Scholar]
- 28.Nicewonger, J., G. Suck, D. Bloch, and S. Swaminathan. 2004. Epstein-Barr virus (EBV) SM protein induces and recruits cellular Sp110b to stabilize mRNAs and enhance EBV lytic gene expression. J. Virol. 78:9412-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertel, P. E., P. G. Spear, and R. Longnecker. 1998. Human herpesvirus-8 glycoprotein B interacts with Epstein-Barr virus (EBV) glycoprotein 110 but fails to complement the infectivity of EBV mutants. Virology 251:402-413. [DOI] [PubMed] [Google Scholar]
- 30.Ramos, A., D. Hollingworth, and A. Pastore. 2003. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA 9:1198-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruvolo, V., E. Wang, S. Boyle, and S. Swaminathan. 1998. The Epstein-Barr virus nuclear protein SM is both a posttranscriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. USA 95:8852-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 78:4389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandri-Goldin, R. M., and G. E. Mendoza. 1992. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 63:848-863. [DOI] [PubMed] [Google Scholar]
- 36.Sun, R., S. F. Lin, L. Gradoville, and G. Miller. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 93:11883-11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swaminathan, S. 2005. Post-transcriptional gene regulation by gamma herpesviruses. J. Cell. Biochem. 95:698-711. [DOI] [PubMed] [Google Scholar]
- 38.Toth, Z., P. Lischka, and T. Stamminger. 2006. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 34:1237-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams, B. J., J. R. Boyne, D. J. Goodwin, L. Roaden, G. M. Hautbergue, S. A. Wilson, and A. Whitehouse. 2005. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387:295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]