Abstract
Epstein-Barr virus (EBV) infection of primary B cells causes B-cell activation and proliferation. Activation of B cells requires binding of antigen to the B-cell receptor and a survival signal from ligand-bound CD40, signals that are provided by the EBV LMP1 and LMP2A latency proteins. Recently, Toll-like receptor (TLR) signaling has been reported to provide a third B-cell activation stimulus. The interaction between the EBV and TLR pathways was therefore investigated. Both UV-inactivated and untreated EBV upregulated the expression of TLR7 and downregulated the expression of TLR9 in naive B cells. UV-inactivated virus transiently stimulated naive B-cell proliferation in the presence of the TLR7 ligand R837, while addition of the TLR7 antagonist IRS 661 impaired cell growth induced by untreated EBV. Interferon regulatory factor 5 (IRF-5) is a downstream mediator of TLR7 signaling. IRF-5 was induced following EBV infection, and IRF-5 was expressed in B-cell lines with type III latency. Expression of IRF-5 in this setting is surprising since IRF-5 has tumor suppressor and antiviral properties. B-cell proliferation assays provided evidence that EBV modulates TLR7 signaling responses. Examination of IRF-5 transcripts identified a novel splice variant, V12, that was induced by EBV infection, was constitutively nuclear, and acted as a dominant negative form in IRF-5 reporter assays. IRF-4 negatively regulates IRF-5 activation, and IRF-4 was also present in type III latently infected cells. EBV therefore initially uses TLR7 signaling to enhance B-cell proliferation and subsequently modifies the pathway to regulate IRF-5 activity.
The innate immune system is the cell's first defense against viruses. Detection of microbial pathogens is mediated through recognition of pathogen-associated molecular patterns by cellular pattern recognition receptors that include cytosolic molecules such as RIG-I, which senses single-stranded RNA from negative-strand viruses and has also been reported to recognize the polymerase III EBERs synthesized by Epstein-Barr virus (EBV) (57) and Toll-like receptors (TLRs) (12, 24, 32). Herpes simplex virus (HSV), varicella-zoster virus, and the human and mouse cytomegaloviruses (CMVs) are recognized by the plasma membrane TLR2 receptor (19). In the case of human CMV, the recognition was mediated by glycoproteins B and H (11). Protection against murine CMV and HSV type 2 infections is also mediated by endosomal TLR9 (46, 64). The responses induced by virus recognition of TLRs are dependent on the cell type, as well as whether or not the virus has mechanisms to counter TLR signaling. For example, TLR signaling in plasmacytoid dendritic cells elicits the secretion of large amounts of type I interferons, while peritoneal macrophages secrete predominantly inflammatory cytokines (19).
Relatively little is known about the interaction of EBV with the TLR pathway or about the mechanisms of EBV evasion of the innate immune response. More than a decade ago, it was observed that B-cell lines expressing the full complement of latency genes (type III latency) were resistant to the effects of interferon treatment and this resistance required the presence of intact EBNA2 and EBNA-LP genes (2, 29). In addition to the recent description of an interaction with RIG-I, the EBERs have been reported to inhibit interferon-stimulated gene activity by binding to PKR (double-stranded RNA-dependent protein kinase) and inhibiting its activation (50). LMP1 interrupts the interferon pathway by preventing TYK2 and STAT2 phosphorylation after interferon treatment (20), but enigmatically, LMP1 has also been reported to establish an antiviral state in cells by promoting the expression of interferon-stimulated genes such as those for oligoadenylate synthetase and ISG15 (74). Conditional expression of EBNA2 has also been found to induce the expression of interferon-stimulated genes (30).
While TLRs serve as sensors of pathogen invasion, the activation of pathways such as the MAPK, PI3K, and NF-κB pathways (1) by TLR signaling can be highjacked by viruses to activate their own gene expression and establish infection. The naive B cell is the target of EBV infection in vivo (28, 67). Human B cells express cell surface TLR1, -2, and -6 and endosomal TLR7, -9, and -10. Two studies found TLR6, -7, -9, and -10 to be more highly expressed on memory B cells than on naive B cells (8, 26). Activation of B cells is stimulated by the binding of antigen to the B-cell receptor and by survival signals produced by CD40 signaling. Stimulation of the TLR7 and TLR9 pathways provides additional signals that drive B cells to proliferate. Unmethylated, CpG-rich bacterial DNA induces splenic B-cell proliferation through TLR9 (1, 38). Synthetic ligands of TLR7, R837, and R848, which are members of the imidazoquinoline family, also activate B cells and induce their proliferation (68). R848-induced signaling was found to be similar to that induced by CD40 ligand in that NF-κB and c-Jun were both activated (9, 10). The effect of TLR signaling on B-cell activation is also apparent in situations where chromatin-immunoglobulin complexes inappropriately activate B cells through synergistic BCR and TLR stimulation and lead to autoimmune disease (40).
EBV infection of B cells induces B-cell activation and proliferation. The EBV latency membrane protein LMP1 is essential for B-cell immortalization in vitro (33). LMP1 functions as a constitutively active, ligand-independent tumor necrosis factor receptor to activate the NF-κB, MAPK, PI3K, and JAK3/STAT pathways (16, 18, 21, 27, 45) and provide signals equivalent to those induced by CD40 ligand (35, 69). The LMP2 latency membrane protein is not essential for in vitro B-cell immortalization (60) but provides important in vivo survival signals that mimic those arising from B-cell receptor activation (55, 63). The recent recognition that TLR signaling may serve as a third signal for optimal B-cell activation, along with antigen binding to the B-cell receptor and CD40 stimulation (56), led us to investigate interactions between the EBV and TLR pathways. We found upregulation of interferon stimulated genes driven by EBV contact with the B cell. Among the upregulated cell genes were TLR7 and MyD88 (myeloid differentiation factor 88), a key participant in TLR signal transduction. EBV infection also upregulated the downstream TLR7 effector IRF-5. Follow-up experiments provided evidence for a contribution of TLR7 signaling to the initial stages of EBV-induced B-cell proliferation and for subsequent virus-induced downmodulation of the IRF-5 arm of the pathway. EBV is thus able to take advantage of the proproliferative effects of TLR signaling while limiting IRF-5-mediated negative effects that would compromise the ability to establish a persistent EBV infection.
MATERIALS AND METHODS
Cell lines and B-cell isolation.
The Akata-4E3, Akata-BX1, Akata, Daudi, Mutu I, Mutu III, Rael, Ramos, EREB2-5 (in 1 μM β-estradiol), Snu719 gastric carcinoma (34), LCL (Akata immortalized primary B cells; gift from C. Farrell, Johns Hopkins University), P3HR-1, BC-2, and 1852 (containing LMP-1 under the control of a tetracycline-responsive promoter) (35) cell lines were maintained in RPMI medium supplemented with 10% fetal bovine serum and antibiotics (penicillin and streptomycin). The HeLa and 293 adherent cell lines were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Primary B cells were selected from peripheral blood mononuclear cells (PBMC) after infection with the MACS CD19 positive-selection kit (Miltenyi Biotec). Naive B cells were isolated from PBMC prior to infection by negative selection (Cambrex) with MACS naive B-cell isolation kit II. After separation, cell purity was checked by fluorescence-activated cell sorter (FACS) with a FACScalibur (Becton Dickinson) and phycoerythrin (PE)-CD27 and fluorescein isothiocyanate (FITC)-CD19 antibodies (eBioscience).
Reverse transcriptase PCR (RT-PCR).
RNA was extracted from cell lines or EBV-infected B cells with the RNeasy kit (QIAGEN) and DNase treated with the DNA-free kit (Ambion) according to the manufacturers' instructions. Reverse transcription was carried out with avian myeloblastosis virus reverse transcriptase (Promega) by following the manufacturer's protocol. The amount of cDNA added to each PCR mixture was normalized relative to the levels of TATA binding protein (TBP). PCRs were performed with PCR Master Mix (Promega) or Platinum PCR Master Mix (Invitrogen). Real-time reactions were performed with SYBR green master mix (AB). All primers were checked for specificity with BLAST. RT-negative samples were used to check for DNA contamination. Real-time products were analyzed by a melting curve or agarose gel electrophoresis to confirm product size and the presence of a unique band. PCR products were run on 2% agarose gels and visualized by ethidium bromide staining. The primers used were for TBP (sense, 5′ CACGAACCACCGGCACTGATT; antisense, 5′ TTTTCTTGCTGCCAGTCTGGAC), IRF-5 (LGH 5091 sense, 5′ AGGAAGAGCTGCAGAGGATG; LGH 5176 antisense, 5′ CCTCCAGCTGGCTGTAGAAG), V12 IRF-5 (LGH 6228 sense, 5′ GAAGAGGAAGAGAGGATGTCA; LGH 5176 antisense, see above), IRF-3 (sense, 5′ CCAAAGCCACGGATCCTG; antisense, 5′ CTGGGTATCAGAAGTACTGC), IRF-3A (sense, 5′ GTTGGAGCTGCAGGGTTACG; antisense, same as IRF-3), TLR7 (sense, 5′ TGTTTCCAATGTGGACACTGAA; antisense, 5′ TGTTCGTGGGAATACCTCCAG), TLR9 (sense, 5′ CTGCCACATGACCATCGAG; antisense, 5′ GGACAGGGATATGAGGGATTTGG), TLR10 (sense, 5′ GGTTATTATGCTAGTTCTGGGGTTGG; antisense, 5′ GTGCATTGACCTAGCATCCTGAGATA), EBNA2 (quantitative PCR) (sense, 5′ CATAGAAGAAGAAGAGGATGAAGA; antisense, 5′ GTAGGGATTCGAGGGAATTACTGA) (28), EBNA2 (semiquantitative RT-PCR) (sense, 5′ AGAGGAGGTGGTAAGCGGTTC 3′; antisense, 5′ TGACGGGTTTCCAAGACTATCC 3′) ISG15 (sense, 5′ GAGAGGCAGCGAACTCATCT; antisense, 5′ GCCAATCTTCTGGGTGATCT), PKR (sense, 5′ ACGCTTTGGGGCTAATTCTT; antisense 5′ TTCTCTGGGCTTTTCTTCCA), IFIT4 (sense, 5′ CGGAACAGCAGAGACACAGA; antisense, 5′ TGCTTGGTCAGCATGTTCTT), MXI (sense, 5′ GTGCATTGCAGAAGGTCAGA; antisense, 5′ TTCAGGAGCCAGCTGTAGGT), RANTES (sense, 5′ CGCTGTCATCCTCATTGCTA; antisense, 5′ GAGCACTTGCCACTGGTGTA), MYD88 (sense, 5′ AGAGCAAGGAATGTGACTTCCAGA; antisense, 5′ GTGTAGTCGCAGACAGTGATGAACC), tumor necrosis factor alpha (sense, 5′ CCCCTCAGCTTGAGGGTTTG; antisense, 5′ TGCTGCACTTTGGAGTGATC), OAS2 (sense, 5′ ACAGCTGAAAGCCTTTTGGA; antisense, 5′ AAGTTTCGCTGCAGGACTGT), STAT1 (sense, 5′ CCGTTTTCATGACCTCCTGT; antisense, 5′ TGAATATTCCCCGACTGAGC), OASL (sense, 5′ TTTCTGAGGCAGGAGCATTAG; antisense, 5′ CTCCTGGAAGCTGTGGAAAC), and IFITM1 (sense, 5′ ATGGTAGACTGTCACAGAGC; antisense, 5′ CTGGTCCCTGTTCAACACCC).
5′ Rapid amplification of cDNA ends (RACE).
Total RNA was extracted from Mutu III cells with the RNeasy kit (QIAGEN). RNA was DNase treated with DNA-free (Ambion). After phenol-chloroform extraction, an adaptor was added to the 5′ end of the mRNA in three steps with the First Choice RNA ligase-mediated (RLM)-RACE kit (Ambion) by following the manufacturer's protocol. Reverse transcription was carried out on the resulting RNA with an antisense IRF-5 primer (5′ CAGCTGGTTCGTGTAGAAGCGCTG 3′) and Thermoscript RT (Invitrogen) as recommended by the manufacturer. The first step of the nested PCR was carried out on the cDNA with a sense outer primer complementary to the adaptor and antisense IRF-5-specific outer primer LGH 5176 (5′ CCTCCAGCTGGCTGTAGAAG 3′). The second step of the nested PCR was carried out with a sense inner primer complementary to the adaptor and antisense V12 IRF-5-specific primer LGH 6816 (5′ GCGGCCACTTGACATCCTCTCTTCCTCTTCTTC 3′). Platinum Taq DNA polymerase High Fidelity (Invitrogen) was used in all of the PCRs, and DNA product bands were extracted from an agarose gel with a Minelute kit (QIAGEN). PCR products were cloned into pCR2.1 with the Topo TA cloning kit (Invitrogen) and sequenced by the Johns Hopkins Sequencing Core.
EBV infection.
Cells were infected with green fluorescent protein (GFP)-BX1 virus (42) in a small volume, after which they were diluted 5- to 10-fold into medium. Cells were harvested at time points following dilution. Virus stocks were divided into two with one-half being treated with Trioxsalen (150 μM; Calbiochem) for 10 to 15 min at room temperature. This stock was transferred to a FEP bag (Vuelife) and UV irradiated at 4 J/cm2 with a UV irradiator (Illuminator; Cerus). Unbound Trioxsalen was removed by washing twice with 40 volumes of RPMI medium with a Centricon-Plus 70 filter. Both irradiated and untreated virus samples were washed. The virus was then filtered with a 45-μm-pore-size filter and stored at 4°C.
Reporter assays.
We seeded 1 × 105 293 adherent cells per well in a six-well plate and transfected them with calcium phosphate the next day. IFNA1 promoter luciferase reporter (1.0 μg per well; gift from Y. Yuan, University of Pennsylvania) was transfected with 0.3 μg of a Renilla luciferase construct which was used to normalize the transfection efficiency. IRF-5 constructs were cotransfected starting with 0.5 μg/well, and SG5 plasmid was used to equalize the amount of DNA transfected. Cells were harvested 24 h after transfection with 300 μl of Glo lysis buffer (Promega) per well. Luciferase assays were performed with 20 μl of lysate and a Lumat (Berthoid) luminometer. Results shown are the average of two independent assays each done in duplicate.
B-cell proliferation assays.
Naive B cells were infected immediately after separation from PBMC in the presence or absence of R837 (2 to 10 μg/ml; Invivogen). After 3 h, the cell culture was diluted and aliquoted into a 96-well plate for a final concentration of 2.64 × 104 cells/300 μl/well. R837 or inhibitor treatments were supplemented every 2 to 3 days as noted in the figure legends. Synthetic oligonucleotides with phosphorothioate backbones IRS 661 (5′-TGCTTGCAAGCTTGAAGCA-3′) and a control oligonucleotide (5′-TCCTGCAGGTTAAGT-3′) were described by Barrat et al. (6) and synthesized by Integrated DNA Technologies Inc. The B-cell proliferation in each well was measured in duplicate with the Cell Titer Glo assay (Promega). Each sample type was set up in duplicate for every time point. The results shown are representative of two experiments. Error bars represent standard deviations.
Isoform analysis.
IRF-5 variants were identified from a pool of PCR products resulting from two consecutive rounds of PCR performed on cDNA made from RNA harvested from EBV-infected primary B cells with primers LGH 5474 (sense, 5′ TTATTCTGCATCCCCTGGAG 3′) and LGH 5176 (antisense, see above). PCR products were TA cloned into pCR2.1 (Invitrogen) and sequenced.
Western blotting.
Endogenous IRF-5 was analyzed in cells that were washed in phosphate-buffered saline and lysed in lysis buffer (50 mM Tris [pH 7.6], 50 mM NaCl, 0.5 mM EDTA, 0.2% NP-40, and 1% glycerol with protease inhibitors) at a concentration of 1 × 107 cells per ml. Cells were lysed at 4°C for 15 min, sonicated, and then centrifuged for 30 min at top speed at 4°C. β-Tubulin was used to normalize the amount of whole-cell lysate loaded onto a 12% sodium dodecyl sulfate-polyacrylamide electrophoresis gel and transferred to a Trans-blot nitrocellulose membrane (Bio-Rad). The membrane was probed with antibodies to IRF-5 (Protein Tech Group), IRF-3 (Santa Cruz), β-tubulin (Sigma), IRF-4 (Cell Signaling), and LMP-1 (DAKO). The secondary antibodies used were horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies (Amersham Biosciences). Protein bands were visualized with the ECL system (Amersham Biosciences). Membranes were stripped for reprobing with Restore Western blot stripping buffer (Pierce). IRF-5 variants were in vitro translated with the Quik Coupled Transcription/Translation System (Promega). Samples were subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and after being blocked in 5% milk-phosphate-buffered saline-Tween 20, the membrane was probed with an anti-hemagglutinin (HA) antibody (rabbit; Santa Cruz) and a horseradish peroxidase-conjugated anti-rabbit secondary antibody (Amersham Biosciences).
Plasmids.
HA-IRF-5 (variant 3) was constructed by ligating a PCR product containing the IRF-5 N-terminal 300 bp amplified from an IRF-5 cDNA (ImageClone) with a cDNA obtained from a B-cell Gal4ACT library (Clontech) into an SG5 vector (Invitrogen) modified to contain a Gateway cassette and an HA tag. Other variants were cloned by digestion of PCR products generated with IRF-5 primers LGH5474 and LGH5176 with SmaI, which has recognition sites flanking the IRF-5 variable region. The individual variable regions were then inserted into HA-IRF-5 between the SmaI sites. Phosphomimetic IRF-5, variant 3 (PV3), was made in the Gateway Entry vector with the Quick Change kit (Stratagene) to convert IRF-5 serines 425, 427, and 430 into glutamic acid. The phosphomimetic IRF-5 protein was then HA tagged with the Gateway system.
Immunofluorescence.
HeLa cells seeded at 1.2 ×104 per well were transfected with 0.5 μg of each DNA by calcium phosphate precipitation. Two days after transfection, cells were fixed in methanol and probed with an anti-HA rabbit primary antibody (Upstate) and a rhodamine-conjugated goat anti-rabbit secondary antibody (Jackson). Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) in Vectashield mounting solution (Vector Laboratories), and cells were photographed with a fluorescence microscope and Image-Pro Plus software.
RESULTS
UV-inactivated EBV induces the expression of interferon-responsive genes in naive B cells.
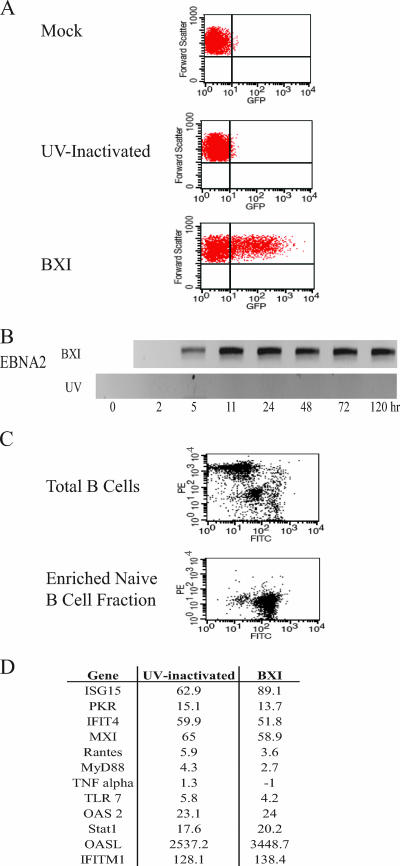
Virus infection induces a cellular immune response. Studies with CMV have found that virus binding and entry are sufficient to induce interferon and proinflammatory cytokine responses (59). To evaluate the consequences of EBV infection in the absence of viral gene expression, virus prepared from lytically induced Akata-BX1 cells was aliquoted and half of the stock was subjected to UV inactivation. To validate the effectiveness of the UV treatment, EBV-negative 4E3 B cells were infected with UV-treated or untreated virus and 24 h after infection the cells were analyzed by FACS for GFP expressed from the recombinant BX1 virus. A GFP-positive cell population was observed after infection with untreated EBV, but the GFP signal in the cells infected with UV-treated virus did not differ from that seen in mock-infected 4E3 cells (Fig. 1A). Expression of the EBV EBNA2 gene was also examined by real-time RT-PCR (Fig. 1B). EBNA2 transcripts appeared 5 h after infection of 4E3 cells with untreated virus, and expression continued for the 5-day period of the experiment. No EBNA2 transcripts were detected in 4E3 cells infected with UV-treated virus. The UV treatment was therefore effective in preventing expression from the EBV genome.
FIG. 1.
UV-inactivated EBV induces ISG expression in naive B cells. (A, B) Confirmation of EBV inactivation. 4E3 cells were infected with EBV-BXI from untreated or UV-inactivated stocks. (A) Cells were harvested at 24 h and analyzed by FACS. GFP expression levels were compared in mock-infected (top), UV-inactivated-EBV-infected (middle), and untreated-EBV-infected (lower) cells. (B) RNA was harvested from infected 4E3 cells, and RT-PCR was performed to detect transcript levels of EBNA2. (C) Naive B cells were separated from PBMC by negative selection. PBMC and the naive B-cell fractions were stained with PE-labeled anti-CD27 antibody (memory B-cell marker) and FITC-labeled anti-CD19 antibody (pan-B-cell marker) and analyzed by FACS. (D) Interferon-stimulated genes are induced in naive B cells by UV-inactivated and untreated EBV infection. RNA isolated from naive B cells 5 h after infection with UV- inactivated or untreated EBV was amplified by real-time PCR. Values are relative to those of uninfected naive B cells. Real-time assays were performed in triplicate, and results are representative of at least two experiments.
The predominant target for EBV infection in vivo is believed to be the naive B cell (28, 67). Naive B cells were separated from PBMC by negative selection with the MACS naive B-cell isolation kit. FACS analyses of PBMC and the naive B-cell fraction stained with PE-labeled anti-CD27 (memory B-cell marker) and FITC-labeled anti-CD19 (pan-B-cell marker) antibodies was used to verify successful enrichment of the naive B-cell population (Fig. 1C). Naive B cells were then infected with UV-inactivated or untreated virus, and 5 h later RNA was isolated and the expression of a panel of interferon-stimulated genes was examined by real-time RT-PCR with the assays being performed in triplicate. Eleven of 12 tested cell genes were induced by both UV-inactivated and untreated virus. The exception was the proinflammatory cytokine tumor necrosis factor alpha, which did not respond to either infection at this early time point (Fig. 1D). The data indicate that contact between EBV and the cell in the absence of EBV gene expression is sufficient to induce the cell to upregulate interferon-stimulated genes. Interestingly, two of the induced genes, TLR7 and MyD88, are participants in the TLR signaling pathway.
TLR7 is specifically induced by EBV contact with B cells.
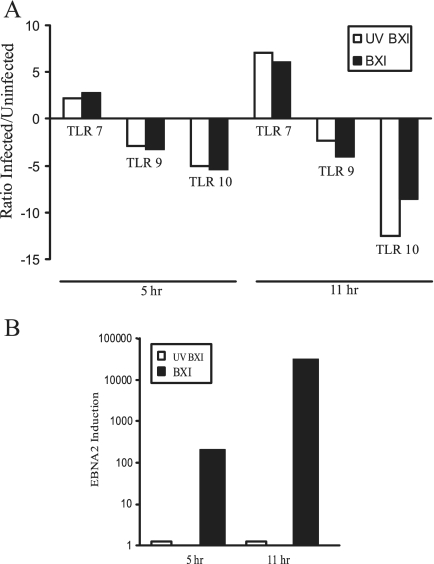
Naive B cells express detectable amounts of TLR1, TLR2, TLR6, TLR7, TLR9, and TLR10, although expression is reported to be low compared to that seen in memory B cells (8, 48). Naive B cells were infected with UV-inactivated or untreated EBV, and TLR expression was examined by real-time RT-PCR at 5 h and 11 h after virus addition. TLR7 was the only one of these TLRs that was induced by EBV. UV-inactivated (open bars) and untreated virus (filled bars) stimulated TLR7 expression two- to threefold at 5 h and six- to sevenfold at 11 h (Fig. 2A). In contrast, TLR9 and TLR10 expression decreased in comparison to that in uninfected cells (Fig. 2A) and TLR1 and TLR6 expression showed no significant change (data not shown). Monitoring of EBNA2 expression in the same infections confirmed that EBNA2 was expressed at 5 and 11 h after infection with untreated virus and was not expressed in cells incubated with UV-inactivated virus (Fig. 2B).
FIG. 2.
EBV induces TLR7. EBV infection induces TLR7 but represses TLR9 and -10 expression in naive B cells. Cells were infected with EBV (filled bars) or UV-inactivated EBV (open bars), and RNA was isolated at the indicated times after infection. (A) TLR expression. (B) EBV EBNA2 expression. RNA was quantified by real-time RT-PCR. The results are representative of two independent experiments performed in triplicate.
EBV interaction with the TLR7 pathway enhances B-cell proliferation.
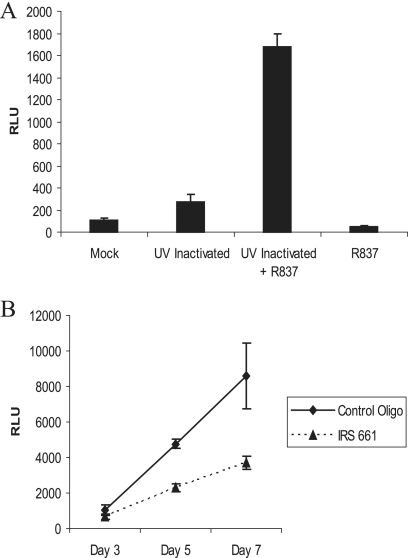
Two approaches were taken to evaluate whether TLR7 signaling contributed to the early stages of EBV-induced B-cell proliferation. Firstly, the growth of naive B cells was compared before and after induction of TLR7 by UV-inactivated virus and in the presence or absence of the TLR7 ligand R837. Cells were harvested after 3 days in culture, and the cell number was assayed with Cell-Titer Glo (Fig. 3A). Addition of R837 (2 μg/ml) to naive B cells reduced cell numbers twofold. Addition of UV-inactivated EBV produced a threefold growth stimulation. However, addition of inactivated virus plus R837 resulted in an augmented 17-fold increase in growth. In a complementary approach, naive B cells were infected with untreated EBV in the presence or absence of the TLR7 inhibitory oligonucleotide IRS 661 (Fig. 3B). EBV-induced proliferation was decreased in the presence of IRS 661 (filled triangles). These experiments indicate that the EBV-induced TLR7 is functional and that stimulation of TLR7 signaling enhances EBV-driven growth of naive B cells.
FIG. 3.
EBV-mediated stimulation of TLR7 enhances B-cell proliferation. (A) Naive B cells were incubated with UV-inactivated virus in the presence or absence of R837 (2 μg/ml) for 3 days. (B) Naive B cells were infected with BXI virus in the presence of a control oligonucleotide or IRS 661 (TLR7 inhibitory oligonucleotide) at 2.8 μM. Cells were treated at days 0, 3, and 5. Cell growth was quantitated in quadruplicate with Cell Titer Glo (Promega). Results are representative of two independent experiments. The standard deviation is shown. RLU, relative light units.
Expression of TLR7 and its downstream effector IRF-5 in B-cell lines.
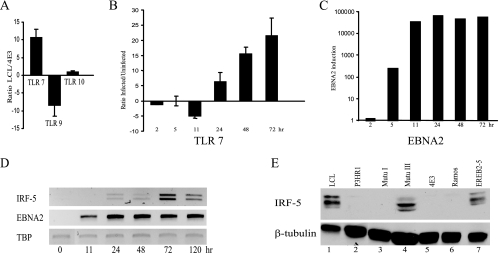
To evaluate whether TLR7 signaling might play a role at later times after EBV infection, TLR expression was compared in an EBV-positive LCL cell line with type III latency versus EBV-negative 4E3 cells. The two cell lines showed equal expression of TLR10, but TLR7 was overexpressed and TLR9 was underexpressed in the LCL cell line relative to 4E3 cells (Fig. 4A). 4E3 cells were then infected with EBV, and the expression of TLR7 and its downstream effector IRF-5 was examined by real-time RT-PCR. EBV infection of 4E3 cells induced TLR7 and IRF-5 expression (Fig. 4B and D). (The IRF-5 RT-PCR products reflect the presence of alternatively spliced IRF-5 variants.) The time courses of induction of TLR7 and IRF-5 were similar and were delayed relative to EBNA2 expression (Fig. 4C) and to the time course of TLR7 induction in primary B cells.
FIG. 4.
Expression of TLR7 and IRF-5 is associated with EBV primary infection, as well as type III latency. (A) TLR7 is upregulated in type III LCLs. Comparison of TLR7, TLR9, and TLR10 expression in the type III latently infected LCL cell line versus the EBV-negative 4E3 cell line. RNAs were quantified by real-time RT-PCR and gene-specific primers. The results are an average of two independent experiments performed in triplicate. (B, C) Induction of TLR7 after infection of EBV-negative 4E3 B cells. TLR7 (B) and EBV EBNA2 (C) RNAs were quantified by real-time RT-PCR. RNA was isolated at the indicated times after infection. The results are an average of two independent experiments performed in triplicate. (D) EBV induces IRF-5 transcripts. Shown are ethidium bromide-stained gels of electrophoretically separated RT-PCR products amplified from EBV-negative 4E3 B cells before (0 h) and at the indicated times after infection with Akata-BX1 virus. Primers were specific for IRF-5 (upper), EBV EBNA2 (middle), and TBP, which served as a loading control (lower). The results are representative of two independent experiments. (E) IRF-5 is expressed in type III latently infected cell lines. Western blot probed with anti-IRF-5 antibody comparing IRF-5 protein expression in type III latently infected cell lines (LCL, Mutu III, and EREB2-5), the EBV-positive EBNA2 deletion-containing cell line P3HR-1, a type I latently infected cell line (Mutu I), and EBV-negative B-cell lines (4E3 and Ramos). Equality of protein loading was determined by probing the membrane with anti-β-tubulin antibody.
IRF-5 protein expression was also examined in EBV-positive cell lines. Western blotting with anti-IRF-5 antibody revealed strong expression of IRF-5 proteins in the type III latently infected B-cell lines LCL, Mutu III, and EREB2-5 (Fig. 4E). The detection of multiple protein bands may reflect expression from differentially spliced IRF-5 transcripts. IRF-5 protein was either not present or barely detectable in the type I Burkitt's lymphoma cell line Mutu I, which is the parent line of Mutu III; in the EBV-negative Burkitt's lymphoma cell lines 4E3 and Ramos; or in P3HR-1, in which the gene for EBNA2 is deleted. β-Tubulin was used as a loading control for the Western blot analysis.
EBV gene expression modifies TLR7 signaling.
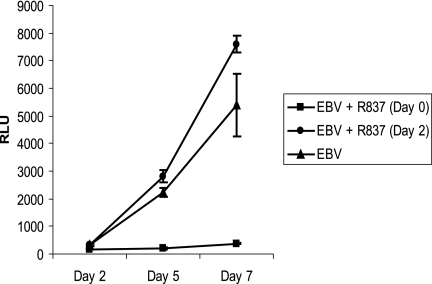
The expression of IRF-5 protein in type III latently infected cell lines was surprising given that IRF-5 has been described as having tumor suppressor and proapoptotic activities (4, 15). It seemed possible that EBV might be modulating the IRF-5 arm of the TLR7 response in such a way as to minimize negative outcomes. To evaluate this scenario, the effects of overstimulation of the TLR7 pathway with increased doses of the TLR7 ligand R837 were investigated. With EBV-infected naive B cells, we found that treatment with a 10-fold dose (20 μg/ml) of R837 at the time of EBV infection was growth inhibitory and prevented EBV-induced B-cell proliferation (Fig. 5, filled squares versus triangles). However, when the naive B cells were infected with EBV and left for 2 days to allow time for expression of EBV latency III genes prior to the addition of 20 μg/ml R837, this growth inhibition was completely overcome (Fig. 5, circles). This result suggests that the functioning of the TLR7 pathway is modified subsequent to the initial infection.
FIG. 5.
EBV ameliorates negative growth effects following overstimulation of TLR7. Naive primary B cells were infected with EBV in medium without or with R837 (20 μg/ml) added initially at the time of infection (day 0) or 2 days postinfection (day 2) and subsequently supplemented on days 2 and 5. Cell proliferation was measured in quadruplicate with the Cell Titer Glo assay. The data shown are representative of two independent assays. The standard deviation is shown. RLU, relative light units.
EBV induction of a novel negative regulatory IRF-5 splice variant, V12.
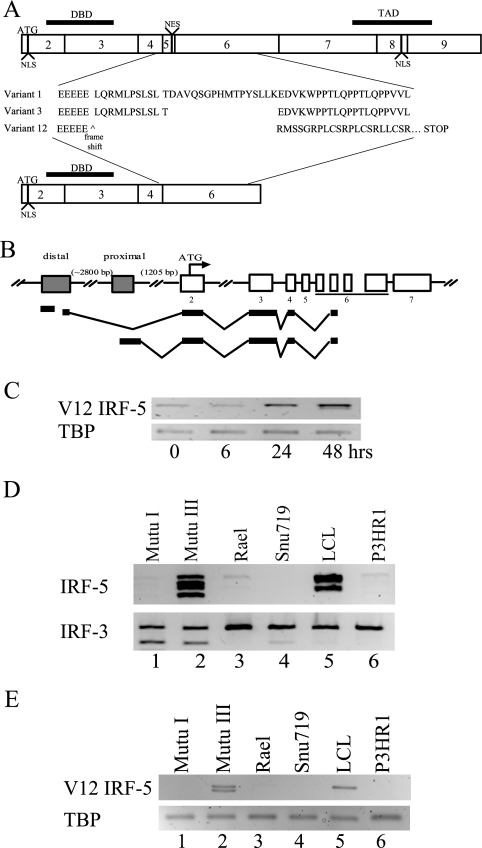
IRF-5 is expressed as multiple splice variants, and expression of particular splice variants has recently been linked to the development of systemic lupus erythematosus (22). IRF-5 splice variant expression in EBV-infected primary B cells was investigated by amplifying IRF-5 transcripts with primers from either exon 2 or exon 4/5 to exon 7. The PCR products obtained were cloned and sequenced. No evidence was found for the splice variant associated with systemic lupus erythematosus, but a previously undescribed splice variant, V12, was identified. Two forms of V12 were isolated that differed by 30 bp in the exon 6 variable region. A comparison of the predicted amino acid sequence of the common region of V12 with the previously described V1 and V3 IRF-5 variants (Fig. 6A) shows that V12 would encode a DNA binding domain in the absence of an activation domain and hence could function as a negative modulator of IRF-5 transcriptional activity.
FIG. 6.
EBV infection induces a novel alternative splice variant of IRF-5, V12. (A) Exon structure of IRF-5 showing the predicted amino acid sequence of the variable exon 4 to 6 region of the V1, V3, and V12 cDNAs amplified by RT-PCR from EBV-infected cells. NLS, nuclear localization signal (3). NES, nuclear export signal (15, 44). DBD, DNA binding domain. TAD, transactivation domain. (B) Diagram of the structure of V12 cDNAs amplified by 5′ RLM RACE from Mutu III cells and cloned and sequenced. Shaded box, promoter region. (C to E) Ethidium bromide-stained, electrophoretically separated RT-PCR products amplified with primers specific for the V12 variant showing induction of V12 transcripts after EBV infection of Ramos cells (C) and expression of V12 transcripts in type III EBV-infected cell lines (Mutu III and LCL) but not in P3HR-1 or the type I latently infected B-cell lines (Mutu I, Rael) or gastric carcinoma cells (Snu719) (E). (D) Generic IRF-5 primers were used to amplify IRF-5 transcripts in the cell lines used in panel C. Amplification of cellular TBP or the ubiquitously expressed IRF-3 protein was used as a loading control. Results are representative of at least two experiments.
It has been suggested that splice variant expression is dependent on promoter usage (47). Four potential promoter regions have been described for IRF-5. Two of these were found to have activity when cloned into reporter vectors (47), and another was identified as the source of transcripts in systemic lupus erythematosus patients (22). To determine the structure of the V12 IRF-5 transcripts in Mutu III cells, 5′ RLM RACE was performed with a 3′ primer located across the exon 4-to-6 splice site. The RLM RACE procedure specifically amplifies 5′-capped mRNAs. Sequencing of the cloned cDNA products identified V12 transcripts that were derived almost equally from the distal and proximal IRF-5 promoters (Fig. 6B). Interestingly, the five sequenced cDNAs initiating from the proximal promoter all had the identical 5′ start point at −1,408 bp from the IRF-5 ATG initiator codon, while the three sequenced cDNAs arising from the distal promoter initiated at two different positions within a 314-bp region. The 5′ termini were located 4,366 and 4,052 bp from the IRF-5 ATG. This type of stuttered 5′ initiation is typical of RNAs arising from TATA-less promoters. From the RACE analysis, we obtained promoter usage and coding sequence information for the V12 variant.
The PCR primers used to examine IRF-5 mRNA induction in Fig. 4 would not have detected the V12 splice variant. V12-specific primers were designed, and V12 expression was examined following EBV infection of Ramos cells (Fig. 6C). EBV-negative Ramos cells expressed low levels of the V12 transcript prior to infection and V12 was upregulated between 6 and 24 h after infection. Since the V12 transcript was upregulated by EBV infection, expression of V12 was also examined in established B-cell lines. The Mutu III and LCL cell lines that express IRF-5 mRNA and protein also expressed the V12 mRNA (Fig. 6D and E, lanes 2 and 5), with Mutu III expressing both V12 forms. The available IRF-5 antibody did not recognize the V12 protein, and so endogenous V12 protein expression could not be examined.
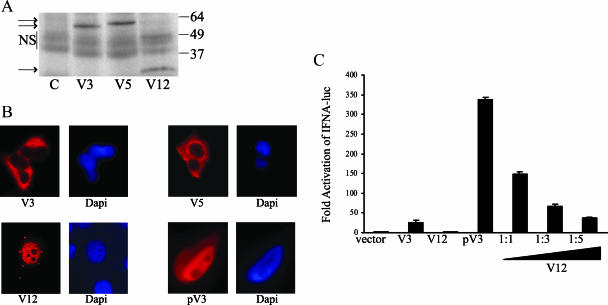
To verify that V12 encodes a truncated IRF-5 protein, in vitro transcription-translation of the HA-tagged V12 cDNA was performed. A smaller 22-kDa polypeptide was generated by the V12 cDNA compared to the approximately 60- to 63-kDa polypeptides generated by HA-tagged V3 and V5 cDNAs (Fig. 7A). To examine whether the V12-encoded protein had the properties of a negative modulator of IRF-5 transactivation, V12 IRF-5 was further characterized. V12 lacks the nuclear export signal encoded in exon 5 (44), and as seen in transfected cells, V12 was constitutively nuclear (Fig. 7B). This contrasts with the constitutively cytoplasmic localization of the V3 and V5 variants but is similar to the nuclear localization of a modified constitutively activated phosphomimetic V3 (PV3, Fig. 7B). In this phosphomimetic V3 version, the serine residues at positions 425, 427, and 430 were mutated to glutamic acid. A similar phosphomimetic V1 IRF-5 version has been described in which these residues were changed to aspartic acid (44).
FIG. 7.
V12 IRF-5 is constitutively nuclear and negatively regulates IRF-5 transactivation. (A) V12 encodes a truncated IRF-5 protein. Western blot probed with anti-HA antibody comparing the electrophoretic mobility of polyacrylamide gel electrophoresis-separated, HA-tagged, in vitro-translated V3, V5, and V12 IRF-5 polypeptides. Lane C, control with no added template. NS, nonspecific bands. The values on the right are molecular sizes in kilodaltons. (B) Immunofluorescence assays comparing the intracellular localization of HA-tagged IRF-5 V3, V5, and V12 proteins and the phosphomimetic V3 protein (pV3) in transfected HeLa cells. Cells were stained with an anti-HA primary antibody and a rhodamine-labeled secondary antibody. Nuclei were stained with DAPI. (C) Reporter assay in which 293 cells were transfected with an IFNA1 promoter-luciferase reporter in the presence of V3 IRF-5, V12 IRF-5, pV3 IRF-5, or pV3 IRF-5 plus increasing amounts of V12 IRF-5 as indicated. The data shown are averages from two assays.
Finally, the functional activity of V12 was compared with that of V3 and PV3 in a reporter assay with the IFNA1 promoter which is known to be IRF-5 responsive (5, 43). V3 activated expression of the promoter-luciferase reporter 20-fold, and constitutively activated PV3 gave a 340-fold activation (Fig. 7C). V12 did not affect reporter expression when transfected alone, but cotransfection of V12 with PV3 resulted in dose-dependent downregulation of PV3 activation (Fig. 7C). Thus, induction of V12 upon EBV infection and V12 expression in type III latently EBV-infected cells could modulate IRF-5 functional activity in these cells.
IRF-4, a negative regulator of IRF-5 activity, is upregulated in type III latency.
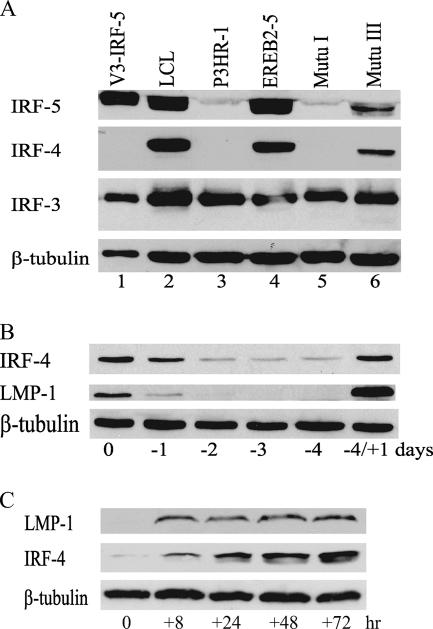
IRF-4 is an IRF family member that negatively modulates TLR-mediated IRF-5 activation by competing with IRF-5 for interaction with MyD88 (51). IRF-4 was among the cellular genes found by gene array analysis to be upregulated in EBV type III latency (13) and upregulated by EBNA2 in EREB2-5 cells (61), but IRF-4 expression in EBV-infected cells was not further characterized. Expression of IRF-4 in IRF-5-expressing and -nonexpressing B-cell lines was therefore examined by Western blotting (Fig. 8A). IRF-4 was strongly expressed in the same type III cell lines, LCL, EREB2-5, and Mutu III, that showed high-level expression of IRF-5 and was not detected in the non-IRF-5-expressing P3HR-1 and Mutu I cell lines. In contrast to IRF-5 and IRF-4, IRF-3 was uniformly expressed in the cell lines tested (Fig. 8A).
FIG. 8.
EBV induces expression of the IRF-5 negative regulator IRF-4. (A) Western blot assay comparing the expression of IRF-5 with that of IRF-4 and IRF-3 in different B-cell lines. Lane 1, 293 cells transfected with V3 IRF-5. Lanes 2, 4, and 6, type III latently infected cell lines. Lane 3, EBNA2 deletion-containing P3HR-1 cells. Lane 5, type I latently infected cell line. β-Tubulin served as a control for protein loading. (B) Western blot assay showing the dependence of IRF-4 expression on EBV latency gene expression. EREB2-5 cells were deprived of β-estradiol and harvested at the time points noted. Four days after withdrawal, β-estradiol was added back into the medium at 1 μM and cells were grown for a further 1 day (−4/+1). The membrane was probed with anti-IRF-4 and LMP1 antibodies. β-Tubulin was used as a loading control. (C) LMP-1 regulates IRF-4 expression. 1852, a tetracycline-on LMP-1, type III cell line, was grown in medium without tetracycline for 5 days, and then tetracycline was added back to the medium (time zero). A Western blot of the cell extract was probed with anti-IRF-4 and LMP1 antibodies. β-Tubulin was used as a loading control. The results are representative of two experiments.
To demonstrate that EBV latency gene expression induced IRF-4, we monitored the expression of IRF-4 in type III EREB2-5 cells grown in the presence or absence of estrogen. EREB2-5 cells contain estrogen-regulated EBNA2, and in the absence of estrogen, EBNA2 is cytoplasmic and transcriptionally inactive (34). Removal of estrogen therefore leads to loss of expression of all EBNA2-regulated latency genes, including LMP-1. After estrogen withdrawal, IRF-4 protein expression was downregulated with the same kinetics as LMP1 protein expression and only trace amounts of IRF-4 were expressed in the absence of LMP1 (Fig. 8B). IRF-4 expression also followed LMP1 expression upon resupplementation of the medium with estrogen, with the expression of both proteins rebounding rapidly (Fig. 8B, −4/+1 days). This result confirms the induction of IRF-4 by EBNA2 or EBNA2-regulated EBV genes expressed in type III latently infected cells. The reduced levels of IRF-4 seen in EBNA2 deletion-containing P3HR-1 cells (Fig. 8A) may consequently reflect the loss of EBNA2 in this cell line.
LMP1 is one of the latency genes that are regulated by EBNA2. To determine whether LMP1 contributed to IRF-4 induction, the 1852 cell line was used. This cell line was generated by infecting primary B cells with a recombinant mini-EBV in which LMP1 is expressed from a tetracycline-regulated promoter (35). After 3 days of washing to remove tetracycline from the culture medium, no LMP1 was detected (Fig. 8C, time zero). Within 8 h of addition of tetracycline to the medium, LMP1 protein expression increased and expression continued throughout the 3-day period of the experiment. IRF-4 protein expression was minimal at time zero, but IRF-4 appeared at 8 h and continued to be expressed at subsequent time points. LMP1 therefore contributes to the induction of IRF-4. Thus, in addition to modulation of IRF-5 DNA binding, IRF-5 activation in response to TLR signaling would be negatively modulated in type III latent infection.
DISCUSSION
The changes in B-cell growth induced by EBV infection are thought to mimic those that occur during the natural process of B-cell activation and differentiation (67). Two of the key natural signals for B-cell activation, namely, ligand interactions with the CD40 receptor and the BCR, are supplied by the constitutive signaling activity of the EBV LMP1 and LMP2 proteins. We now provide evidence that a third proliferative signal provided by TLR signaling (56) is induced by EBV infection and contributes to the initial B-cell growth response.
Contact between naive B cells and UV-inactivated EBV upregulated the expression of a number of interferon-responsive genes, including those for TLR7 and MyD88. The virus-cell interaction provoking the initial interferon response was not pursued here. However, the described stimulation of TLR2 by HSV, CMV, and varicella-zoster virus glycoproteins suggests that stimulation of TLR2, which is expressed on the plasma membrane of B cells, is a good candidate. TLR7 was the only TLR observed to be upregulated and, in contrast TLR9 expression, was downregulated by contact with untreated or UV-inactivated EBV. The repression of TLR9 initially seems surprising since the HSV and CMV herpesviruses stimulate TLR9 signaling through their CpG-rich genomes (39, 46, 64) and stable expression of LMP2A in B cells of transgenic mice moderately upregulated TLR9 expression in a BCR-dependent manner (70). NF-κB p65 suppresses TLR9 transcription (66). EBV binding to the CD21 cell surface receptor activates NF-κB (62), and a principal function of LMP1 is NF-κB activation. Thus, EBV-induced NF-κB activity may be the source of the TLR9 downregulation. TLR7 and TLR9 also modulate each other's activity. TLR7 signaling itself mediates NF-κB activation, creating a negative feedback loop for TLR9 expression, and conversely, TLR9 can physically interact with TLR7 and inhibit TLR7 function in a dose-dependent manner (71). MyD88 is an adaptor molecule that, upon TLR stimulation, is recruited to the cytoplasmic tail of all TLRs except TLR3 (17) and is necessary for the subsequent recruitment of the tumor necrosis receptor-associated factors TRAF3 and TRAF6 and the kinases IRAK4 and IRAK1. The upregulation of TLR7 and its adaptor MyD88, in concert with the downregulation of TLR9, would establish conditions suited for TLR7 pathway activity.
Naive B cells, the target of EBV infection in vivo, have low TLR expression compared to memory B cells. Proliferation of naive B cells in response to TLR7 ligand required costimulation with alpha interferon to upregulate TLR7 expression (7). We also saw no stimulation when naive B cells were treated with the TLR7 ligand R837 alone, but naive B-cell growth was stimulated by the combination of R837 and UV-inactivated EBV, presumably because of the induction of TLR7 and MyD88 as part of the interferon-stimulated gene response that followed initial contact between EBV and the B cell. A contribution of TLR7 activity to initial B-cell outgrowth was supported by the reduced growth observed when the EBV-infected naive B-cell cultures were treated with IRS 661, a TLR7-specific inhibitory oligonucleotide. While TLR7 expression is upregulated by EBV contact with the cell in the absence of viral gene expression, EBV gene expression may be a source of the single-stranded RNA that normally activates TLR7 signaling.
Two interferon regulatory factors, IRF-5 and IRF-7, are recruited to MyD88 and activated by TLR7 signaling. IRF-7 was first described in type III latently infected B cells, where it was identified as a transcription factor that bound and repressed the EBV Qp promoter that serves as an alternative promoter for driving EBNA1 expression (75). Subsequently, LMP1 induction and activation of IRF-7 and a positive feedback loop of IRF-7 induction of LMP1 were described previously (52). IRF-7 activation leads to nuclear translocation and induction of all of the alpha interferon gene promoters, as well as the beta interferon gene promoter (25). Despite this activity, EBV has been able to incorporate IRF-7 into its own biology in type III latently infected cells. We found that IRF-5 expression was also induced by EBV infection. Stimulation of TLR7 signaling induces IRF-5 (58), and we noted that IRF-5 induction after infection of 4E3 B cells coincided temporally with the time at which TLR7 was upregulated. IRF-5 was initially identified as a transcription factor expressed as multiple splice variants that is active on type I interferon promoters (5, 47). Irf5−/− mice have a reduced type I interferon response to HSV type 1 infection and enhanced viral propagation (72). Alpha and beta interferon production in response to TLR7 stimulation is also impaired in Irf5−/− mice (73). IRF-5 has also been recognized as a key mediator of cytokine induction after TLR stimulation and has been proposed to activate cytokine promoters synergistically with NF-κB (49, 65). While an initial induction of cytokines such as interleukin-6 and -12 may be beneficial for EBV infection, continued interferon production and IRF-5 activity would be expected to have a negative impact on EBV-induced B-cell expansion. For example, IRF-5 induces apoptosis in response to DNA damage (72) and BJAB B cells stably expressing IRF-5 formed fewer and smaller tumors in mice than did control BJAB cells (4). The continued expression of IRF-5 in type III cell lines therefore suggested that, as is the case for IRF-7, EBV must modulate IRF-5 activity to balance the positive and negative downstream consequences. Initially to test this hypothesis, EBV infection of naive B cells was performed in the presence of a 10-fold dose of R837 to overstimulate the TLR7 pathway. R837 at 20 μg/ml completely blocked B-cell growth when supplied concurrently with virus infection. However, if EBV latent gene expression was allowed to initiate prior to the addition of the R837, then there was no negative effect on growth compared to that of untreated, EBV-infected B cells, implying that one or more of the EBV latency gene products was modifying signaling downstream of TLR7.
We explored ways in which IRF-5 activity could be modified in EBV-infected cells and obtained evidence for two separate mechanisms. IRF-5 is expressed as multiple splice variants, and alterations in IRF-5 splicing have been linked to the development of systemic lupus erythematosus, suggesting that different splice variants may differ in their functional properties (22, 23). An examination of IRF-5 splice variant expression in EBV-infected cells led to the identification of a novel IRF-5 splice variant, V12, that was induced by EBV infection and was expressed in all type III latently infected cells expressing IRF-5. The V12 sequence encoded a truncated IRF-5 protein that retained the DNA binding domain but lacked the nuclear export signal and transactivation domain. In transient expression assays, V12 was constitutively nuclear and negatively regulated IRF-5 transactivation of an alpha interferon promoter. The amount of V12 IRF-5 mRNA in LCL cells was 6.9-fold less than the sum of the other IRF-5 transcripts, as measured by quantitative RT-PCR (data not shown). However, the V12 protein is constitutively nuclear and by occupying IRF binding sites on responsive promoters could attenuate binding by IRF-5 entering the nucleus in response to signaling. A non-DNA binding, activation domain-only IRF-5 protein that acted as a functional negative mutant form in reporter assays has previously been described (47), as have splice variants of IRF-1 (41), IRF-2 (37), and IRF-3 (31, 36) that have altered functional properties.
We further observed that IRF-4 was consistently expressed in type III cell lines expressing IRF-5. IRF-4 negatively modulates IRF-5 activity after TLR stimulation by competing with IRF-5 for binding to MyD88 (51). EBV genes expressed in type III latency have been shown to upregulate IRF-4 (13, 61). Experiments with B-cell lines with conditional expression of EBNA2 or LMP1 revealed firstly that EBNA2 or EBNA2-regulated EBV genes were necessary for IRF-4 expression and secondly that LMP1 was one of the genes responsible for the induction. The combination of V12 and IRF-4 induction would provide negative regulation at both the IRF-5 protein activation and IRF-5 promoter upregulation steps. Further, since IRF-4 does not compete with IRF-7 binding to MyD88, the induction of IRF-4 could bias the downstream signal from TLR7 activation away from an IRF-5 gene expression signature and toward an IRF-7 signature.
The extent to which IRF-5 is integrated into the regulation of EBV gene expression remains to be investigated. The only reports to date have shown that IRF-5 heterodimerization with IRF-7 negatively affects IRF-7 activation of LMP1 expression (53) and that IRF-5 is able to downregulate a promoter for the BamHI-A rightward transcripts (14) that generate EBV-encoded microRNAs (54). The induction early after infection of TLR7 and the TLR7 downstream adaptor MyD88 and effector IRF-5, along with the effects of the TLR7 ligand R837 and the TLR7 inhibitor IRS 661 on the proliferation of EBV-infected naive B cells, indicates that EBV uses TLR7 signaling to promote the initial phase of B-cell activation and expansion. The induction of the negative regulators V12 IRF-5 and IRF-4 implies that subsequent modulation of the TLR7 pathway is necessary for successful establishment of a latent infection.
Acknowledgments
We thank B. Kempkes, W. Hammerschmidt, L. Hutt-Fletcher, and C. Farrell for the EREB2-5, 1852, Akata-BX1, and LCL cell lines, respectively, and R. Ambinder for help with virus inactivation.
This work was funded by Public Health Service grant R37 CA42245 to S.D.H.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499-511. [DOI] [PubMed] [Google Scholar]
- 2.Aman, P., and A. von Gabain. 1990. An Epstein-Barr virus immortalization associated gene segment interferes specifically with the IFN-induced anti-proliferative response in human B-lymphoid cell lines. EMBO J. 9:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, B. J., M. J. Kellum, A. E. Field, and P. M. Pitha. 2002. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol. Cell. Biol. 22:5721-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, B. J., M. J. Kellum, K. E. Pinder, J. A. Frisancho, and P. M. Pitha. 2003. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 63:6424-6431. [PubMed] [Google Scholar]
- 5.Barnes, B. J., P. A. Moore, and P. M. Pitha. 2001. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J. Biol. Chem. 276:23382-23390. [DOI] [PubMed] [Google Scholar]
- 6.Barrat, F. J., T. Meeker, J. Gregorio, J. H. Chan, S. Uematsu, S. Akira, B. Chang, O. Duramad, and R. L. Coffman. 2005. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 202:1131-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bekeredjian-Ding, I. B., M. Wagner, V. Hornung, T. Giese, M. Schnurr, S. Endres, and G. Hartmann. 2005. Plasmacytoid dendritic cells control TLR7 sensitivity of naive B cells via type I IFN. J. Immunol. 174:4043-4050. [DOI] [PubMed] [Google Scholar]
- 8.Bernasconi, N. L., N. Onai, and A. Lanzavecchia. 2003. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood 101:4500-4504. [DOI] [PubMed] [Google Scholar]
- 9.Bishop, G. A., Y. Hsing, B. S. Hostager, S. V. Jalukar, L. M. Ramirez, and M. A. Tomai. 2000. Molecular mechanisms of B lymphocyte activation by the immune response modifier R-848. J. Immunol. 165:5552-5557. [DOI] [PubMed] [Google Scholar]
- 10.Bishop, G. A., L. M. Ramirez, M. Baccam, L. K. Busch, L. K. Pederson, and M. A. Tomai. 2001. The immune response modifier resiquimod mimics CD40-induced B cell activation. Cell. Immunol. 208:9-17. [DOI] [PubMed] [Google Scholar]
- 11.Boehme, K. W., M. Guerrero, and T. Compton. 2006. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J. Immunol. 177:7094-7102. [DOI] [PubMed] [Google Scholar]
- 12.Bowie, A. G., and I. R. Haga. 2005. The role of Toll-like receptors in the host response to viruses. Mol. Immunol. 42:859-867. [DOI] [PubMed] [Google Scholar]
- 13.Cahir-McFarland, E. D., K. Carter, A. Rosenwald, J. M. Giltnane, S. E. Henrickson, L. M. Staudt, and E. Kieff. 2004. Role of NF-κB in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, H., J. Huang, F. Y. Wu, G. Liao, L. Hutt-Fletcher, and S. D. Hayward. 2005. Regulation of expression of the Epstein-Barr virus BamHI-A rightward transcripts. J. Virol. 79:1724-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng, T. F., S. Brzostek, O. Ando, S. Van Scoy, K. P. Kumar, and N. C. Reich. 2006. Differential activation of IFN regulatory factor (IRF)-3 and IRF-5 transcription factors during viral infection. J. Immunol. 176:7462-7470. [DOI] [PubMed] [Google Scholar]
- 16.Dawson, C. W., G. Tramountanis, A. G. Eliopoulos, and L. S. Young. 2003. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J. Biol. Chem. 278:3694-3704. [DOI] [PubMed] [Google Scholar]
- 17.Dunne, A., M. Ejdeback, P. L. Ludidi, L. A. O'Neill, and N. J. Gay. 2003. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J. Biol. Chem. 278:41443-41451. [DOI] [PubMed] [Google Scholar]
- 18.Eliopoulos, A. G., N. J. Gallagher, S. M. Blake, C. W. Dawson, and L. S. Young. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274:16085-16096. [DOI] [PubMed] [Google Scholar]
- 19.Finberg, R. W., J. P. Wang, and E. A. Kurt-Jones. 2007. Toll like receptors and viruses. Rev. Med. Virol. 17:35-43. [DOI] [PubMed] [Google Scholar]
- 20.Geiger, T. R., and J. M. Martin. 2006. The Epstein-Barr virus-encoded LMP-1 oncoprotein negatively affects Tyk2 phosphorylation and interferon signaling in human B cells. J. Virol. 80:11638-11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, R. R., S. V. Kozyrev, E. C. Baechler, M. V. Reddy, R. M. Plenge, J. W. Bauer, W. A. Ortmann, T. Koeuth, M. F. Gonzalez Escribano, B. Pons-Estel, M. Petri, M. Daly, P. K. Gregersen, J. Martin, D. Altshuler, T. W. Behrens, and M. E. Alarcon-Riquelme. 2006. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 38:550-555. [DOI] [PubMed] [Google Scholar]
- 23.Graham, R. R., C. Kyogoku, S. Sigurdsson, I. A. Vlasova, L. R. Davies, E. C. Baechler, R. M. Plenge, T. Koeuth, W. A. Ortmann, G. Hom, J. W. Bauer, C. Gillett, N. Burtt, D. S. Cunninghame Graham, R. Onofrio, M. Petri, I. Gunnarsson, E. Svenungsson, L. Ronnblom, G. Nordmark, P. K. Gregersen, K. Moser, P. M. Gaffney, L. A. Criswell, T. J. Vyse, A. C. Syvanen, P. R. Bohjanen, M. J. Daly, T. W. Behrens, and D. Altshuler. 2007. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. USA 104:6758-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda, K., and T. Taniguchi. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644-658. [DOI] [PubMed] [Google Scholar]
- 25.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 26.Hornung, V., S. Rothenfusser, S. Britsch, A. Krug, B. Jahrsdorfer, T. Giese, S. Endres, and G. Hartmann. 2002. Quantitative expression of Toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 168:4531-4537. [DOI] [PubMed] [Google Scholar]
- 27.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 28.Joseph, A. M., G. J. Babcock, and D. A. Thorley-Lawson. 2000. Cells expressing the Epstein-Barr virus growth program are present in and restricted to the naive B-cell subset of healthy tonsils. J. Virol. 74:9964-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanda, K., T. Decker, P. Aman, M. Wahlstrom, A. von Gabain, and B. Kallin. 1992. The EBNA2-related resistance towards alpha interferon (IFN-α) in Burkitt's lymphoma cells effects induction of IFN-induced genes but not the activation of transcription factor ISGF-3. Mol. Cell. Biol. 12:4930-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanda, K., B. Kempkes, G. W. Bornkamm, A. von Gabain, and T. Decker. 1999. The Epstein-Barr virus nuclear antigen 2 (EBNA2), a protein required for B lymphocyte immortalization, induces the synthesis of type I interferon in Burkitt's lymphoma cell lines. Biol. Chem. 380:213-221. [DOI] [PubMed] [Google Scholar]
- 31.Karpova, A. Y., P. M. Howley, and L. V. Ronco. 2000. Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes Dev. 14:2813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawai, T., and S. Akira. 2007. TLR signaling. Semin. Immunol. 19:24-32. [DOI] [PubMed] [Google Scholar]
- 33.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilger, E., A. Kieser, M. Baumann, and W. Hammerschmidt. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 17:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, T. Y., K. H. Lee, S. Chang, C. Chung, H. W. Lee, J. Yim, and T. K. Kim. 2003. Oncogenic potential of a dominant negative mutant of interferon regulatory factor 3. J. Biol. Chem. 278:15272-15278. [DOI] [PubMed] [Google Scholar]
- 37.Koenig Merediz, S. A., M. Schmidt, G. J. Hoppe, J. Alfken, D. Meraro, B. Z. Levi, A. Neubauer, and B. Wittig. 2000. Cloning of an interferon regulatory factor 2 isoform with different regulatory ability. Nucleic Acids Res. 28:4219-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieg, A. M., A. K. Yi, S. Matson, T. J. Waldschmidt, G. A. Bishop, R. Teasdale, G. A. Koretzky, and D. M. Klinman. 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546-549. [DOI] [PubMed] [Google Scholar]
- 39.Krug, A., G. D. Luker, W. Barchet, D. A. Leib, S. Akira, and M. Colonna. 2004. Herpes simplex virus type 1 activates murine natural interferon-producing cells through Toll-like receptor 9. Blood 103:1433-1437. [DOI] [PubMed] [Google Scholar]
- 40.Leadbetter, E. A., I. R. Rifkin, A. M. Hohlbaum, B. C. Beaudette, M. J. Shlomchik, and A. Marshak-Rothstein. 2002. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 416:603-607. [DOI] [PubMed] [Google Scholar]
- 41.Lee, E. J., M. Jo, J. Park, W. Zhang, and J. H. Lee. 2006. Alternative splicing variants of IRF-1 lacking exons 7, 8, and 9 in cervical cancer. Biochem. Biophys. Res. Commun. 347:882-888. [DOI] [PubMed] [Google Scholar]
- 42.Li, H., J. Zhang, A. Kumar, M. Zheng, S. S. Atherton, and F. S. Yu. 2006. Herpes simplex virus 1 infection induces the expression of proinflammatory cytokines, interferons and TLR7 in human corneal epithelial cells. Immunology 117:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, R., L. Yang, M. Arguello, C. Penafuerte, and J. Hiscott. 2005. A CRM1-dependent nuclear export pathway is involved in the regulation of IRF-5 subcellular localization. J. Biol. Chem. 280:3088-3095. [DOI] [PubMed] [Google Scholar]
- 45.Luftig, M., T. Yasui, V. Soni, M. S. Kang, N. Jacobson, E. Cahir-McFarland, B. Seed, and E. Kieff. 2004. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-κB activation. Proc. Natl. Acad. Sci. USA 101:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lund, J., A. Sato, S. Akira, R. Medzhitov, and A. Iwasaki. 2003. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 198:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mancl, M. E., G. Hu, N. Sangster-Guity, S. L. Olshalsky, K. Hoops, P. Fitzgerald-Bocarsly, P. M. Pitha, K. Pinder, and B. J. Barnes. 2005. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms: multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J. Biol. Chem. 280:21078-21090. [DOI] [PubMed] [Google Scholar]
- 48.Månsson, A., M. Adner, U. Hockerfelt, and L. O. Cardell. 2006. A distinct Toll-like receptor repertoire in human tonsillar B cells, directly activated by PamCSK, R-837 and CpG-2006 stimulation. Immunology 118:539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moynagh, P. N. 2005. TLR signalling and activation of IRFs: revisiting old friends from the NF-κB pathway. Trends Immunol. 26:469-476. [DOI] [PubMed] [Google Scholar]
- 50.Nanbo, A., K. Inoue, K. Adachi-Takasawa, and K. Takada. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 21:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Negishi, H., Y. Ohba, H. Yanai, A. Takaoka, K. Honma, K. Yui, T. Matsuyama, T. Taniguchi, and K. Honda. 2005. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. USA 102:15989-15994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ning, S., A. M. Hahn, L. E. Huye, and J. S. Pagano. 2003. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. J. Virol. 77:9359-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ning, S., L. E. Huye, and J. S. Pagano. 2005. Interferon regulatory factor 5 represses expression of the Epstein-Barr virus oncoprotein LMP1: braking of the IRF7/LMP1 regulatory circuit. J. Virol. 79:11671-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 55.Portis, T., and R. Longnecker. 2004. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene 23:8619-8628. [DOI] [PubMed] [Google Scholar]
- 56.Ruprecht, C. R., and A. Lanzavecchia. 2006. Toll-like receptor stimulation as a third signal required for activation of human naive B cells. Eur. J. Immunol. 36:810-816. [DOI] [PubMed] [Google Scholar]
- 57.Samanta, M., D. Iwakiri, T. Kanda, T. Imaizumi, and K. Takada. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 25:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoenemeyer, A., B. J. Barnes, M. E. Mancl, E. Latz, N. Goutagny, P. M. Pitha, K. A. Fitzgerald, and D. T. Golenbock. 2005. The interferon regulatory factor, IRF5, is a central mediator of Toll-like receptor 7 signaling. J. Biol. Chem. 280:17005-17012. [DOI] [PubMed] [Google Scholar]
- 59.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Speck, P., K. A. Kline, P. Cheresh, and R. Longnecker. 1999. Epstein-Barr virus lacking latent membrane protein 2 immortalizes B cells with efficiency indistinguishable from that of wild-type virus. J. Gen. Virol. 80:2193-2203. [DOI] [PubMed] [Google Scholar]
- 61.Spender, L. C., W. Lucchesi, G. Bodelon, A. Bilancio, C. E. Karstegl, T. Asano, O. Dittrich-Breiholz, M. Kracht, B. Vanhaesebroeck, and P. J. Farrell. 2006. Cell target genes of Epstein-Barr virus transcription factor EBNA-2: induction of the p55alpha regulatory subunit of PI3-kinase and its role in survival of EREB2.5 cells. J. Gen. Virol. 87:2859-2867. [DOI] [PubMed] [Google Scholar]
- 62.Sugano, N., W. Chen, M. L. Roberts, and N. R. Cooper. 1997. Epstein-Barr virus binding to CD21 activates the initial viral promoter via NF-κB induction. J. Exp. Med. 186:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swanson-Mungerson, M., R. Bultema, and R. Longnecker. 2006. Epstein-Barr virus LMP2A enhances B-cell responses in vivo and in vitro. J. Virol. 80:6764-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tabeta, K., P. Georgel, E. Janssen, X. Du, K. Hoebe, K. Crozat, S. Mudd, L. Shamel, S. Sovath, J. Goode, L. Alexopoulou, R. A. Flavell, and B. Beutler. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 101:3516-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takaoka, A., H. Yanai, S. Kondo, G. Duncan, H. Negishi, T. Mizutani, S. Kano, K. Honda, Y. Ohba, T. W. Mak, and T. Taniguchi. 2005. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434:243-249. [DOI] [PubMed] [Google Scholar]
- 66.Takeshita, F., K. Suzuki, S. Sasaki, N. Ishii, D. M. Klinman, and K. J. Ishii. 2004. Transcriptional regulation of the human TLR9 gene. J. Immunol. 173:2552-2561. [DOI] [PubMed] [Google Scholar]
- 67.Thorley-Lawson, D. A., and A. Gross. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350:1328-1337. [DOI] [PubMed] [Google Scholar]
- 68.Tomai, M. A., L. M. Imbertson, T. L. Stanczak, L. T. Tygrett, and T. J. Waldschmidt. 2000. The immune response modifiers imiquimod and R-848 are potent activators of B lymphocytes. Cell. Immunol. 203:55-65. [DOI] [PubMed] [Google Scholar]
- 69.Uchida, J., T. Yasui, Y. Takaoka-Shichijo, M. Muraoka, W. Kulwichit, N. Raab-Traub, and H. Kikutani. 1999. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 286:300-303. [DOI] [PubMed] [Google Scholar]
- 70.Wang, H., M. W. Nicholas, K. L. Conway, P. Sen, R. Diz, R. M. Tisch, and S. H. Clarke. 2006. EBV latent membrane protein 2A induces autoreactive B cell activation and TLR hypersensitivity. J. Immunol. 177:2793-2802. [DOI] [PubMed] [Google Scholar]
- 71.Wang, J., Y. Shao, T. A. Bennett, R. A. Shankar, P. D. Wightman, and L. G. Reddy. 2006. The functional effects of physical interactions among Toll-like receptors 7, 8, and 9. J. Biol. Chem. 281:37427-37434. [DOI] [PubMed] [Google Scholar]
- 72.Yanai, H., H. M. Chen, T. Inuzuka, S. Kondo, T. W. Mak, A. Takaoka, K. Honda, and T. Taniguchi. 2007. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc. Natl. Acad. Sci. USA 104:3402-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yasuda, K., C. Richez, J. W. Maciaszek, N. Agrawal, S. Akira, A. Marshak-Rothstein, and I. R. Rifkin. 2007. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF) 5 and IRF7 dependent and is required for IL-6 production. J. Immunol. 178:6876-6885. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, J., S. C. Das, C. Kotalik, A. K. Pattnaik, and L. Zhang. 2004. The latent membrane protein 1 of Epstein-Barr virus establishes an antiviral state via induction of interferon-stimulated genes. J. Biol. Chem. 279:46335-46342. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]