Abstract
Human immunodeficiency virus type 1 (HIV-1) transcription is subject to substantial fluctuation during the viral life cycle. Due to the low frequencies of HIV-1-infected cells, and because latently and productively infected cells collocate in vivo, little quantitative knowledge has been attained about the range of in vivo HIV-1 transcription in peripheral blood mononuclear cells (PBMC). By combining cell sorting, terminal dilution of intact cells, and highly sensitive, patient-specific PCR assays, we divided PBMC obtained from HIV-1-infected patients according to their degree of viral transcription activity and their cellular phenotype. Regardless of a patient's treatment status, the bulk of infected cells exhibited a CD4+ phenotype but transcribed HIV-1 provirus at low levels, presumably insufficient for virion production. Furthermore, the expression of activation markers on the surface of these CD4+ T lymphocytes showed little or no association with enhancement of viral transcription. In contrast, HIV-infected T lymphocytes of a CD4−/CD8− phenotype, occurring exclusively in untreated patients, exhibited elevated viral transcription rates. This cell type harbored a substantial proportion of all HIV RNA+ cells and intracellular viral RNAs and the majority of cell-associated virus particles. In conjunction with the observation that the HIV quasispecies in CD4+ and CD4−/CD8− T cells were phylogenetically closely related, these findings provide evidence that CD4 expression is downmodulated during the transition to productive infection in vivo. The abundance of viral RNA in CD4−/CD8− T cells from viremic patients and the almost complete absence of viral DNA and RNA in this cell type during antiretroviral treatment identify HIV+ CD4−/CD8 T cells as the major cell type harboring productive infection in peripheral blood.
The longevity of latently infected resting memory T-helper lymphocytes is widely perceived as the major obstacle to the eradication of human immunodeficiency virus type 1 (HIV-1) by current antiretroviral therapy (ART) (4, 10, 16, 17, 73). In addition, ongoing viral replication and reactivation of latently infected cells may cause continuous replenishment of HIV-1 reservoirs (12, 27, 64). To assess the contributions of productively infected cells and latently HIV-infected cells to viral persistence in different subsets of peripheral blood mononuclear cells (PBMC), we dissected HIV-infected cells in peripheral blood according to their degree of viral-transcription activity and their cellular phenotype.
Although dendritic cells (35), natural killer cells (68), and B cells (14, 45) have also been reported to harbor infectious provirus, T-helper lymphocytes play a pivotal role as sites of both HIV-1 production and viral persistence in PBMC. Resting memory CD4+ T cells, the predominantly infected cellular subset (5), represent the major latent reservoir harboring replication-competent provirus (13, 17, 73), whereas activated CD4+ T cells are the main targets for HIV-1 infection and show enhanced levels of viral gene expression in lymph nodes (77). CD4−/CD8− T cells, a relatively small CD3+ T-cell population of heterogeneous lineage and function (65), have been reported to harbor HIV RNA and replication-competent provirus (8, 42). Although certain HIV-1 isolates may infect T cells in a CD4-independent manner (51), it appears plausible that the HIV-1-expressing fraction of CD4−/CD8− T cells descend from CD4+ T cells by CD4 down-modulation. In vitro, this has been demonstrated to be the concerted action of the viral proteins Vpu, Env, and, mainly, Nef (7, 37, 53, 58). CD8+ T cells may be susceptible to HIV-1 infection (23, 51), but they probably constitute a viral reservoir of minor importance (5).
A further PBMC subset susceptible to HIV-1 are CD14+ peripheral blood monocytes (31, 44), which were reported to harbor replication-competent virus in patients on successful ART (38, 61, 79) even though they may be greatly resistant to in vitro infection (60).
Among HIV-infected PBMC from individuals on ART, only a minor fraction can be induced to release replication-competent virus (10), a majority of viral genomes being defective or permanently silenced (28, 54). Nevertheless, unspliced (usRNA), as well as low levels of multiply spliced (msRNA), HIV-1 RNAs can be detected consistently in the circulating PBMC of successfully treated individuals (18, 19, 25, 76). It is still under debate whether such HIV RNAs mirror viral replication, eventually resulting in replenishment of HIV-1 reservoirs, or whether they reflect residual transcription of latent provirus (2, 11, 12, 19, 29, 66). The latter view is supported by the finding that effective ART results in the partial reduction of intracellular viral transcripts but prompts exhaustive depletion of HIV RNA enclosed in cell-associated viral particles (vRNAex). We have previously shown that the presence of such virions, presumably residing at the cell surface, marks predominantly autochthonous virion production and is therefore a marker for ongoing productive infection (19, 20, 22). In contrast, latently infected cells were devoid of virion-enclosed HIV RNA (20, 21, 22), but there is ample evidence that latently infected CD4+ T cells may detain usRNA and msRNA (19, 39, 40). Interestingly, the levels of multiply spliced, Nef-encoding mRNA in patients on therapy have been shown to predict the extent of rebounding viral replication in PBMC during structured treatment interruption, while HIV usRNA levels did not (19). This suggested that at least two classes of HIV RNA+ PBMC may be present in patients undergoing ART, one class expressing usRNA only and a second category of cells containing msRNA species in addition to usRNA. Furthermore, single-cell analyses and limiting dilution data indicate that such low-expressing cells also constitute the majority of HIV RNA+ cells in lymphocytes from untreated individuals, whereas productively infected cells are much rarer (22, 28).
The present study was designed to assess virological activity in different PBMC subpopulations by combining patient-matched real-time PCR assays for HIV usRNA and msRNA with limiting-dilution analysis of intact cells. This novel approach provides insights into HIV transcription on a single-cell level and allows the differentiation of productive infection and basal transcription in latently infected cells.
MATERIALS AND METHODS
Patients and specimens.
Twelve chronically HIV-1-infected patients followed in our infectious disease outpatient clinic were enrolled. Six patients were on long-term combination ART and six had never been on treatment or had been off treatment for at least 8 months. All patients provided written informed consent, according to the guidelines of the ethics committee of the University Hospital of Zürich. For the inclusion of untreated patients, a CD4 count of >150 cells per μl and a plasma HIV RNA level of >50,000 copies per ml was required. Treated patients showing HIV-1 RNA levels below 50 copies/ml during the 6 months preceding sampling were selected. However, patient 04 exhibited a short time period of intermittent viremia on the day of sampling. Patient and virological characteristics are listed in Table 1. In the first series of sampling, cells obtained from 150 ml venous blood were sorted according to protocol 1 (see below). Since analyses revealed that none of the collected PBMC subsets were likely to include productively infected cells at substantial frequencies, additional samples of PBMC, CD4+ T cells, and CD4−/CD8− T cells (protocol 2) were collected from the blood of nine patients disposed to repeated blood donation. Because the patients had been in stable condition (patients on ART were receiving unaltered drug regimens and untreated patients had no clinical events calling for the initiation of ART) since previous sampling and no evidence for substantial changes in plasma HIV RNA levels (P = 1.0) and CD4 counts (P ≥ 0.605) was observed, data sets were subsequently pooled to augment their statistical power (Table 1).
TABLE 1.
Patient characteristics
| Group | Patient | Age (yr) | Gendera | Antiretroviral treatmentb | Plasma viremia (RNA copies/ml) in sample:
|
Absolute CD4 count (%) in sample:
|
Cell fractions collectedc:
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 2 | 1 | 2 | |||||
| A | 03 | 42 | m | d4T, NFV, 3TC | <6 | 956 (35) | 1, 2, 5 | |||
| 04 | 48 | m | ABC, d4T, LPV/r | 2280 | 90d | 372 (13) | 430 (16)d | 1-5 | 1, 2, 6 | |
| 07 | 52 | m | AZT, 3TC, RTV | <8 | <2d | 587 (29) | 444 (27)d | 1-5 | 1, 2, 6 | |
| 08 | 50 | m | AZT, 3TC, LPV/r | <12 | 575 (32) | 1-5 | ||||
| 13 | 42 | f | TDF, FTC, ATV/r | <1 | 48 | 437 (25) | 523 (29) | 1-5 | 1, 2, 6 | |
| 14 | 33 | f | ddI, TDF, NVP | <2 | <2 | 426 (26) | 236 (28)d | 1-5 | 1, 2, 6 | |
| B | 05 | 37 | m | 76,300 | 16,900d | 358 (29) | 429 (27)d | 1-5 | 1, 2, 6 | |
| 06 | 38 | m | 62,000 | 83,000d | 423 (24) | 249 (15)d | 1-5 | 1, 2, 6 | ||
| 10 | 34 | m | 121,000 | 101,000d | 474 (24) | 433 (22) | 1-5 | 1, 2, 6 | ||
| 11 | 73 | f | 1,225,000 | 177 (9) | 1-5 | 1, 2, 6 | ||||
| 15 | 45 | m | 66,500 | 60,500d | 295 (13) | 266 (14)d | 1-5 | |||
| 16 | 33 | m | 92,800 | 111,000d | 403 (38) | 328 (37)d | 1-5 | 1, 2, 6 | ||
m, male; f, female.
Abbreviations used for antiretroviral drugs: d4T, stavudine; NFV, nelfinavir; 3TC, lamivudine; ABC, abacavir; LPV/r, lopinavir/ritonavir; AZT, zidovudine; RTV, ritonavir; TDF, tenofovir; FTC, emtricitabine; ATV/r, atazanavir-ritonavir; ddI, didanosine; NVP, nevirapine.
1, PBMC; 2, CD4+ T cells; 3, resting CD4+ T cells; 4, activated CD4+ T cells; 5, monocytes; 6, CD4−/CD8− T cells.
Value extrapolated from closely adjacent time points.
Cell sorting and storage of samples.
PBMC were isolated from anticoagulated blood by Ficoll gradient purification and washed three times in phosphate-buffered saline. Resting (CD25−, CD69−, and HLA-DR−) CD4+ T cells, activated (CD25+, CD69+, or HLA-DR+) CD4+ T cells, αβ T-cell receptor (TCR)-expressing double-negative T cells, and peripheral blood monocytes were isolated by magnetic cell sorting and sampled in replicate dilution series. The cells were purified according to protocol 1 (first sampling) or protocol 2 (second sampling). Magnetic cell sorting was performed using monoclonal mouse antihuman antibodies (MAbs) coupled to colloidal paramagnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). Except for the modifications indicated below, the labeling and separation of cells was carried out using type LS positive selection columns (Miltenyi) according to the supplier's instructions. The relative proportions of the PBMC subpopulations, as well as the purity of the sorted fractions, were assessed by flow cytometry (FACSCalibur system; BD Biosciences, San Jose, CA) on unfixed cells labeled with MAbs against surface lineage markers (CD3, -4, -8, and -14) and activation markers (CD25, CD-69, and HLA-DR) coupled to fluorescein isothiocyanate, phycoerythrin (PE), or phycoerythrin-cyanin 5 fluorescent dye (all conjugates were obtained from Caltag Laboratories, Burlingame, CA, except CD25-PE, which was from Sigma, St. Louis, MO).
Protocol 1.
CD4+ T cells were isolated by using anti-CD4 MAbs coupled to detachable paramagnetic beads (CD4 MultiSort kit; Miltenyi Biotec). After removal of the paramagnetic beads, the cells were stained with PE-labeled anti-CD25, -CD69, or -HLA-DR. Subsequently, the cells were sorted according to activation marker expression by means of paramagnetic beads coupled to a secondary antibody specific for an epitope on PE. To enhance the specificity of the detection of the cell fractions, resting CD4+ T cells were negatively selected by using depletion columns (type LD), followed by a second step of positive selection for activation marker-positive cells. Monocytes were isolated by using a two-step procedure including a negative selection step (monocyte isolation kit II; Miltenyi Biotec) followed by a positive selection step using CD14 MicroBeads.
The proportions of the cellular subsets (Table 2) and the purity of the fractions (Table 3) were assessed by flow cytometry.
TABLE 2.
Percentage of cell subsets in total PBMC
| Group | Patient | % in PBMC of:
|
|||||
|---|---|---|---|---|---|---|---|
| CD4+ T cells in first sampling set
|
Total CD4+ T cells in second sampling set | CD4−/CD8− CD3+ cells in second sampling seta | Monocytes (CD14+) in first sampling set | ||||
| Total | CD25−, CD69−, HLA-DR− | CD25+, CD69+, HLA-DR+ | |||||
| A | 03 | 30 | 15 | 15 | 8 | ||
| 04 | 13 | 8 | 5 | 10 | 12 | 10 | |
| 07 | 23 | 6 | 16 | 27 | 3 | 19 | |
| 08 | 26 | 13 | 13 | 2 | 18 | ||
| 13 | 19 | 10 | 9 | 32 | 3 | 26 | |
| 14 | 24 | 8 | 16 | 22 | 3 | 19 | |
| Median | 23 | 9 | 14 | 25 | 3 | 19 | |
| B | 05 | 24 | 12 | 12 | 20 | 7 | 18 |
| 06 | 26 | 17 | 9 | 19 | 17 | 12 | |
| 10 | 28 | 17 | 11 | 22 | 4 | 13 | |
| 11 | 11 | 5 | 6 | 20 | |||
| 15 | 13 | 5 | 8 | 15 | 6 | 21 | |
| 16 | 26 | 14 | 12 | 34 | 10 | 22 | |
| Median | 25 | 13 | 10 | 20 | 7 | 19 | |
αβ TCR+ CD4−/CD8− T cells were assumed to represent 50% of total CD4−/CD8− T cells (45).
TABLE 3.
Purities of cell fractions as analyzed by flow cytometry
| Group | Patient | Purity (%)a of CD4+ T cells in sample:
|
Resting CD4+ T cells
|
Activated CD4+ T cells
|
CD4−/CD8− T cells
|
Monocytes
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | Purity (%)a | Activated CD4+ T cells (%)b | Purity (%)a | Resting CD4+ T cells (%)b | Purity (%)a | CD4+ T cells (%)b | Purity (%)a | CD4+ | ||
| A | 03 | 99.2 | 90.0 | 0.0 | |||||||
| 04 | 99.4 | 98.4 | 89.9 | 5.9 | 79.8 | 13.6 | 97.8 | 0.1 | 83.7 | 0.9 | |
| 07 | 99.7 | 99.3 | 91.1 | 5.0 | 95.2 | 4.0 | 99.0 | 0.1 | 96.3 | 0.2 | |
| 08 | 99.7 | 92.7 | 6.4 | 96.0 | 3.6 | 97.3 | 0.2 | ||||
| 13 | 99.6 | 99.0 | 85.0 | 13.9 | 93.3 | 6.4 | 94.4 | 0.0 | 97.0 | 0.1 | |
| 14 | 99.5 | 96.2 | 89.5 | 7.00 | 92.7 | 5.9 | 35.2 | 0.1 | 99.1 | 0.01 | |
| B | 05 | 99.1 | 98.6 | 91.5 | 3.2 | 95.4 | 3.9 | 98.6 | 0.1 | 71.6 | 3.9 |
| 06 | 99.4 | 99.3 | 96.5 | 3.7 | 94.6 | 4.5 | 93.4 | 0.3 | 67.4 | 3.5 | |
| 10 | 99.5 | 99.4 | 96.9 | 2.3 | 91.2 | 7.8 | 59.4 | 0.4 | 96.9 | 0.1 | |
| 11 | 98.6 | 82.0 | 10.3 | 88.7 | 4.6 | 92.8 | 0.2 | ||||
| 15 | 97.3 | 97.7 | 74.5 | 10.0 | 86.3 | 12.5 | 68.0 | 2.7 | 98.0 | 0.1 | |
| 16 | 99.5 | 96.4 | 90.3 | 8.7 | 85.8 | 13.8 | 81.3 | 0.1 | 99.0 | 0.0 | |
| Median | 99.5 | 98.6 | 90.3 | 6.4 | 92.7 | 5.9 | 93.4 | 0.1 | 96.6 | 0.2 | |
Percentage of living cells.
Levels of cross contamination with relevant cell subsets.
Protocol 2.
CD4+ and CD8+ T cells were isolated using CD4 or CD8 MicroBeads, respectively. Double-negative T cells positive for αβ TCR were enriched by magnetic depletion using a cocktail of biotin-conjugated antibodies against CD8, CD14, CD16, CD19, CD36, CD56, CD123, γ/δ TCR, and CD235a subsequently linked to streptavidin-coupled paramagnetic beads (CD4+ T-cell isolation kit II). To remove residual CD4+ cells, the antibodies were supplemented with additional CD4 MicroBeads. Finally, CD4−/CD8− T cells were purified by using fluorescence-activated cell sorting (FACS) (MoFlo high perfomance sorter; Dako, Glostrup, Denmark) of CD3+ CD4−/CD8− live cells or by using CD3 MicroBeads when FACS was precluded for technical reasons. For calculating the frequencies of CD4−/CD8− T cells in total PBMC, the proportion of αβ TCR-expressing cells to total CD4−/CD8− T cells was assumed to be approximately 50% (43).
Limiting dilution of patients' cells.
Total undiluted (D0) PBMC and purified cell subsets were counted in a COULTER Ac·T diff analyzer (Beckman Coulter, Nyon, Switzerland) and aliquots (n = 8 to 16) ranging from 50,000 (CD4−/CD8− T cells) to 1.5 million (monocytes) cells per tube were prepared. Further serial 5-fold dilutions of patients' cells (n = 8 to 16 per dilution step; D1, D2, D3, D4, and D5 were diluted 5-, 25-, 125-, 625-, and 3,125-fold, respectively) were prepared by mixing patients' cells with PBMC (105 cells per aliquot) from healthy uninfected donors (blood bank of the Swiss Red Cross, Zurich, Switzerland) aliquoted in replicates (n = 8 to 16), in 1- to 5-fold-dilution series. The cells were stored at −80°C as dry pellets.
Extraction of HIV nucleic acids.
For DNA PCR, cells were lysed in 150 μl cell lysis buffer (9) (kindly provided by Roche Molecular Diagnostics) supplemented with 0.5% QIAGEN protease (QIAGEN) for 60 min at 60°C, followed by 60 min of incubation at 95°C. Total cellular RNA was extracted with a MagNA Pure automatic preparation device and a high-performance extraction kit (Roche, Rotkreuz, Switzerland) using external cell lysis in 800 μl lysis buffer and elution of RNA in a final volume of 0.1 ml.
Selective isolation of extracellular, vRNAex associated with PBMC was performed as previously described (19, 20, 22, 33). The procedure included the disintegration of cells by repeated freezing and thawing followed by nuclease and protease digestion of intracellular RNAs and relied on the physical resistance of small viral envelopes to this treatment (22, 30, 33). In brief, pellets were thawed and resuspended in 130 μl phosphate-buffered saline containing 10 mM Tris HCl, pH 7.5, 1 mM MgCl2, 1 mg/ml DNase I (Roche), and 1.8 mg/ml RNase A (QIAGEN). The samples underwent two cycles of freezing on dry ice and thawing at 37°C, followed by incubation for 60 min at 37°C under constant agitation. To inactivate the nucleases, protease (10 μl, 50 mg/ml; QIAGEN) was added and incubation continued for 45 min. Finally, viral RNA was recovered by automatic extraction as described above.
Adjustment of patient-matched PCR primers.
The HIV primers used in the PCR assays were individually adjusted to each patient's predominant PBMC quasispecies. Total cellular RNA, or in cases when viral cDNAs could not be amplified, total DNA, was extracted from each patient's sample. For primer identification in genomic HIV-1 usRNA and HIV-1 DNA, a stretch of sequences in the gag gene was scanned for amplifiable sequences (positions 1273 to 1658; nucleotide numbers are in respect to the HXB2 genome, as calculated by the SEQUENCE LOCATOR software provided by the Los Alamos HIV database, www.hiv.lanl.gov). For the identification of primer sites for the amplification of HIV-1 msRNA, spliced cDNA (spanning nucleotides 626 to 743, 5778 to 6045, and 8379 to 9096) was scanned. Alternatively, when msRNA levels were insufficient for the amplification of spliced cDNA, usRNAs in exon 4/5 (positions 5889 to 6252) in the env and in exon 7 (positions 8079 to 9096) in the gp41/nef coding regions were assessed. PCR-based sequence scanning was performed by a nested approach using batteries of amplification primers with highly conserved 3′ ends (Table 4) as follows. In a first round, sequences ranging from 360 to 1,020 bases in length were amplified by a one-tube, two-phase reverse transcription (RT)-PCR approach as described earlier (33) using 1 to 10 separate or combined primer combinations per patient. In a second step, pools of 1 to 5 first-round amplification products were further amplified by nested DNA PCR, utilizing HotStarTaq polymerase (QIAGEN) and 1 to 20 separate combinations of inner primers unused in the previous RT-PCR. Genuine PCR products were identified based on their mobility in agarose gel electrophoresis, gel purified, and sequenced. The primers used for real-time PCR quantification were adjusted to each patient's isolate (Table 5). To allow for reliable quantification, synthetic RNA and DNA standards differing significantly in their primer binding sites (PCR primers with mismatches proximal to their 3′ ends and fluorescent probes with mismatches proximal to their 3′ and 5′ ends) were prepared for patients' isolates. For this purpose, the PCR products were tagged with a T7 promoter sequence and transcribed in vitro (33). For DNA quantitation, purified PCR products were used directly as external quantification standards in a range of 2 to 106 copies/PCR. For RNA quantification in vitro, transcribed RNAs (33) were used as external standards in a range of 2 to 106 copies/PCR.
TABLE 4.
PCR primers for isolation of patient-specific RT-PCR primer binding sites
| Sequence target(s) | Primer (reference) | Geneb | Strandc | Positiond
|
Sequencee | |
|---|---|---|---|---|---|---|
| 5′ | 3′ | |||||
| usRNA, vRNAex | Mf219 | gag | + | 1273 | 1304 | AAGGCTTTCAGCCCAGAAGTAATACCCATGTT |
| Mf06 | gag | + | 1296 | 1325 | ACCCATGTTTTCAGCATTATCAGAAGGAGC | |
| Mf118 | gag | + | 1309 | 1332 | GCATTATCAGAAGGAGCCACCCCA | |
| Mf119 | gag | + | 1326 | 1354 | CACCCCACAAGATTTAAACACCATGCTAA | |
| Mf117 (46) | gag | − | 1571 | 1544 | ATTTCTCCTACTGGGATAGGTGGATTAT | |
| Mf218 | gag | − | 1623 | 1597 | GCTATACATTCTTACTATTTTATTTAA | |
| Sk39 (46) | gag | − | 1658 | 1631 | TTTGGTCCTTGTCTTATGTCCAGAATGC | |
| Spliced msRNA | CR5 (70) | U5/PBS | + | 626 | 648 | TCTCTAGCAGTGGCGCCCGAACA |
| Mf104 | psi/env | + | 721 | 5780 | CAAGAGGCGAGGGGAGGCGACTGAAT | |
| Mf103 | vpr/tat | + | 5833 | 5862 | GGAGCCAGTAGATCCTAGACTAGAGCCCTG | |
| Pr1 (71) | tat | + | 5853 | 5877 | TAGAGCCCTGGAAGCATCCAGGAAG | |
| Mf221 | tat | + | 5889 | 5914 | CTGCTTGTAACAAGTGTTATTGTAAA | |
| Mf236 | tat | + | 5907 | 5931 | ATTGTAAAAAGTGTTGCTTTCATTG | |
| msRNA exon4/5a | Mf238 | vpu | − | 6117 | 6087 | GCTATTATTGCTGCTACTACTAATGCTACTA |
| Mf239 | vpu | − | 6152 | 6120 | TATATTCTATGAATACTATGGACCACACAACTA | |
| Mf240 | vpu | − | 6201 | 6167 | CTTATTCTATCAATTAACCTGTCTATTTTTCTTTG | |
| Mf237 | vpu | − | 6251 | 6225 | ATTCTTCCTGATCCCCTTCACTCTCAT | |
| msRNA exon7a | M159 (50) | gp41 | + | 8079 | 8106 | CTGGATGAGATTTGGGATAACATGACCT |
| Mf161 | gp41 | + | 8121 | 8152 | AGAGAAATTGACAATTACACAAGCTTAATATA | |
| Mf160 | gp41 | + | 8148 | 8172 | ATATACACCTTAATTGAAGAATCGC | |
| Mf214 | gp41 | + | 8317 | 8349 | TTTTTGCTGTACTTTCTATAGTGAATAGAGTTA | |
| Mf222 | gp41 | + | 8350 | 8379 | GGCAGGGATATTCACCATTATCGTTTCAGA | |
| Mf224 | gp41 | + | 8361 | 8388 | TCACCATTATCGTTTCAGACCCACCTCC | |
| Spliced msRNA | Mf223 | gp41 | − | 8517 | 8493 | GGCACAGGCTCCGCAGATCGTCCCA |
| Mf220 | gp41 | − | 8533 | 8510 | CGGTGGTAGCTGAAGAGGCACAGG | |
| Mf215 | gp41 | − | 8552 | 8526 | CAAGAGTAAGTCTCTCAAGCGGTGGTA | |
| Mf156 (50) | gp41 | − | 8567 | 8542 | AATCCTCGTTACAATCAAGAGTAAGT | |
| Mf116 | gp41 | − | 8619 | 8589 | GATTCCACCAATATTTGAGGGCTTCCCACCC | |
| Mf121 | gp41 | − | 8681 | 8652 | GGCTGTGGCATTGAGCAAGCTAACAGCACT | |
| Mf105 | gp41 | − | 8706 | 8682 | TATCTGTCCCCTCAGCTACTGCTAT | |
| mascM (24) | nef | − | 9096 | 9068 | TAGCCCTTCCAGTCCCCCCTTTTCTTTTA | |
Exon numbering of spliced RNA is according to the work of Schwartz et al. (58).
Genes and landmarks of the HIV-1 genome are given as compiled in the Los Alamos HIV database (www.hiv.lanl.gov).
+, coding strand of the proviral genome; −, noncoding strand.
Numbering on the Hxb2 genome according to the software provided by the Los Alamos HIV database (SEQUENCE LOCATOR; www.hiv.lanl.gov).
Sequences are given 5′ to 3′ from left to right.
TABLE 5.
PCR primers used for patient-specific real-time PCR
| PCR target | Primer (reference)a | Functionb | Strandc | Positiond
|
Sequencee | Patient(s)f | |
|---|---|---|---|---|---|---|---|
| 5′ | 3′ | ||||||
| usRNA vRNAex | Ts5gag (3) | Sense | + | 1372 | 1397 | CAAGCAGCCATGCAAATGTTAAAAGA | 03, 04, 06-08, 13-16 |
| HIV-DNA | CAAGCAGCTATGCAGATGTTAAAAGA | 05 | |||||
| CAGGCAGCTATGCAAATGTTAAAAGA | 10 | ||||||
| CAGGCAGCTATGCAAATGTTAAAAGA | 11 | ||||||
| Boe3tq (3) | Probe | − | 1430 | 1405 | f-CTATCCCATTCTGCAGCTTCCTCATT-q | 03-08, 13, 15, 16 | |
| f-CCTGTCCCATTCTGCAGCTTCCTCATT-q | 10 | ||||||
| f-CCTGTCCCATTCTGCAGCTTCCTCATT-q | 11 | ||||||
| f-CCTATCCCATTCTGCAGCCTCCTCATT-q | 14 | ||||||
| Boe2 (3) | Antisense | − | 1488 | 1467 | TCCCCTTGGTTCTCTCATCTGG | 03, 05-08, 13 | |
| TCCCCTTGGTTCTCTCATCTGA | 04 | ||||||
| TCCCCTTGGTTCTCTCATCTGA | 10 | ||||||
| TCCCCTTGGTTCCCTTATCTGG | 11 | ||||||
| Skcc1b (46) | Antisense | − | 1513 | 1486 | TACTAGTAGTTCCTGCTATGTCACTTCC | 14-16 | |
| msRNA | Ts5allspl (19) | Sense | + | 5978 | 6001 | AAGAAGCGGAGACAGCGACGAAGA | 04-07 |
| AAGAAGCGAAGACAGCGACGAAGA | 03 | ||||||
| Mf84 (19, 50) | Sense | + | 6012 | 6045 | ACAGTCAGACTCATCAAGTTTCTCTATCAAAGCA | ||
| ACAGTAAGACTCATCAAGCTTCTCTATCAAAGCA | 08 | ||||||
| GCAGTGAAGATCATCAAAATCCTATATCAAAGCA | 10 | ||||||
| GCAATAAGGATCATCAAAATCCTATA | 11 | ||||||
| ACCGTCAGACTAATCAAGATTCTCTATCAAAGCA | 13 | ||||||
| GCAGTGAGGATCATCAAAATCCTATATCAAAGCA | 14 | ||||||
| ACAATCAGACTCATCAGGCTTCTCTATCAAAACA | 15 | ||||||
| ACAGTCAGATTCATCAAGTTTCTCTATCAAAGCA | 16 | ||||||
| mf2tq (19, 52) | Probe | − | 8421 | 8399 | f-TTCCTTCGGGCCTGTCGGGTCCC-q | 04, 07 | |
| f-TTCCTCCGGGCCTGTCGGGTTCCC-q | 03 | ||||||
| f-TTCCTCCGGGCCTGTCGGGTCCC-q | 05 | ||||||
| f-TTCCTTCGGGCCCGTCGGGTCCC-q | 06 | ||||||
| Mf226tq | Probe | + | 8397 | 8414 | f-AGGGGACCCGACAGGCCC-q | 08, 13, 15, 16 | |
| f-AGGGGTCTCGACAGGCCC-q | 10 | ||||||
| f-AGGGATCTCGACAGGCCC-q | 11 | ||||||
| f-AGGGGACCCGACAGGCTC-q | 14 | ||||||
| Mf83 (19, 50) | Antisense | − | 8459 | 8433 | GGATCTGTCTCTGTCTCTCTCTCCACC | 03, 06, 07, 13 | |
| GGATCTGCCGCTGTCTCTCTCTCCACC | 04 | ||||||
| GGATCTGTCTCTGTCTCTTTCTCCACC | 05 | ||||||
| TGATGTGTCTCTGTCTCTCTCTCCACC | 08 | ||||||
| TGATCTGTCTCTGTCTTGCTCTCCACC | 10 | ||||||
| CGATCTGCTTCTGTCTTGCTCTCCACC | 11 | ||||||
| GGATCTGTCTCTGCCTTGCTCTCCACC | 14 | ||||||
| GGATCTGTCTCTGTCTCTGTCCCCACC | 15 | ||||||
| GGATGTGTCTCTGTCTCTCTCTCCACC | 16 | ||||||
Primers are denominated according to internal nomenclature in the authors' laboratory and not necessarily by previously used designations in the indicated references.
Functions of oligonucleotides: sense, primer of positive polarity used for PCR; antisense, primer of negative polarity used for cDNA synthesis and PCR; probe, fluorogenic probe used for detection of genuine amplification products.
+, coding strand of the proviral genome; −, noncoding strand.
Numbering on the HXB2 genome according to the software provided by the Los Alamos HIV database (SEQUENCE LOCATOR; www.hiv.lanl.gov).
Sequences are given 5′ to 3′ from left to right. Underlined positions show deviation from the standard sequence (HXB2 or clade B consensus) which is indicated in bold letters. f, fluorescein moiety attached to the DNA probe; q, fluorescence quenching moiety attached to the DNA probe.
Primers were adjusted to isolates from the patient(s) listed.
Real-time PCR assays.
Viral RNAs, usRNA, msRNA, and vRNAex were quantified in duplicate by one-tube real-time RT-PCR approaching single-copy sensitivity using PCR primers and probes as outlined in Table 5 under the conditions described previously (20, 22, 33). HIV DNA quantification was performed in duplicate in a total volume of 50 μl with 10 μl DNA template using HotStarTaq master mix (QIAGEN) supplemented with PCR primers (1 μM each) and probes (0.3 μM), listed in Table 5, and additional MgCl2 (1.5 μM). Amplification was performed in a real-time thermocycler (iCycler; Bio-Rad) as follows: 15 min at 95°C and 60 cycles of 10 s at 95°C and 60 s at 60°C. Calibration was performed by using iCycler software.
Cloning and sequencing of env sequences for phylogenetic analysis.
A 401-bp fragment spanning the C2 to C3 region of HIV-1 env was amplified from RNA and DNA samples obtained from CD4+ and CD4−/CD8− T cells (patients 05 and 06) using primers mf170 (5′-AAATGTCAGCACAGTACAATGTACACATGG-3′) and V3Bin (5′-GCGTTAAAGCTTCTGGGTCCCCTCCTGAG-3′) (32). RNA was amplified by a one-tube RT-PCR as described previously (20, 22, 33). DNA was amplified using HotStarTaq master mix (QIAGEN). Cycling was performed as follows: 30 min at 50°C for RT and 15 min at 95°C for activation of Taq polymerase, followed by 60 cycles (10 s at 95°C, 10 s at 60°C, and 60 s at 72°C) for amplification of cDNAs and DNA. The pooled and purified PCR products were ligated into pDrive cloning vector (PCR cloning kit; QIAGEN) according to the supplier's instructions. Individual clones were picked after bacterial transformation and resuspended in 100 μl selective growth medium. To rule out contamination with a free PCR product, DNase I (Roche) (0.2 mg/ml) was added and the clones were incubated at 37°C for 3 to 5 h. Subsequently, 5 μl of bacterial suspension was lysed in 0.5 ml water (5 min at 95°C) and inserts were amplified from 1 μl of lysate in a volume of 20 μl using HotStarTaq master mix (QIAGEN) and primers T7 (5′-TAAGCCTTAATACGACTCACTATAGGGAGA-3′) and m13 (5′-GTAAACGACGGCCAGT-3′) as follows: the polymerase was activated for 15 min at 95°C and amplified for 35 cycles (10 s at 95°C, 10 s at 56°C, and 60 s at 72°C). One microliter of each clonal PCR product containing approximately 20 to 30 ng DNA was sequenced in both directions with primers mf170 and V3Bin using BigDye chain terminator chemistry and an ABI 31000 automated sequencer (Applied Biosystems, Rotkreuz, Switzerland) as described previously (32).
Phylogenetic analyses.
Sequences were edited and aligned with Lasergene software version 5.08 (DNASTAR Inc., Madison, WI). The alignments were manually corrected to adjust sequence gaps to the reading frame. Phylogenetic analyses were conducted using MEGA version 3.1 (36). Genetic distance estimates were obtained using the Tamura-Nei six-parameter model. Neighbor-joining phylogenetic trees were constructed by MEGA using a sequence (GenBank U63632) from HIV-1 isolate JRFL as the reference sequence and bootstrapping (1,000 replications).
Calculations and statistics.
Statistical analyses were performed using GraphPad Prism 4.0 software (GraphPad Software, Inc, San Diego, CA), using nonparametric tests throughout the analyses because the data lacked normal distribution. The Mann-Whitney signed-rank test was used for the comparison of unpaired data, and the Wilcoxon signed-rank test for matched paired analysis.
The numbers of HIV RNA+ cells were calculated by 50% endpoint analysis (49). Global analysis of all limiting dilutions performed for PBMC and T lymphocytes revealed that the dilutions calculated to contain on average 1 to 0.5 HIV RNA+ cells were HIV RNA+ with a median frequency of 33%, which was statistically not different from 0.37% as predicted by Poissonian distribution (P = 0.63; Wilcoxon signed-rank test). Dilutions containing smaller numbers of HIV-infected cells were positive with median frequencies significantly lower than 37% (P < 0.0001). This verifies that the dilutions were prepared properly, without unforeseen loss of the HIV-infected patients' cells.
The specific viral RNA contents per cell were calculated by normalization of the RNA copies quantified in each specimen to the frequencies of HIV RNA+ cells as follows: specific (msRNA) content in class II, copies msRNA/msRNA+ cells; specific (vRNAex) content in class III, copies vRNAex/vRNAex+ cells; and specific (usRNA) content in class I, (copies usRNA/usRNA+ cells) ∉ msRNA > ⊘. In order to exclude class II cells expressing usRNA, the latter calculation was performed using only specimens negative for msRNA.
Nucleotide sequence accession numbers.
The clonal env sequences have been deposited in GenBank under accession numbers EU023940 to EU024109.
RESULTS
Three expression patterns of HIV-1 RNA in PBMC.
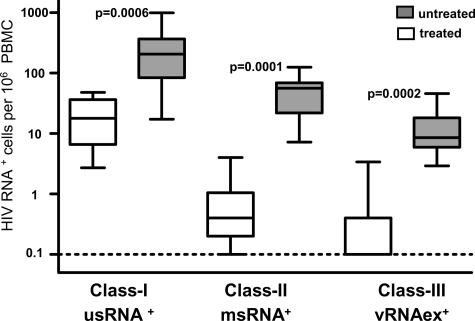
Unfractionated PBMC, total CD4+ T cells, resting or activated CD4+ T cells, CD8/CD4 double-negative T cells, and blood monocytes were isolated by magnetic cell sorting and sampled in replicate dilution series. HIV DNA, HIV usRNA mapping to the gag gene, HIV msRNA coding for tat, rev, or nef, and vRNAex (19, 20, 22) were measured quantitatively in each dilution (Table 6). Terminal dilution analysis allowed for the calculation of the frequencies of cells positive for each HIV RNA species. Overall, this revealed three categories of HIV-infected cells expressing viral RNA which could be distinguished by virtue of their dissimilar frequencies; one class expressing solely usRNA (referred to as class I), a category of cells expressing msRNA and presumably also usRNA (class II), and cells positive for vRNAex (class III). Irrespective of cell type, cells assigned to class I were more frequent than class II cells (P < 0.0001; n = 55; Wilcoxon signed-rank test). In turn, cells of class III, expressing vRNAex, were consistently less frequent than cells of class II (P < 0.0001; n = 45; Wilcoxon signed-rank test).
TABLE 6.
Viral nucleic acids in PBMC and cellular subsetsa
| Cell subset | HIV-1 nucleic acid | No. of HIV nucleic acid copies/106 cells from:
|
No. of HIV nucleic acid-positive cells/106cells fromb:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Untreated patients
|
Treated patients
|
Untreated patients
|
Treated patients
|
||||||
| Median | Min-max | Median | Min-max | Median | Min-max | Median | Min-max | ||
| Total PBMC | DNA | 696 | 491-1,431 | 123 | 15-997 | ||||
| usRNA | 2,029 | 160-13,082 | 56 | 3.7-301 | 206 | 17-990 | 18 | 3-48 | |
| msRNA | 598 | 106-2,679 | 0.3 | <0.1-9 | 56 | 7-125 | 0.4 | <0.1-4 | |
| vRNAex | 33 | 3-201 | <0.1 | <0.1-0.5 | 10 | 3-46 | <0.1 | <0.1-3 | |
| Total CD4+ | DNA | 4,823 | 2,232-10,379 | 727 | 123-2,869 | ||||
| T cells | usRNA | 4,020 | 379-21,554 | 342 | 44-1,410 | 473 | 64-9,278 | 111 | 8-247 |
| msRNA | 1,259 | 85-5,158 | 4.1 | <0.1-121 | 144 | 13-655 | 2 | 0.5-23 | |
| vRNAex | 1.3 | <0.1-108 | 0.2 | <0.1-19 | 3 | <0.1-36 | 0.2 | <0.1-1 | |
| Resting CD4+ | DNA | 2,198 | 253-8,962 | 585 | 146-1,898 | ||||
| T cells | usRNA | 6,007 | 2,206-41,858 | 8.3 | 5-488 | 428 | 161-6,720 | 34 | 8-352 |
| msRNA | 1,319 | 471-3,814 | 8.3 | <0.1-118 | 96 | 39-269 | 8 | <0.1-41 | |
| vRNAex | 0.5 | <0.1-54 | <0.1 | <0.1-<0.1 | 1 | <0.1-24 | <0.1 | <0.1-<0.1 | |
| Activated CD4+ | DNA | 6,388 | 2,240-29,054 | 1,248 | 690-2,989 | ||||
| T cells | usRNA | 9,809 | 2,198-29,769 | 434 | 249-2,618 | 1,402 | 315-34,771 | 102 | 42-5,172 |
| msRNA | 2,576 | 401-10,239 | 16 | <0.1-59 | 256 | 33-590 | 3 | <0.1-46 | |
| vRNAex | 2.3 | <0.1-4 | <0.1 | <0.1-<0.1 | 15 | <0.1-26 | <0.1 | <0.1-<0.1 | |
| CD4−/CD8− | DNA | 166 | 55-541 | <0.1 | <0.1-47 | ||||
| T cells | usRNA | 14,782 | 6,049-59,411 | <0.1 | <0.1-6 | 427 | 14-1,485 | <0.1 | <0.1-6 |
| msRNA | 7,775 | 370-14,617 | <0.1 | <0.1-<0.1 | 474 | 2-1,485 | <0.1 | <0.1-<0.1 | |
| vRNAex | 192 | 23-1,737 | <0.1 | <0.1-0.2 | 101 | 9-282 | <0.1 | <0.1-0.2 | |
| Monocytes | DNA | <0.1 | <0.1-8 | <0.1 | <0.1-5 | ||||
| usRNA | 21 | 0.7-233 | <0.1 | <0.1-3 | 5 | 0.5-35 | 0.3 | <0.1-0.7 | |
| msRNA | <0.1 | <0.1-191 | <0.1 | <0.1-0.6 | <0.1 | <0.1-7 | 0.2 | <0.1-0.7 | |
| vRNAex | 1 | <0.1-8 | <0.1 | <0.1-0.3 | 0.7 | <0.1-3 | <0.1 | <0.1-0.4 | |
If no HIV RNA was detected, the limit of detection (0.1 copy per 106 cells or 0.1 positive cell per 106 cells of each subset) was used for statistical analyses. Min-max, minimum to maximum.
HIV DNA was not assessed by limiting dilution analysis.
The numbers of HIV RNA+ cells were smaller in treated (group A) than in untreated (group B) patients in all three transcriptional categories (Fig. 1; Table 6). In agreement with this, the levels of HIV DNA, indicative of the total number of HIV-infected cells, were markedly higher in viremic patients than in patients on ART (Table 6).
FIG. 1.
HIV-1 RNA+ cells in total PBMC. Frequencies of cells positive for usRNA (class I), msRNA (class II), and vRNAex (class III) were calculated by limiting dilution analysis. Open bars depict data from treated patients (group A) and shaded bars frequencies in untreated patients (group B). P values indicate comparison of group A to B (Mann-Whitney test). “Box and whisker” bars depict medians, quartiles, and ranges. The broken line indicates the lowest detection threshold for PBMC expressing HIV-1 RNA.
HIV-1 RNA in CD4+ T cells comprises only a fraction of viral transcripts.
To dissect HIV-1 expression according to both viral transcriptional patterns and cellular phenotype, we assessed the levels of HIV-1 nucleic acids in total PBMC, CD4+ T cells, and monocytes (Table 6).
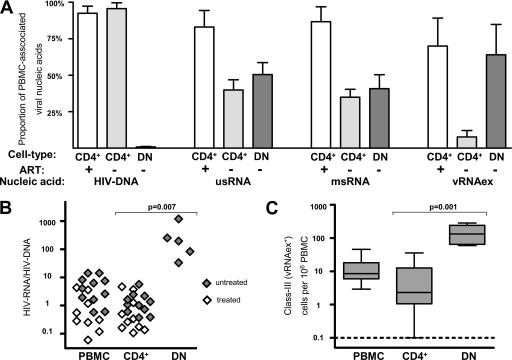
HIV DNA recovered from total PBMC could be attributed predominantly to CD4+ T cells in either patient group. Similarly, in treated patients' samples, the usRNA and msRNA quantified in total PBMC were almost entirely attributable to the CD4+ fraction (Fig. 2A). In marked contrast, generally less than 50% of the PBMC-associated HIV RNAs were recovered from CD4+ cells in untreated patients' samples (group B) (Fig. 2A). Furthermore, vRNAex was detected only at basal levels in CD4+ T cells from both patient groups (Fig. 2A), while it was present at substantial levels in PBMC from untreated patients (Table 6). These findings, showing a pronounced disparity between HIV RNA levels in CD4+ T cells compared to the total PBMC population, indicated that in viremic patients, a major fraction of HIV-infected cells expressing viral RNA may be associated with a cell type expressing no or low levels of surface CD4.
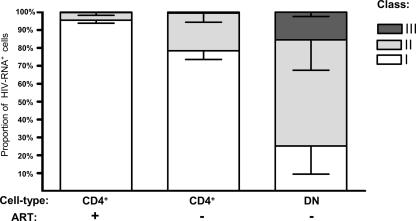
FIG. 2.
High levels of HIV-1 production in CD4−/CD8− T cells from untreated patients. (A) Proportions of HIV-1 nucleic acids in PBMC attributed to CD4+ T cells (CD4+; gray bars, untreated patients; open bars, on-treatment patients) and CD4−/CD8− T cells (DN [double negative]; dark bars, untreated patients; no analysis during ART was possible due to almost-complete depletion of viral DNA and RNA). Error bars show standard errors of the means. +, on-treatment patients; −, untreated patients. (B) HIV RNA expression normalized to HIV DNA content in PBMC, CD4+ T cells (CD4+), and CD4−/CD8− T cells (DN) in untreated and treated patients. HIV RNA expression was calculated as the sum of the average msRNA and usRNA expression at each time point. (C) Frequencies of cells positive for vRNAex (category III) in PBMC, CD4+ T cells (CD4+), and CD4−/CD8− T cells (DN) of untreated patients. The broken line indicates the lowest detection threshold for cells expressing HIV-1 RNA. “Box and whisker” bars depict medians, quartiles, and ranges. Analyses A to C comprised one data point per visit, resulting in duplicate measurements for PBMC and CD4+ T cells for all patients except for patients 03, 08, and 11. (B and C) Results of nonparametric comparison of CD4+ versus CD4−/CD8− T cells using Mann-Whitney testing are indicated by P values.
In CD14+ monocytes of all untreated and in five of six treated individuals, HIV-infected cells were present. However, they harbored minute levels of viral nucleic acids (Table 6) and their frequencies were 100- to 1,000-fold lower than those of HIV-infected CD4+ T cells. In particular, msRNA expression in CD14+ cells was very low and detectable in only 50% of samples in both groups. Thus, monocytes appear unlikely to play a major role for virus production in peripheral blood.
Abundant viral transcription in CD8/CD4 double-negative T lymphocytes.
Considering that HIV can downmodulate CD4 surface expression of its host cells (7, 37, 53) and that CD8+ T cells were not expected to contribute significantly to PBMC viral load (5), we examined CD4/CD8 double-negative CD3+ T lymphocytes for their content of viral nucleic acids. For untreated patients, this analysis revealed that 50% ± 8% (mean ± standard error) of usRNA, 41% ± 10% of msRNA, and 64% ± 21% of PBMC-associated vRNAex was attributable to this minor cellular subset (Fig. 2A), whereas HIV DNA was present at lower levels than in CD4+ T cells (Table 6). In accord with this, we observed higher viral RNA/DNA ratios in CD4−/CD8− T cells than in total PBMC and CD4+ T cells (Fig. 2B), indicating increased viral transcription and/or reduction of defective or transcriptionally silent proviral DNA in this cell subset. Furthermore, cells of class III, positive for vRNAex, were approximately 10 times more frequent in CD4−/CD8− T cells than in CD4+ T lymphocytes (Fig. 2C).
In contrast, HIV-1 nucleic acids in CD4−/CD8− T cells from patients undergoing antiretroviral treatment were almost completely depleted (Table 6). Among the CD4−/CD8− T-cell specimens from the four patients assessed, HIV DNA was present only in patient 04 and usRNA was detected in patient 13 in one of four RNA extracts tested. Thus, HIV-infected CD4−/CD8− T cells expressing viral RNA were at least 500 times less frequent in patients on ART than in viremic patients.
T cells losing CD4 expression due to HIV-1 infection are expected to express CD4 mRNA because the virus mediates CD4 down-modulation at a posttranslational level (1). Thus, preliminary experiments to measure CD4 mRNA were performed (data not shown). CD4 mRNA expression was readily detectable in CD4−/CD8− T cells, consistent with the hypothesis that the double-negative T-cell subset indeed may harbor cells derived from previously CD4-positive helper T lymphocytes. However direct analysis of CD4 mRNA expression in HIV-infected CD4−/CD8− T cells was precluded by the low frequencies (<0.1%) of HIV-infected cells within this cell subset.
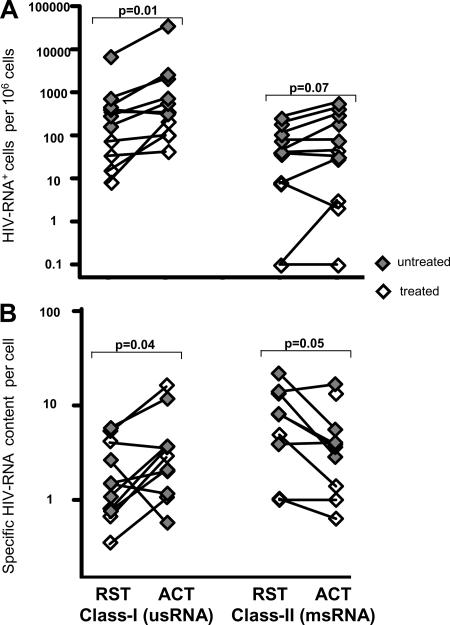
Similar HIV-1 transcription in resting and activated CD4+ T cells.
Although CD4+ T cells harbored only 30 to 50% of all HIV-1 transcripts in PBMC of untreated patients, this nevertheless considerable contribution to the cellular viral burden warranted further investigation. Contrary to initial expectations, only subtle differences between resting and activated CD4+ T cells were observed with regard to viral transcriptional activity (Fig. 3) and HIV DNA content (Table 6). HIV RNA+ cells appeared, at most, slightly enriched in the activated fraction in comparison to the resting population (P = 0.01 for usRNA+ class I cells and P = 0.07 for msRNA+ class II cells) (Fig. 3A). As assessed by specific per-cell HIV RNA burdens, a moderate tendency for enhanced transcription in the activated CD4+ subset was observed in class I cells (P = 0.04), whereas class II cells expressing msRNA showed an opposite trend (P = 0.05), harboring lower levels of msRNA in the activated than in the resting CD4+ subset (Fig. 3B). Assessment of vRNAex revealed that this RNA species, indicative of ongoing virus production, was only occasionally present, at low levels, in activated and resting CD4+ T cells (Table 6).
FIG. 3.
Similar HIV-1 transcriptional activity in resting and activated CD4+ T cells from treated and untreated patients. (A) Frequencies of HIV RNA+ cells in resting (RST) and activated (ACT) CD4+ T cells normalized to 106 cells of the corresponding cell types. (B) Geometric means of HIV RNA contents per cell in RST and ACT CD4+ T cells normalized to numbers of HIV RNA+ cells. Analyses comprised one data point per visit, resulting in duplicate measurements for PBMC and CD4+ T cells for all patients except for patients 03, 08, and 11. Results of nonparametric paired analyses of CD4+ versus CD4−/CD8− T cells using Wilcoxon signed-rank testing are indicated by P values.
In summary, the lack of evidence for the association of activation markers CD25, CD69, or HLA-DR with enhanced HIV transcription or virus production suggests that HIV transcription may be similar in activated and resting T-helper lymphocytes carrying the CD4 receptor at the cell surface.
Differences in HIV-1 single-cell transcriptional activity in T-helper-cell subsets.
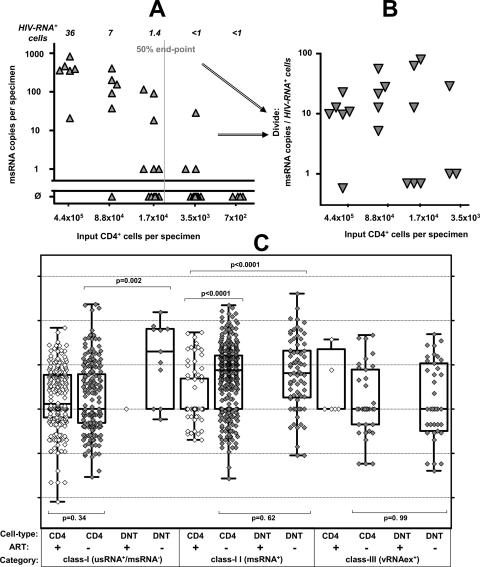
To assess viral transcription at the single-cell level, and to further explore whether the categorization of HIV RNA+ cells into three transcriptional classes represented a valid model of HIV transcription in PBMC, the per-cell viral RNA content of CD4+ T cells was compared with that of CD4−/CD8− T cells in an analysis using pooled PCR-positive data from all patients (Fig. 4; CD4+ T cells, n = 826; CD4−/CD8− T cells, n = 81).
FIG. 4.
Specific HIV contents per cell of CD4+ and CD4−/CD8− T cells. (A and B) Schematic outline of determination of specific cell contents as exercised by measurements of msRNA (class II expression) in CD4+ T cells obtained from patient 10. (A) Primary measurements of msRNA levels in serial five-fold dilutions of purified CD4+ T cells are shown in panel A. Triangles depict individual measurements and the vertical gray line shows the calculated 50% end point of specimens positive for msRNA. Italic numbers above the panel show the calculated numbers of HIV RNA+ cells in each dilution series. (B) Normalization of measured HIV msRNA copies to numbers of HIV RNA+ cells results in specific HIV RNA contents in each sample with detectable msRNA (triangles). (C) Per-cell HIV RNA expression data comprising per-cell expression in resting, activated, and total CD4+ T cells (CD4+) and CD4−/CD8− T cells (DNT [double negative]) from all patients and time points is displayed by single data points (open symbols, on-treatment patients; gray symbols, untreated patients) and “box and whisker” bars showing medians, quartiles, and ranges. Note that viral RNA expression in CD4−/CD8− T cells obtained from patients on ART, remaining below the threshold of detection, could not be displayed except for one sample (left panel, class I expression). Results of nonparametric comparisons using Mann-Whitney testing are indicated by P values.
In CD4+ T cells, class I (usRNA) expression in treated and viremic patients was similar (Mann-Whitney test; P = 0.34), suggesting that basal usRNA transcription in CD4+ cells was unaffected by treatment status and ongoing viral replication. Class I expression in CD4−/CD8− T cells was almost completely absent in treated patients and rare but perceptible in viremic individuals. Interestingly, in untreated patients, we observed a markedly higher specific per-cell usRNA expression than in class I cells of CD4+ cells (Mann-Whitney test; P < 0.002). Thus, even the HIV-infected CD4−/CD8− T cells with basal RNA transcriptional activity displayed an increment in viral RNA expression.
The median class II (msRNA) expression in CD4+ T cells was about 10-fold higher in viremic patients than in individuals on ART (Mann-Whitney test; P < 0.0001). Equivalent trends (Mann-Whitney test; P ≤ 0.08) for increased per-cell expression of msRNA in viremic patients were observed when activated, resting, and unfractionated CD4+ cells were analyzed separately (data not shown). The levels of class II-specific per-cell expression in CD4−/CD8− T cells were similar to the boosted RNA expression in CD4+ cells in viremic patients (Mann-Whitney test; P = 0.62) and completely undetectable in treated patients.
The specific per-cell expression in class III was similar in the CD4+ T-cell and the CD4−/CD8− T-cell subsets (P = 0.99) in untreated patients. A comparison of class III expression in treated and viremic patients was not performed, since vRNAex expression was profoundly depleted in CD4+ T cells of treated patients (eight positive specimens) and completely absent in their CD4−/CD8− T cells. Thus, it was conceivable that class III-positive cells in treated patients did not reflect productively infected lymphocytes, but rather uninfected CD4+ T cells carrying residual virions picked up from the plasma at their surfaces (22).
Distribution of viral transcriptional classes in T-helper lymphocytes.
Our analysis of specific per-cell transcription implied that in viremic patients, class II HIV-infected cells may comprise subpopulations with elevated rates of expression of msRNA. Because active virus production requires the expression of high levels of msRNA and usRNA (19, 48), it is conceivable that class III cells carrying vRNAex may reflect such a subpopulation (19, 20, 22), actively expressing viral particles as well as viral regulatory and structural genes. However, a direct determination of whether the expression of vRNAex in a given specimen coincided with elevated expression of msRNA was precluded since the extraction procedures for total cellular RNA and vRNAex are mutually exclusive.
Nevertheless, given the assumption that class III expression required the expression of msRNA, calculations of the relative frequencies of the different transcriptional classes were performed in CD4−/CD8− T cells and CD4+ T cells (Fig. 5). This analysis revealed that in the CD4+ T cells of treated patients, the vast majority (95% ± 2%) of HIV RNA+ cells could be assigned to viral transcriptional class I, while the remaining cells showed mostly class II expression (4% ± 1.5%) and, very rarely, class III transcription (0.3% ± 0.17%). In viremic individuals, the relative contributions of class II were elevated both in CD4+ and in CD4−/CD8− T cells, to 21% ± 5% and 59% ± 17%, respectively. Class III cells remained scarce in CD4+ T cells (0.57 ± 0.25%); whereas in CD4−/CD8− T cells, these cells, expressing vRNAex, represented a sizable fraction (15% ± 2%) of all HIV RNA+ cells.
FIG. 5.
Distribution of cellular transcription categories in CD4+ and CD4−/CD8− T cells. Average class I, class II, and class III contributions to total HIV-1 RNA+ cells were calculated for CD4+ T cells (CD4+) from untreated and treated patients and for CD4−/CD8− T cells (DN [double negative]) from untreated patients. Analyses comprised one data point per visit, resulting in duplicate measurements for CD4+ T cells for all patients except for patients 03, 08, and 11. Error bars depict standard errors of the means.
Taken together, these observations confirm that viral transcription is greatly enhanced in HIV-infected CD4−/CD8− T cells compared to the level in CD4+ T cells.
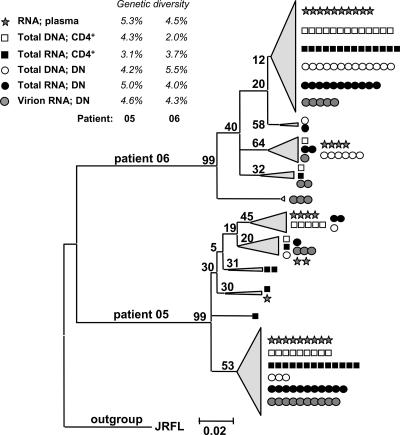
Comparison of T-cell- and plasma-derived HIV-1 quasispecies.
In order to assess a potential compartmentalization between HIV-1 quasispecies in different cell types and plasma virus, molecular clones spanning the C2 to C3 region of the env gene (32) were isolated from plasma RNA, from CD4+ and CD4−/CD8− T cells (DNA and total RNA), and from the vRNAex fraction of CD4−/CD8− T cells of two viremic patients (Fig. 6). Within each patient's sequence cluster, one major and several minor phylogenetic groups were present. However, there was no evidence for compartmentalization between PBMC subsets or different nucleic acid fractions, as we observed clones from all fractions in every major phylogenetic group. Furthermore, the genetic diversities were similar in plasma and in CD4+ and CD4−/CD8− T cells. These findings suggest that HIV-infected CD4+ and CD4−/CD8− T cells in PBMC contain quasispecies which are closely related to each other and to the dominant viral sequences prevailing in plasma.
FIG. 6.
Uniform distribution of HIV-1 env quasispecies in plasma and CD4+ and CD4−/CD8− T cells. Neighbor-joining phylogenetic tree of env C2-V3-C3 sequences obtained from patients 05 and 06 constructed using MEGA 3.1. Single molecular clones (mean n = 14; range, 5 to 16) were obtained from plasma RNA, from CD4+ T cells (CD4+; total RNA and total DNA), and from CD4−/CD8− T cells (DN [double negative]; total RNA, vRNAex, and total DNA). Symbols indicate individual clones and numbers above branches show bootstrap values. Mean diversities for each cell type are indicated.
DISCUSSION
In the present study, we assessed HIV RNA expression in the major host cell types of peripheral blood using highly sensitive, patient-matched RT-PCR combined with terminal dilution. This novel approach allowed for discrimination between the viral transcription levels in latently versus productively infected cells. Most noteworthy, our data suggest that, contrary to current belief, productive HIV-1 infection in peripheral blood may be restricted to a major extent to T-helper lymphocytes of a CD4/CD8 double-negative phenotype.
As demonstrated by bulk RNA analysis, by assessment of viral transcriptional per-cell expression patterns, by determination of the frequencies of infected cells expressing HIV RNA, and by quantification of HIV DNA, the lion's share of active HIV-1 in PBMC could be assigned to the T-lymphocyte population. Thus, resting and activated CD4+ T cells were subjected to a detailed investigation, which revealed that both subsets exhibited similar viral transcriptional activities and frequencies. Although we observed an accumulation of HIV-1 RNA+ CD4+ T cells in the activated cell fraction, this was only moderate and may reflect preferential infection of activated cells (10) and, possibly, stimulation of formerly transcriptionally silent provirus.
Such a lack of association of activation markers with viral transcriptional strength was unexpected, since activated CD4+ T cells are widely considered to be primarily responsible for viral propagation (6, 56, 63, 75). Possibly the ambiguity of cell surface markers in designating a cell's potential to support HIV-1 replication (11, 15, 34, 67, 77, 78) might have confounded the analysis of activated CD4+ T cells to some extent. In particular, the inclusion of CD25, in addition to CD69 and HLA-DR as in similar studies (11, 12, 79), may have resulted in the inclusion of primary CD25+ regulatory T cells (59) lacking a classical activation profile.
Notwithstanding, the apparent lack of viral transcriptional enhancement in the population of HIV-infected activated CD4+ T cells, compared to that in resting lymphocytes, may be primarily related to the HIV-1-induced downregulation of CD4 surface expression (7, 53). Hence, activated CD4+ T cells with transcriptionally active proviral integrants may have an extremely short half-life, simply because they may rapidly switch to a CD4− phenotype and would not be identifiable as T-helper cells in cell-sorting experiments using the CD4 marker. In agreement with this assumption, we have gathered strong evidence that productively infected T lymphocytes in vivo are characterized by down-modulation of their CD4 receptor in vivo and that HIV-infected CD4−/CD8− T cells exhibit generally augmented viral transcription, as reflected by the following findings.
(i) HIV-infected cells with elevated viral transcription (class II and class III) contributed 74% of all cells expressing viral RNA in CD4−/CD8− T cells, in stark contrast to CD4+ T cells, which were dominated (>78%) by cells displaying basal HIV transcription.
(ii) Expression of vRNAex, a strong correlate of ongoing viral replication in PBMC (19, 20, 22), was predominantly confined to CD4−/CD8− T cells.
(iii) The specific per-cell expression of usRNA in cells with basal viral expression (class I) was significantly higher than in CD4+ T cells of untreated patients (group B). In addition, the specific per-cell expression of msRNA in virus-infected cells of transcriptional class II, albeit equal to that of CD4+ cells of untreated patients, was substantially augmented compared to the expression in CD4+ cells of patients on ART.
Very likely, HIV-infected CD4−/CD8− T cells originate from infected CD4+ T cells, as supported by our phylogenetic analysis showing similar genetic diversities in env clones derived from CD4+ and CD4−/CD8− T cells and suggesting an absence of compartmentalization.
In concordance with this concept, CD4−/CD8− T cells harbored all viral transcriptional categories also found in CD4+ lymphocytes. The finding that, besides the accumulated class II and class III cells, some class I HIV-infected cells with basal transcriptional activity also resided in the CD4−/CD8− T-cell population, could indicate that this fraction of infected CD4−/CD8− T cells represents cells which reduce HIV transcription following a burst of virion production to recuperate to viral latency (47). Conversely, it is likely that class II expression in CD4−/CD8− T cells may in part reflect latently infected cells in transition to productive infection, because infected cells may lose surface CD4 receptors before active production of viral structural proteins and shedding of virions sets in (7, 53). This assumption is supported by our observation that cells harboring vRNAex (class III), even though markedly accumulated in the CD4−/CD8− T-cell subset, were still much less frequent than cells expressing msRNA (class II). Furthermore, there is evidence that the expression of msRNA (19, 74) may result in the expression of the Nef protein in latently infected cells, which may predispose these cells to rapid induction of virus production (19, 62) and to CD4 downregulation.
Thus, HIV-1-infected cells of a CD4−/CD8− phenotype appear to be a dynamic population closely connected with HIV-1-infected CD4+ T cells. Extending the observations of Marodon et al. (42), and supported by a recent report demonstrating the occurrence of HIV-1-infected CD4−/CD8− T cells during incomplete response to antiretroviral treatment (8), our findings suggest that HIV RNA+ CD4−/CD8− T cells may be the predominant cell type supporting ongoing HIV-1 production in peripheral blood. Given that studies of HIV-1 infection in lymphoid tissues using combinations of in situ hybridization and in situ histochemistry have reported productive infection of CD4+ T cells (55, 69), the role of HIV-1-infected CD4−/CD8− T-helper lymphocytes requires further study in other body compartments.
Complementary to the notion that ongoing HIV-1 production may be exclusively associated with CD4−/CD8− T cells in peripheral blood, our analysis suggests that the vast majority of circulating infected CD4+ T cells transcribing the proviral genome does so with an efficiency insufficient for virion production. This is primarily supported by the vast predominance of class I HIV RNA+ cells in CD4+ T lymphocytes. Moreover, even in a viremic milieu, only a minority of infected CD4+ T cells were found to express HIV-1 msRNA. The bulk of these class II CD4+ T cells may reflect latently infected cells with subthreshold expression of msRNA, insufficient to mediate the expression of structural viral proteins (19, 40, 62). A further indication that HIV-infected CD4+ T cells are mainly of a latent or subproductive type was the finding that the presence of viral particles (vRNAex) on CD4+ T cells was extremely low and may reflect virions randomly attached to surface CD4 rather than virus emerging from productively infected CD4−/CD8− T cells. Furthermore, the expression of the classical activation markers CD25, CD69, and HLA-DR in CD4+ T cells appeared to be associated with susceptibility to infection rather than with vigorous transcriptional activity, which further supports the concept that productive infection may be initiated in CD4+ T cells but actually takes place, after downmodulation of the CD4 receptors, in CD4−/CD8− T-helper lymphocytes.
Despite its sophisticated design, using PCR assays with primers adjusted to the predominant viral isolates in combination with limiting dilution analysis, which allowed the decipherment of the numbers and the activities of infected cells, the present study has some limitations.
First, PBMC subsets were collected by using magnetic cell sorting, which is less specific than FACS, in order to allow rapid and gentle isolation of cells and reliable RNA extraction.
Second, the division of HIV RNA+ cells into three transcriptional categories provides only an approximate model, with limited resolution, which needs to be further refined by long-term longitudinal assessment of ART, by the determination of different splice variants of msRNA (19), and by using smaller dilution steps.
Third, due to the use of end-point dilution in the present study, comprising in total almost 6,000 PCR analyses, only a limited number of patients in each group could be studied.
In conclusion, using a novel approach combining limiting dilution with ultrasensitive patient-matched real-time RT-PCR, this study is the first to quantify HIV-1 RNA expression at a single-cell level and to determine the frequency and distribution of both latently and productively infected cells in PBMC subsets ex vivo. The fact that productive HIV-1 infection in peripheral blood appeared to be largely restricted to the distinct CD4−/CD8− cellular phenotype suggests that specific quantification of ongoing HIV-1 replication during ART may be attempted by the analysis of viral transcripts in this cell type. Furthermore, our findings imply that CD3+ lymphocytes, rather than CD4+ T cells, should be used for monitoring cellular HIV-1 replication ex vivo in order to include the complete range of virus-expressing T-helper lymphocytes.
Acknowledgments
We thank the participating patients for their commitment; Christine Grube, Christine Schneider, Milo Huber, Doris Baumann, and Roland Hafner for excellent patient care; Eva Niederer and Mustafa Oezbas (Institute for Biomedical Engineering, ETH Zürich) for dedicated support in flow cytometry; Alexandra Trkola, Amapola Manrique, Michael Huber, Viktor von Wyl, and Annette Audigé for advice and helpful discussions; Joseph Wong for critical review of the manuscript; and Ingrid Nievergelt and Christine Vögtli for administrative assistance.
Financial support was provided by the Novartis Foundation (grant 02A03), the Roche Foundation (grant 281-2005), the Bonizzi Theler Stiftung, Stiftung für Wissenschaftliche Forschung der Universität Zürich, Jubiläumsstiftung Swiss Life, Olga Maienfisch Stiftung, the Swiss National Science Foundation (grant 3100A0-112670), Hermann Klaus Stiftung, Abbott Inc. (an unrestricted educational grant, SWIT-02-002), and the Swiss HIV Cohort Study (grant 477).
The members of the Swiss HIV Cohort Study (www.shcs.ch) are M. Battegay, E. Bernasconi, J. Böni, HC Bucher, P. Bürgisser, S. Cattacin, M. Cavassini, R. Dubs, M. Egger, L. Elzi, P. Erb, M. Fischer, M. Flepp, A. Fontana, P. Francioli, H. Furrer, M. Gorgievski, H. Günthard, H. Hirsch, B. Hirschel, I. Hösli, C. Kahlert, L. Kaiser, U. Karrer, C. Kind, T. Klimkait, B. Ledergerber, G. Martinetti, B. Martinez, N. Müller, D. Nadal, M. Opravil, F. Paccaud, G. Pantaleo, M. Rickenbach, C. Rudin, P. Schmid, D. Schultze, J. Schüpbach, R. Speck, P. Taffé, P. Tarr, A. Telenti, A. Trkola, P. Vernazza, R. Weber, and S. Yerly.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853-864. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J. R., A. R. Sedaghat, T. Kieffer, T. Brennan, P. K. Lee, M. Wind-Rotolo, C. M. Haggerty, A. R. Kamireddi, Y. Liu, J. Lee, D. Persaud, J. E. Gallant, J. Cofrancesco, Jr., T. C. Quinn, C. O. Wilke, S. C. Ray, J. D. Siliciano, R. E. Nettles, and R. F. Siliciano. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80:6441-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisset, L. R., S. Bosbach, Z. Tomasik, H. Lutz, J. Schüpbach, and J. Böni. 2001. Quantification of in vitro retroviral replication using a one-tube real-time RT-PCR system incorporating direct RNA preparation. J. Virol. Methods 91:149-155. [DOI] [PubMed] [Google Scholar]
- 4.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 5.Brenchley, J. M., B. J. Hill, D. R. Ambrozak, D. A. Price, F. J. Guenaga, J. P. Casazza, J. Kuruppu, J. Yazdani, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 78:1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B. K., R. T. Gandhi, and D. Baltimore. 1996. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 70:6044-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheney, K. M., R. Kumar, A. Purins, L. Mundy, W. Ferguson, D. Shaw, C. J. Burrell, and P. Li. 2006. HIV type 1 persistence in CD4−/CD8− double negative T cells from patients on antiretroviral therapy. AIDS Res. Hum. Retrovir. 22:66-75. [DOI] [PubMed] [Google Scholar]
- 9.Christopherson, C., Y. Kidane, B. Conway, J. Krowka, H. Sheppard, and S. Kwok. 2000. PCR-based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38:630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 11.Chun, T. W., J. S. Justement, R. A. Lempicki, J. Yang, G. Dennis, Jr., C. W. Hallahan, C. Sanford, P. Pandya, S. Liu, M. McLaughlin, L. A. Ehler, S. Moir, and A. S. Fauci. 2003. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc. Natl. Acad. Sci. USA 100:1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, T. W., D. C. Nickle, J. S. Justement, D. Large, A. Semerjian, M. E. Curlin, M. A. O'Shea, C. W. Hallahan, M. Daucher, D. J. Ward, S. Moir, J. I. Mullins, C. Kovacs, and A. S. Fauci. 2005. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J. Clin. Investig. 115:3250-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Silva, F. S., D. S. Venturini, E. Wagner, P. R. Shank, and S. Sharma. 2001. CD4-independent infection of human B cells with HIV type 1: detection of unintegrated viral DNA. AIDS Res. Hum. Retrovir. 17:1585-1598. [DOI] [PubMed] [Google Scholar]
- 15.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity 15:671-682. [DOI] [PubMed] [Google Scholar]
- 16.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 17.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, M., H. F. Günthard, M. Opravil, B. Joos, W. Huber, L. R. Bisset, P. Ott, J. Böni, R. Weber, and R. W. Cone. 2000. Residual HIV-RNA levels persist for up to 2.5 years in peripheral blood mononuclear cells of patients on potent antiretroviral therapy. AIDS Res. Hum. Retrovir. 16:1135-1140. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, M., B. Joos, B. Hirschel, G. Bleiber, R. Weber, and H. F. Günthard. 2004. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J. Infect. Dis. 190:1979-1988. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, M., B. Joos, J. K. Wong, P. Ott, M. Opravil, B. Hirschel, R. Weber, and H. F. Günthard. 2004. Attenuated and nonproductive viral transcription in the lymphatic tissue of HIV-1-infected patients receiving potent antiretroviral therapy. J. Infect. Dis. 189:273-285. [DOI] [PubMed] [Google Scholar]
- 21.Fischer, M., A. Trkola, B. Joos, R. Hafner, H. Joller, M. A. Muesing, D. R. Kaufman, E. Berli, B. Hirschel, R. Weber, and H. F. Günthard. 2003. Shifts in cell-associated HIV-1 RNA but not in episomal HIV-1 DNA correlate with new cycles of HIV-1 infection in vivo. Antivir. Ther. 8:97-104. [PubMed] [Google Scholar]
- 22.Fischer, M., J. K. Wong, D. Russenberger, B. Joos, M. Opravil, B. Hirschel, A. Trkola, H. Kuster, R. Weber, and H. F. Günthard. 2002. Residual cell-associated unspliced HIV-1 RNA in peripheral blood of patients on potent antiretroviral therapy represents intracellular transcripts. Antivir. Ther. 7:91-103. [PubMed] [Google Scholar]
- 23.Flamand, L., R. W. Crowley, P. Lusso, S. Colombini-Hatch, D. M. Margolis, and R. C. Gallo. 1998. Activation of CD8+ T lymphocytes through the T cell receptor turns on CD4 gene expression: implications for HIV pathogenesis. Proc. Natl. Acad. Sci. USA 95:3111-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fultz, P. N., L. Yue, Q. Wei, and M. Girard. 1997. Human immunodeficiency virus type 1 intersubtype (B/E) recombination in a superinfected chimpanzee. J. Virol. 71:7990-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Günthard, H. F., J. K. Wong, C. A. Spina, C. Ignacio, S. Kwok, C. Christopherson, J. Hwang, R. Haubrich, D. Havlir, and D. D. Richman. 2000. Effect of influenza vaccination on viral replication and immune response in persons infected with human immunodeficiency virus receiving potent antiretroviral therapy. J. Infect. Dis. 181:522-531. [DOI] [PubMed] [Google Scholar]
- 28.Haase, A. T. 1999. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu. Rev. Immunol. 17:625-656. [DOI] [PubMed] [Google Scholar]
- 29.Hermankova, M., J. D. Siliciano, Y. Zhou, D. Monie, K. Chadwick, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2003. Analysis of human immunodeficiency virus type 1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J. Virol. 77:7383-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber, M., M. Fischer, B. Misselwitz, A. Manrique, H. Kuster, B. Niederost, R. Weber, V. von Wyl, H. F. Günthard, and A. Trkola. 2006. Complement lysis activity in autologous plasma is associated with lower viral loads during the acute phase of HIV-1 infection. PLoS Med. 3:e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Innocenti, P., M. Ottmann, P. Morand, P. Leclercq, and J. M. Seigneurin. 1992. HIV-1 in blood monocytes: frequency of detection of proviral DNA using PCR and comparison with the total CD4 count. AIDS Res. Hum. Retrovir. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 32.Joos, B., A. Trkola, M. Fischer, H. Kuster, P. Rusert, C. Leemann, J. Böni, A. Oxenius, D. A. Price, R. E. Phillips, J. K. Wong, B. Hirschel, R. Weber, and H. F. Günthard. 2005. Low human immunodeficiency virus envelope diversity correlates with low in vitro replication capacity and predicts spontaneous control of plasma viremia after treatment interruptions. J. Virol. 79:9026-9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser, P., B. Niederöst, B. Joos, V. von Wyl, M. Opravil, R. Weber, H. F. Günthard, and M. Fischer. 2006. Equal amounts of intracellular and virion-enclosed hepatitis C virus RNA are associated with peripheral blood mononuclear cells in vivo. J. Infect. Dis. 194:1713-1723. [DOI] [PubMed] [Google Scholar]
- 34.Kinter, A. L., C. A. Umscheid, J. Arthos, C. Cicala, Y. Lin, R. Jackson, E. Donoghue, L. Ehler, J. Adelsberger, R. L. Rabin, and A. S. Fauci. 2003. HIV envelope induces virus expression from resting CD4+ T cells isolated from HIV-infected individuals in the absence of markers of cellular activation or apoptosis. J. Immunol. 170:2449-2455. [DOI] [PubMed] [Google Scholar]
- 35.Knight, S. C., S. E. Macatonia, and S. Patterson. 1990. HIV I infection of dendritic cells. Int. Rev. Immunol. 6:163-175. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 37.Lama, J. 2003. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr. HIV Res. 1:167-184. [DOI] [PubMed] [Google Scholar]
- 38.Lambotte, O., Y. Taoufik, M. G. de Goer, C. Wallon, C. Goujard, and J. F. Delfraissy. 2000. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 23:114-119. [DOI] [PubMed] [Google Scholar]
- 39.Lassen, K. G., J. R. Bailey, and R. F. Siliciano. 2004. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 78:9105-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lassen, K. G., K. X. Ramyar, J. R. Bailey, Y. Zhou, and R. F. Siliciano. 2006. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathogens 2:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Marodon, G., D. Warren, M. C. Filomio, and D. N. Posnett. 1999. Productive infection of double-negative T cells with HIV in vivo. Proc. Natl. Acad. Sci. USA 96:11958-11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathiot, N. D., R. Krueger, M. A. French, and P. Price. 2001. Percentage of CD3+CD4−CD8−γδTCR− T cells is increased by HIV disease. AIDS Res. Hum. Retrovir. 17:977-980. [DOI] [PubMed] [Google Scholar]
- 44.McElrath, M. J., J. E. Pruett, and Z. A. Cohn. 1989. Mononuclear phagocytes of blood and bone marrow: comparative roles as viral reservoirs in human immunodeficiency virus type 1 infections. Proc. Natl. Acad. Sci. USA 86:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moir, S., A. Malaspina, Y. Li, T. W. Chun, T. Lowe, J. Adelsberger, M. Baseler, L. A. Ehler, S. Liu, R. T. Davey, Jr., J. A. Mican, and A. S. Fauci. 2000. B cells of HIV-1-infected patients bind virions through CD21-complement interactions and transmit infectious virus to activated T cells. J. Exp. Med. 192:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou, C. Y., S. Kwok, S. W. Mitchell, D. H. Mack, J. J. Sninsky, J. W. Krebs, P. Feorino, D. Warfield, and G. Schochetman. 1988. DNA amplification for direct detection of HIV-1 in DNA of peripheral blood mononuclear cells. Science 239:295-297. [DOI] [PubMed] [Google Scholar]
- 47.Persaud, D., Y. Zhou, J. M. Siliciano, and R. F. Siliciano. 2003. Latency in human immunodeficiency virus type 1 infection: no easy answers. J. Virol. 77:1659-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pomerantz, R. J., T. Seshamma, and D. Trono. 1992. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: potential implications for latency. J. Virol. 66:1809-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed, I. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 50.Rusert, P., M. Fischer, B. Joos, C. Leemann, H. Kuster, M. Flepp, S. Bonhoeffer, H. F. Günthard, and A. Trkola. 2004. Quantification of infectious HIV-1 plasma viral load using a boosted in vitro infection protocol. Virology 326:113-129. [DOI] [PubMed] [Google Scholar]
- 51.Saha, K., J. Zhang, A. Gupta, R. Dave, M. Yimen, and B. Zerhouni. 2001. Isolation of primary HIV-1 that targets CD8+ T lymphocytes using CD8 as a receptor. Nat. Med. 7:65-72. [DOI] [PubMed] [Google Scholar]
- 52.Saksela, K., E. Muchmore, M. Girard, P. Fultz, and D. Baltimore. 1993. High viral load in lymph nodes and latent human immunodeficiency virus (HIV) in peripheral blood cells of HIV-1-infected chimpanzees. J. Virol. 67:7423-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salmon, P., R. Olivier, Y. Riviere, E. Brisson, J. C. Gluckman, M. P. Kieny, L. Montagnier, and D. Klatzmann. 1988. Loss of CD4 membrane expression and CD4 mRNA during acute human immunodeficiency virus replication. J. Exp. Med. 168:1953-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez, G., X. Xu, J. C. Chermann, and I. Hirsch. 1997. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J. Virol. 71:2233-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schacker, T., S. Little, E. Connick, K. Gebhard, Z. Q. Zhang, J. Krieger, J. Pryor, D. Havlir, J. K. Wong, R. T. Schooley, D. Richman, L. Corey, and A. T. Haase. 2001. Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J. Infect. Dis. 183:555-562. [DOI] [PubMed] [Google Scholar]
- 56.Schnittman, S. M., H. C. Lane, J. Greenhouse, J. S. Justement, M. Baseler, and A. S. Fauci. 1990. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type 1: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proc. Natl. Acad. Sci. USA 87:6058-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz, S., B. K. Felber, E. M. Fenyo, and G. N. Pavlakis. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 64:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shevach, E. M. 2000. Suppressor T cells: rebirth, function and homeostasis. Curr. Biol. 10:R572-R575. [DOI] [PubMed] [Google Scholar]
- 60.Sonza, S., A. Maerz, N. Deacon, J. Meanger, J. Mills, and S. Crowe. 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol. 70:3863-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sonza, S., H. P. Mutimer, R. Oelrichs, D. Jardine, K. Harvey, A. Dunne, D. F. Purcell, C. Birch, and S. M. Crowe. 2001. Monocytes harbour replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS 15:17-22. [DOI] [PubMed] [Google Scholar]
- 62.Spina, C. A., T. J. Kwoh, M. Y. Chowers, J. C. Guatelli, and D. D. Richman. 1994. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spina, C. A., H. E. Prince, and D. D. Richman. 1997. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J. Clin. Investig. 99:1774-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strain, M. C., H. F. Günthard, D. V. Havlir, C. C. Ignacio, D. M. Smith, A. J. Leigh-Brown, T. R. Macaranas, R. Y. Lam, O. A. Daly, M. Fischer, M. Opravil, H. Levine, L. Bacheler, C. A. Spina, D. D. Richman, and J. K. Wong. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. USA 100:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strober, S., L. Cheng, D. Zeng, R. Palathumpat, S. Dejbakhsh-Jones, P. Huie, and R. Sibley. 1996. Double negative (CD4−CD8− alpha beta+) T cells which promote tolerance induction and regulate autoimmunity. Immunol. Rev. 149:217-230. [DOI] [PubMed] [Google Scholar]
- 66.Tobin, N. H., G. H. Learn, S. E. Holte, Y. Wang, A. J. Melvin, J. L. McKernan, D. M. Pawluk, K. M. Mohan, P. F. Lewis, J. I. Mullins, and L. M. Frenkel. 2005. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J. Virol. 79:9625-9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valentin, A., M. Rosati, D. J. Patenaude, A. Hatzakis, L. G. Kostrikis, M. Lazanas, K. M. Wyvill, R. Yarchoan, and G. N. Pavlakis. 2002. Persistent HIV-1 infection of natural killer cells in patients receiving highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 99:7015-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van der Ende, M. E., M. Schutten, B. Raschdorff, G. Grossschupff, P. Racz, A. D. Osterhaus, and K. Tenner-Racz. 1999. CD4 T cells remain the major source of HIV-1 during end stage disease. AIDS 13:1015-1019. [DOI] [PubMed] [Google Scholar]
- 70.Vesanen, M., M. Markowitz, Y. Cao, D. D. Ho, and K. Saksela. 1997. Human immunodeficiency virus type-1 mRNA splicing pattern in infected persons is determined by the proportion of newly infected cells. Virology 236:104-109. [DOI] [PubMed] [Google Scholar]
- 71.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reference deleted.
- 73.Wong, J. K., M. Hezareh, H. F. Günthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 74.Wu, Y., and J. W. Marsh. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J. Virol. 77:10376-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, Z. Q., S. W. Wietgrefe, Q. Li, M. D. Shore, L. Duan, C. Reilly, J. D. Lifson, and A. T. Haase. 2004. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. USA 101:5640-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, T., D. Muthui, S. Holte, D. Nickle, F. Feng, S. Brodie, Y. Hwangbo, J. I. Mullins, and L. Corey. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]