Abstract
Vectors based on the chicken embryo lethal orphan (CELO) avian adenovirus (Ad) have two attractive properties for gene transfer applications: resistance to preformed immune responses to human Ads and the ability to grow in chicken embryos, allowing low-cost production of recombinant viruses. However, a major limitation of this technology is that CELO vectors demonstrate decreased efficiency of gene transfer into cells expressing low levels of the coxsackie-Ad receptor (CAR). In order to improve the efficacy of gene transfer into CAR-deficient cells, we modified viral tropism via genetic alteration of the CELO fiber 1 protein. The αv integrin-binding motif (RGD) was incorporated at two different sites of the fiber 1 knob domain, within an HI-like loop that we identified and at the C terminus. Recombinant fiber-modified CELO viruses were constructed containing secreted alkaline phosphatase (SEAP) and enhanced green fluorescent protein genes as reporter genes. Our data show that insertion of the RGD motif within the HI-like loop of the fiber resulted in significant enhancement of gene transfer into CAR-negative and CAR-deficient cells. In contrast, CELO vectors containing the RGD motif at the fiber 1 C terminus showed reduced transduction of all cell lines. CELO viruses modified with RGD at the HI-like loop transduced the SEAP reporter gene into rabbit mammary gland cells in vivo with an efficiency significantly greater than that of unmodified CELO vector and similar to that of Ad type 5 vector. These results illustrate the potential for efficient CELO-mediated gene transfer into a broad range of cell types through modification of the identified HI-like loop of the fiber 1 protein.
The chicken embryo lethal orphan (CELO) virus is representative of type 1 fowl adenoviruses (Ads) (3, 24, 25, 34). Similar to mammalian Ads, the CELO virus has an icosahedral capsid comprised of hexons and pentons as major subunits (35). The viral capsid contains a double-stranded DNA genome that has substantial homology with mastadenovirus genomes. Like many Ads, CELO uses the fiber proteins of its capsid to gain entry into host cells through binding to the coxsackie-Ad receptor (CAR).
Recent studies have shown that CELO virus can be used as a gene transfer vehicle for gene therapy (24, 34) and gene vaccination (10). However, the usefulness of CELO-based vectors is significantly limited by their poor transduction efficiency of CAR-deficient cells compared to CAR-positive cells (24, 35).
A general approach to solve the problem of gene transfer to specific cell types is genetic alteration of Ad fiber proteins by construction of chimeric fibers or by introducing heterologous receptor binding motifs into the fiber (8, 17, 26, 37, 39). This strategy has been used successfully for a number of applications involving retargeting human Ads to receptors other than CAR (8, 37). Thus, we set out to explore the possibility of improving transduction of CAR-deficient cells by CELO vectors by genetically altering the CELO fiber protein. Unlike other Ads, the CELO virus is characterized by two fibers of different lengths at each vertex of the virion. The CELO long fiber (fiber 1) is responsible for recognition of CAR, and unlike the CELO short fiber (fiber 2), it is dispensable for both virus assembly and penetration into permissive cells (35). Thus, the fiber 1 protein is an ideal target for genetic modification that might result in expanded virus tropism.
To avoid creating significant changes in the CELO virus natural tropism, we chose to modify surface-exposed portions of the fiber 1 protein without significantly disturbing the knob domain. Previous studies suggested that this might be achieved by introducing receptor binding motifs (e.g., the integrin-binding motif RGD or the heparan sulfate-containing receptor binding motif pK7) within a surface-exposed loop of the fiber or at its C terminus (8, 37, 39). However, due to a lack of structural information about CELO fiber domains, it was not obvious where places appropriate for modification were located within the CELO fiber 1 protein. For human Ad vectors, two sites of modification that have been well characterized are the C terminus of the fiber and the HI loop of the fiber knob (1, 8, 19, 37, 39). To determine whether the CELO fiber 1 knob domain contains an HI-like loop, we compared the primary sequence and secondary-structure prediction of the CELO fiber 1 protein with the X-ray crystal structure of the Ad type 5 (Ad5) fiber protein (42). An HI-like surface-exposed loop in the CELO fiber 1 knob was localized to amino acid residues 661 to 666.
Based upon previous studies, we chose to insert the integrin-binding motif RGD (5, 6, 15, 17, 21, 36) as a heterologous receptor binding sequence within the identified HI-like loop, as well as at the C terminus of the fiber 1 protein directly before the stop codon. Functional analysis of these two types of CELO vector mutants demonstrated that insertion of RGD into the HI-like loop of the CELO fiber, but not into the C terminus, dramatically increases the ability of the virus to transduce CAR-negative (MCF-7 and CHO) and CAR-deficient (ACHN and H596) cells, which are normally resistant to CELO transduction. Moreover, CELO virus modified at the HI-like loop provided enhanced gene delivery into primary rabbit mammary gland cells in vitro and provided increased levels of transgene expression in milk when injected into the channel of rabbit mammary glands in vivo.
In summary, the HI-like loop that we have identified in the CELO fiber 1 knob provides an appropriate site for successful incorporation of receptor binding sequences to retarget CELO virus infection. Such fiber-modified CELO vectors greatly broaden the potential usefulness of CELO virus for experimental and clinical applications.
MATERIALS AND METHODS
Construction of recombinant plasmids.
To create mutations in the CELO Ad fiber 1 gene (in the C terminus and in the HI loop), PCR-based mutagenesis was used. Two pairs of primers were designed for mutagenesis in the HI loop: primer pRGDhi1 (5′-CCT GGT CAC GTA TAC CAA G-3′), primer pRGDhi2 (5′-CGT ATT TCG TCG TCT GCA CGC AGA AAC AGT CTC CGC GGC AGT CAC AGG GCG ATT TCG CCG CTG CAA C-3′), primer pRGDhi3 (5′-CTG CAG GAG TTA ACA AGT TC-3′), and primer pRGDhi4 (5′-GGT GTG ACT GCC GCG CAG ACT GTT TCT GCG TGC AGA CGA CGA AAT ACG-3′). The nucleotides encoding the RGD motif are underlined. Primers pRGDhi2 and pRGDhi4 are complementary to the site of the mutation; primers pRGDhi1 and pRGDhi3 are complementary to DNA sequences outside the mutation site and were designed as partners for pRGDhi2 and pRGDhi4, respectively. Generation of the mutation was accomplished via two sequential PCRs. First, primer pairs pRGDhi1-pRGDhi2 and pRGDhi3-pRGDhi4 were used to amplify two DNA fragments from CELO wild-type DNA overlapping at the mutation site. These two fragments were then used as the template for a second PCR with primer pair pRGDhi1-pRGDhi3. The DNA fragment generated via the second PCR contained the inserted RGD motif. To insert the RGD-containing segment of the fiber gene into pCRVA (bp 23830 to 33358 of the CELO genome), the PCR product and pCRVA were digested with Bst1107I and HpaI. The obtained fragment (242 bp) was used to replace the analogous segment in pCRVA. The DNA of the new plasmid, pCRVA-hiRGD, was sequenced to confirm the presence of the mutation.
The plasmid pSCELO-HIRGD was constructed by cloning the CELO fragment (9,548 bp) from the pCRVA-hiRGD plasmid digested with EcoRV into EcoRV-digested plasmid pDL113 containing a ClaI (16,254-bp) and EcoRV (23,830-bp) fragment. The recombinant plasmid pCELO-HIRGD, containing the modified fiber 1 gene, was generated by homologous recombination in Escherichia coli, as described below.
For comparative analyses of virus transduction, secreted alkaline phosphatase (SEAP) or enhanced green fluorescent protein (EGFP) reporter gene expression cassettes were introduced into the region of deletion of the CELO genome (bp 41731 to 43685) by homologous recombination in E. coli between pCELO-HIRGD and a shuttle plasmid containing an expression cassette (pCShCMV-SEAP or pCShCMV-EGFP) as described below. Genomic clones (pCELOSEAP-HIRGD or pCELOEGFP-HIRGD) were sequenced and analyzed by PCR prior to transfection into LMH cells.
For mutagenesis of the C terminus of the CELO fiber 1 protein, three primers were designed: pRGD1 (5′-ACC TGG TCA CGT ATA CCA AG-3′), pRGD2 (5′-CAG GAG TTA ACA AGT TCA CGC AGA AAC AGT CTC CGC GGC AGT CAC ATT GAT AGT ACC CCA GAT AAG TAA ACG-3′), and pRGD3 (5′-GTC GGT CTT GAC CCC GAT CG-3′). The nucleotides encoding the RGD motif are underlined. Primer pRGD2 is complementary to the insertion sequence; primers pRGD1 and pRGD3 are complementary to DNA sequences outside of the insertion site. Generation of the mutation was accomplished via two sequential PCRs. Primer pair pRGD1-pRGD2 was used for amplification of the DNA fragment from the CELO wild-type DNA containing the mutation site. This fragment (263 bp) was then used as a primer for a second PCR with primer pRGD3. The DNA fragment generated by the second PCR contained the RGD motif. The PCR product (242 bp) was digested with Bst1107I and HpaI and cloned into pCRVA digested with the same restriction endonucleases. The plasmid pCRVA-RGD was sequenced to confirm the presence of the mutation.
The plasmids containing the RGD insertion at the C terminus of CELO fiber 1 (pSCELO-RGD, pCELO-RGD, pCELOSEAP-RGD, and pCELOEGFP-RGD) were constructed as described above for generation of pCELOSEAP-HIRGD and pCELOEGFP-HIRGD.
Generation of recombinant Ad genomes by homologous recombination in E. coli.
Recombinant CELO Ad genomes containing the modified fiber 1 gene were generated by homologous recombination in E. coli as described by Michou et al. (25). Briefly, the vector (pSCELO-HIRGD or pSCELO-RGD) was linearized with SpeI and mixed with a plasmid containing the genome of the CELO avian Ad (with the EcoRV fragment, bp 41731 to 43685, deleted) digested with NotI, and the DNA mixture was electroporated into E. coli BJ5183 cells using a MicroPulser (Bio-Rad). Bacterial colonies obtained after 18 to 24 h of growth at 30°C were analyzed by PCR and restriction endonuclease digestions. Recombinant plasmids with the reported genes (pCELOSEAP-HIRGD and pCELOSEAP-RGD or pCELOEGFP-HIRGD and pCELOEGFP-RGD) were generated by homologous recombination in E. coli BJ5183 between a shuttle plasmid containing an expression cassette, pCShCMV-SEAP or pCShCMV-EGFP, linearized with AscI and a plasmid (pCELO-HIRGD or pCELO-RGD) linearized with PacI. Genomic clones were sequenced and analyzed by PCR analysis (see Fig. 2b).
FIG. 2.
Generation of CELO vectors with genetic alteration of fiber 1 protein. (a) Schematic diagram of the CELO fiber 1 (long-fiber) protein and the sites chosen for modification. The fiber 1 protein includes three regions: the tail, the shaft, and the knob. Within the knob domain, the HI loop is indicated by a gray circle and the C terminus by a white circle. The amino acid sequences of insertion sites within the HI loop and the C terminus and of the RGD-containing inserted sequences are shown. (b) Construction of the plasmid pCELOSEAP-HIRGD by homologous recombination in E. coli BJ5183 cells. pCELOSEAP-HIRGD contains the SEAP gene driven by the cytomegalovirus promoter in a CELO vector backbone that contains the RGD insertion within the HI loop of the fiber 1 gene (pCELO-HIRGD). See Materials and Methods for details of construction.
Viruses.
The recombinant Ad5-SEAP and CELO-SEAP or Ad5-EGFP and CELO-EGFP vectors containing expression cassettes [the human cytomegalovirus promoter, the SEAP or EGFP reporter gene, and a poly(A)/stop signal] integrated in nonessential regions of the virus genome were generated in our previous study (24). For the present study, the following viruses were created: CELOSEAP-RGD and CELOEGFP-RGD, with the high-affinity RGD sequence CDCRGDCFC incorporated at the C terminus of the fiber protein, and CELOSEAP-HIRGD and CELOEGFP-HIRGD, with the RGD sequence CDCRGDCFC incorporated at the HI loop of the fiber protein. Avian recombinant CELO Ads were grown in specific-pathogen-free chicken embryos and purified by double cesium chloride gradient centrifugation. The yields of CELO recombinant viruses were not affected by fiber modification. The optical densities (OD) of Ad vector stocks were as follows: (i) 7 OD units/ml and 8.4 OD units/ml for CELO-SEAP and CELO-EGFP, respectively; (ii) 6.8 OD units/ml and 7.3 OD units/ml for CELOSEAP-HIRGD and CELOEGFP-HIRGD, respectively; and (iii) 10.8 OD units/ml and 6.4 OD units/ml for CELOSEAP-RGD and CELOEGFP-RGD, respectively. Each virus was verified by PCR to contain the correct insert. Restriction analysis of DNA from each virus further confirmed the identities of the viruses with unique restriction sites. Virus samples were analyzed for the titer of viral particles (vp) (absorbance at 260 nm) using a conversion factor of 1 OD unit equaling 1012 vp/ml.
Cell culture.
The human kidney 293, chicken hepatoma LMH, human lung cancer H1299, human colon cancer HCT-116, human breast cancer MCF-7, Chinese hamster ovary CHO, human renal carcinoma ACHN, and human adenosquamous bronchogenic carcinoma H596 cell lines were used in the experiments. CHO cells were propagated in a 50:50 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium (DMEM/F-12) supplemented with 10% fetal calf serum (HyClone), l-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 mg/ml). All other cell lines were grown in DMEM supplemented with 10% fetal bovine serum (HyClone), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine at 37°C in a humidified atmosphere with 5% CO2.
Primary cell lines.
Rabbit primary breast cells were cultured in a 50:50 mixture of DMEM/F-12 supplemented with 10% fetal calf serum (HyClone), 10 ng/ml epidermal growth factor (Gibco BRL), 2 mM l-glutamine, and 50 μg/ml gentamicin.
Ad-mediated gene transfer experiments.
To determine the efficiency of reporter gene transfer by Ad vectors into various cell lines, 105 cells per well were seeded in a 24-well plate and incubated overnight at 37°C. On the next day, the cell monolayers were infected for 2 h at a multiplicity of infection (MOI) of either 100, 1,000, or 10,000 vp/cell with either virus in 200 μl of medium. Two hours later, the cells were washed once with medium and added to growth medium. SEAP activity in the culture medium was measured 48 h postinfection using an iEMS Reader MF (Thermo Labsystems, Finland) as described previously (24). Experiments were performed in triplicate.
For Ad-mediated gene transfer into human cell lines involving blocking conditions, Ad5 fiber protein at 10-μg/ml final concentration was incubated with the cells at 37°C in transduction medium for 30 min prior to the addition of the virus. Following infection by Ad5-SEAP, CELO-SEAP, and CELOSEAP-HIRGD for 2 h at 1,000 vp/cell, the cells were maintained at 37°C in an atmosphere of 5% CO2.
In order to determine the number of internalized vp, virus bound to the outsides of cells was removed by incubation in 0.2 M acetic acid-150 mM NaCl for 1 min at room temperature, followed by three washes with PBS prior to cell lysis, DNA extraction, and real-time PCR analysis.
RT-PCR analysis.
Expression of CAR, αv integrin, β3 integrin, and β5 integrin genes in human cell lines was analyzed using reverse transcription (RT)-PCR. Total RNA was isolated using TRIZOL reagent (Invitrogen). RT was performed using an RT System (Promega) according to the instructions of the manufacturer. After the RT reaction, cDNA encoding the human CAR and integrin (αv, β3, and β5) genes was amplified by PCR with the following primers: CAR, forward (5′-AGC CTT CAG GTG CGA GAT GTT ACG-3′) and reverse (5′-TAG GAC AGC AAA AGA TGA TAA GAC-3′); αv integrin, forward (5′-GAC TGT GTG GAA GAC AAT GTC TGT AAA CCC-3′) and reverse (5′-CCA GCT AAG AGT TGA GTT CCA GCC-3′); β3 integrin, forward (5′-GAG GAT GAC TGT GTC GTC AG-3′) and reverse (5′-CTG GCG CGT TCT TCC TCA AA-3′); β5 integrin, forward (5′-GCC TAT CTC CAC GCA CAC TG-3′) and reverse (5′-AGA CTC CGA CCC TTC CTG AC-3′); and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), forward (5′-TCT AGA CGG CAG GTC AGG TCC ACC-3′) and reverse (5′-CCA CCC ATG GCA AAT TCC ATG GCA-3′). The PCR conditions were as follows: human CAR, 30 s at 94°C, 40 s at 65°C, and 60 s at 72°C for 40 cycles; human αv integrin, 40 s at 94°C, 40 s at 55°C, and 40 s at 72°C for 40 cycles; human β3 integrin, 30 s at 94°C, 40 s at 65°C, and 60 s at 72°C for 40 cycles; human β5 integrin, 30 s at 94°C, 40 s at 58°C, and 60 s at 72°C for 40 cycles; and human GAPDH, 30 s at 94°C, 40 s at 65°C, and 60 s at 72°C for 40 cycles.
PCR products were analyzed by electrophoresis in a 1.5% agarose gel. Amplification of the GAPDH gene was used for semiquantitative estimation of reverse-transcribed mRNA levels.
Quantitative RT-PCR for Ad5 and CELO genomes.
To determine the adenoviral copy number, real-time PCR was performed as described previously (34). Briefly, total DNA from cells transduced with Ad5-SEAP, CELO-SEAP, or CELOSEAP-HIRGD was extracted with a DNA purification kit (Promega) according to the manufacturer's instructions. To amplify adenoviral DNA, the following primers and probes were designed: Ad5 forward primer (5′-CTG GGC AAT GGT CGC TAT GT-3′), Ad5 reverse primer (5′-AGA AGG TGG CGT AAA GGC AAA T-3′), and probe for 5′-R6G-CAT CCA GGT GCC TCA GAA GTT CTT TGC CA-BHQ2-3′; CELO forward primer (5′-TCA AGA CCG ACG GAA GCA TT-3′), CELO reverse primer (5′-TTC TTG CCG CTG CTA CCG TC-3′), and probe (5′-FAM [6-carboxyfluorescein]-AGC GCG CCG TCC GCA TTG-BHQ1-3′). Known quantities of Ad5 and CELO were serially diluted and added to DNA isolated from cells. The quantitative PCR was initiated with a hot start at 94°C for 10 min. Amplification occurred during 50 cycles, with each cycle consisting of 20 s at 94°C and 50 s at 62°C in an iQCycler (Bio-Rad).
Quantitative RT-PCR for CAR.
Total RNA was extracted as described above, and RNA (1.0 μg) was converted to cDNA by RT, using an RT System (Promega). Real-time PCR was performed in duplicate to determine the relative quantities of GAPDH and CAR amplicons. Primers for determination of the CAR and GAPDH were as follows: CAR forward (5′-AAA TTT ACG CTT AGT CCC GAA GAC-3′), CAR reverse (5′-CCT TCT GAT TAT CAG CTG GTG ATA TC-3′), and probe (5′-FAM-CCA CTC GAT GTC CAG CGG TCC CT-BHQ1 -3′); GAPDH forward, (5′-GGT TTA CAT GTT CCA ATA-3′), GAPDH reverse, (5′-ATG GGA TTT CCA TTG ATG ACA AG-3′), and GAPDH probe (5′-R6G-CGT TCT CAG CCT TGA CGG TGC CAT-BHQ2 -3′). The CAR mRNA level of H1299 cells was defined as 100% for the analysis of other samples. The CAR mRNA levels in other cell lines are presented in the figures as the calculated percentages of mRNA relative to the levels in H1299 cells (11). GAPDH was included as a control reference gene to normalize the amount and quality of cDNA. CAR mRNA expression was normalized to GAPDH mRNA using the standard-curve method (14). Standard curves were constructed using dilutions of cDNA (50, 25, 10, and 5 ng per reaction). Quantitative real-time RT-PCR was performed using an ANA-32 instrument (Syntol, Russia).
In vivo experiments.
All animal work was carried out in accordance with the Regulation of Laboratory Practice and the Sanitary Epidemic and Veterinary legislation of the Russian Federation. For analysis of Ad-mediated gene transfer in vivo, female rabbits (strain Chinchilla; age, 15 months) were injected with recombinant Ads in the mammary gland, at 1010 vp/rabbit, on the first day of the lactation period. SEAP activities were determined in milk on the third day of the lactation period. Prior to being milked, the rabbits were injected (intravenously) with 3 U ocytocin (alpha hypophamine). The samples of milk were diluted with an equal volume of buffer (10 mM CaCl2, 100 mM sodium acetate, pH 6.0) and centrifuged at 10,000 × g for 10 min to separate the milk sera. SEAP activity was measured in prepared milk sera as described previously (24).
RESULTS
Identification of an HI-like motif in the CELO fiber 1 protein structure.
The first stage of our work was to predict an appropriate place in the CELO fiber 1 knob domain for introduction of the integrin-binding RGD motif. For human Ads, the HI loop of the fiber knob domain has proved to be an ideal site for incorporation of the RGD sequence (the full incorporated sequence is CDCRGDCFC) (8). Unfortunately, there is no primary sequence similarity between the fiber knob domains of CELO and human Ads. Therefore, we used secondary-structure prediction coupled with analysis of limited sequence similarity to identify the amino acid residues of the CELO fiber 1 knob domain that correspond to the HI loop of the human Ad5 fiber knob domain (Fig. 1). The programs PSI-PRED and JPRED (4, 16) were used to predict the secondary structure. Good correspondence was demonstrated for the results of prediction with both programs, as well as X-ray crystallographic analysis of the human Ad5 fiber knob domain (Fig. 1, bottom). The C-terminal 14 amino acid residues of the CELO fiber 1 knob domain showed limited sequence similarity to the corresponding region of the human Ad5 fiber knob domain. The PSI-PRED program predicted a discontinuous beta strand in this region of the CELO fiber 1 protein, which corresponds to the J beta strand of the Ad5 fiber. In human fiber knob domains, the J beta strand plays a role in intersubunit interaction (42). Based upon the identification of the J beta strand, the next predicted beta strands of the CELO fiber 1 knob domain, which are closer to the N terminus, were named I, H, and G. Amino acid residues 661 to 666, located between the predicted H and I beta strands of the CELO fiber 1 knob domain, were determined to correspond to the surface-exposed HI loops of human Ad fiber knob domains.
FIG. 1.
Amino acid sequences of the C-terminal portions of the CELO large fiber (top) and human Ad5 fiber knob (bottom) domains. The numbering of amino acids shown in the upper line is according to Q64761 (top) and P11818 (bottom) GenBank entries. The letters E above the sequences indicate beta strands predicted by both the PSI-PRED and JPRED programs (top) or by both programs and X-ray analysis (bottom). The italicized letters E indicate beta strands predicted only by PSI-PRED (top) or detected only by X-ray analysis (bottom). The letters G, H, I, and J in the line under the numbering indicate the names of the beta strands in the structure of the human Ad5 fiber knob domain (Protein Data Bank accession no. 1KNB) and predicted for the CELO large fiber knob domain. Boldface type indicates amino acid residues that are identical or similar in the two proteins. Pro661 and Val662 residues in the predicted HI loop of the CELO large fiber knob domain are underlined.
Construction of fiber-modified recombinant CELO Ads.
In the next stage of our study, we constructed two types of fiber-modified CELO vectors via PCR-based mutagenesis. One modified virus contained the RGD sequence insertion (CDCRGDCFC) at the C terminus of the fiber 1 protein, and the other contained the RGD sequence insertion within the predicted HI-like loop (between the Pro661 and Val662 residues) of the fiber 1 knob (Fig. 2a). In order to measure the efficiencies of transduction by the modified viruses in comparison to wild-type CELO, SEAP and EGFP genes were used as reporter genes. All viruses were constructed by the method described by Michou et al. (25). Briefly, pCELO-HIRGD (with RGD inserted in the HI-like loop) or pCELO-RGD (with RGD inserted at the C terminus) was generated by homologous recombination in E. coli between the pSCELO-HIRGD or pSCELO-RGD plasmid, which contained a fragment of the CELO genome with the fiber modification (bp 16254 to 33358), and the plasmid pCELOdlRV, containing the CELO genome with a deletion of bp 41731 to 43685 (Fig. 2b). pCELO-RGD and pCELO-HIRGD, containing SEAP or EGFP reporter genes, was generated by homologous recombination in E. coli between the pCELO-HIRGD or pCELO-RGD plasmid, which contained the full-length CELO genome with the modified fiber genes, and a pCShCMV shuttle vector bearing the SEAP or EGFP expression cassette flanked by CELO DNA sequences. The resulting plasmids were transfected into LMH cells for preparation of fiber-modified vectors. The identities of the viruses were confirmed by PCR and restriction endonuclease mapping (data not shown).
A CELO-based vector containing an RGD motif in the HI loop is capable of effective gene transfer into CAR-negative cells.
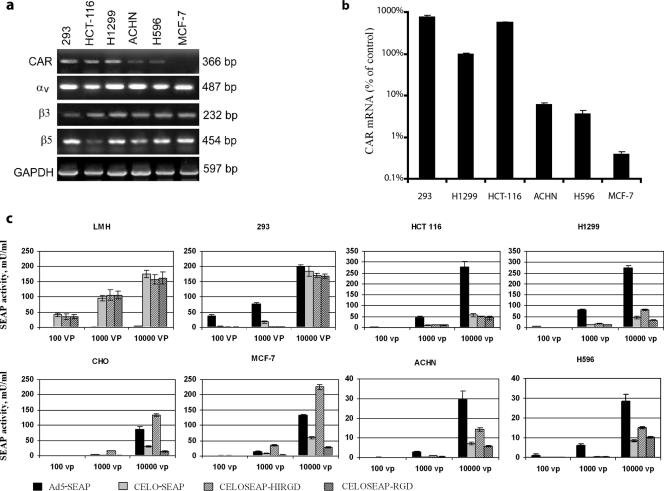
Expansion of Ad5 tropism through incorporation of an RGD motif into the fiber protein has previously been shown to increase virus uptake and transgene expression in CAR-negative cells (37, 41). We therefore examined whether CELO fiber-modified vectors could also effectively transduce genes into CAR-negative cells. We tested four groups of mammalian cell lines that were characterized by different levels of CAR expression and by expression of specific RGD-binding receptors (αvβ5 and αvβ3 integrins): (i) permissive cells that support viral replication (LMH cells for CELO virus and 293 cells for Ad5), (ii) CAR-positive cells (HCT-116 and H1299), (iii) CAR-deficient cells (ACHN and H596), and (iv) CAR-negative cells (CHO and MCF-7). The levels of CAR and integrin expression in 293, HCT-116, H1299, ACHN, H596, and MCF-7 cells were determined via semiquantitive RT-PCR (Fig. 3a). We found that 293 cells express high levels of CAR and of αv and β5 integrins, while expression of β3 is moderate. HCT-116 and H1299 cells express high levels of both CAR and integrins, with the exception of moderate expression of β5 in HCT-116 cells. ACHN and H596 cells demonstrated moderate levels of CAR expression but high levels of integrin expression. MCF-7 cells were CAR negative but showed high levels of αv, β5, and β3 expression. As standard CAR-negative cells, we used the well-characterized CHO cell line (7, 27).
FIG. 3.
Comparison of efficiencies of gene transfer into different mammalian cell lines provided by CELO and Ad5 vectors. (a) RT-PCR analysis of CAR and integrin (αv, β3, and β5) mRNA expression in human cell lines. Expression of GAPDH was used as a positive control to confirm the quality and amount of cDNA in each sample. (b) Quantification of CAR mRNA levels in human cell lines by real-time RT-PCR. CAR mRNA expression was normalized to GAPDH expression and is shown relative to the value obtained for H1299 cells (arbitrarily set at 100%). (c) SEAP activities in the culture media of cells transduced with adenoviral vectors. The indicated cell lines were incubated with SEAP-expressing viruses at an MOI of 100, 1,000, or 10,000 vp/cell for 48 h. SEAP activity in the culture supernatant was determined at the end of the incubation. LMH and 293 cells were incubated with virus for 24 h. The error bars represent standard deviations from the mean.
To more accurately measure CAR expression levels, quantitative real-time RT-PCR was used in selected cell lines. CAR mRNA levels were normalized to GAPDH expression levels, and the results were expressed as the percentage of mRNA, with the CAR mRNA level in H1299 cells defined as 100% (11, 14). Significant variation in CAR mRNA levels between different cell lines was observed by quantitative real-time PCR, and these results were consistent with the standard RT-PCR results shown in Fig. 3a. The quantitative analysis confirmed that CAR mRNA is expressed at high levels in 293, HCT-116, and H1299 cells; at low levels in ACHN and H596 cells; and at nearly undetectable levels in MCF-7 cells (Fig. 3b). Therefore, the set of cell lines chosen for our gene transfer experiments represents a full range of CAR expression profiles with moderate to high levels of integrin αvβ5 and αvβ3 expression.
To determine the efficiency of gene transfer provided by CELO fiber-modified vectors (CELOSEAP-HIRGD and CELOSEAP-RGD), cells were infected at an MOI of 100, 1,000, or 10,000 vp per cell. As controls, we used unmodified Ad5 (Ad5-SEAP) and CELO (CELO-SEAP) vectors. The efficiency of gene transfer was determined by measuring SEAP activity in the culture medium of the transduced cells (Fig. 3c).
All constructed vectors demonstrated a high level of SEAP expression following infection of permissive cells (Ad5 in 293 cells and CELO vectors in LMH cells) (Fig. 3c). At an MOI of 104 vp/cell, all CELO vectors (both unmodified and fiber modified) were about fivefold less effective in transducing CAR-positive cells (HCT-116 and H1299) than Ad5-SEAP was (Fig. 3c). For all vectors tested, infection of CAR-deficient (low CAR expression) cells (ACHN and H596) was significantly lower (P < 0.05) than infection of CAR-positive cells (10-fold for Ad5-SEAP and 4- to 6-fold for CELO vectors) (Fig. 3c). Nevertheless, CELOSEAP-HIRGD was more effective in transducing CAR-deficient cells (ACHN and H596) than either unmodified CELO or CELOSEAP-RGD (P < 0.05) and was only twofold less effective than the Ad5 vector. In contrast, in CAR-negative cells (MCF-7 and CHO), CELOSEAP-HIRGD was 1.5- to 3-fold more efficient than Ad5-SEAP and 4-fold more efficient than the unmodified CELO-SEAP vector. CELOSEAP-RGD, containing the RGD motif insertion at the fiber 1 C terminus, did not improve gene transfer into CAR-negative cells compared to the unmodified CELO-SEAP vector. Transduction with CELOSEAP-RGD actually resulted in less SEAP expression than CELO-SEAP in most cell types.
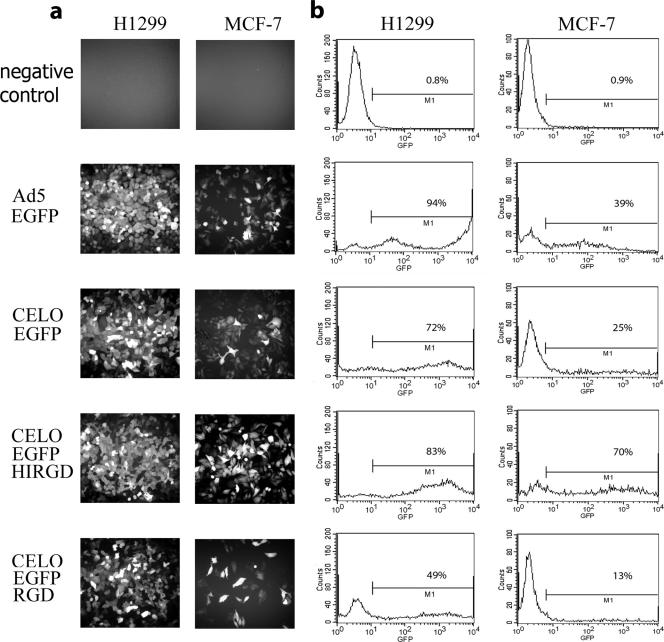
To confirm the data obtained using the SEAP reporter gene, we carried out similar experiments with CELO vectors bearing the EGFP gene. Representative CAR-positive (H1299) and CAR-negative (MCF-7) cell lines were infected with CELOEGFP-HIRGD, CELOEGFP-RGD, CELO-EGFP, and Ad5-EGFP vectors. The efficiency of transduction was measured by fluorescence microscopy (Fig. 4a) and flow cytometry (Fig. 4b). Consistent with our findings using the SEAP reporter, CELOEGFP-HIRGD demonstrated the highest transduction efficacy in CAR-negative MCF-7 cells but provided less efficient gene transfer into CAR-positive H1299 cells than the Ad5-based vector. The significant increase in EGFP expression following infection of MCF-7 cells with CELOEGFP-HIRGD (70%) compared to CELO-EGFP (25%) clearly demonstrates that RGD modification of the HI loop of CELO fiber 1 expands virus tropism to include CAR-negative cells. CELOEGFP-RGD transduced both CAR-positive and CAR-negative cell lines less efficiently than the other CELO vectors, suggesting that the activity of the virus particles was negatively affected by the RGD addition at the C terminus of the fiber protein. Based upon this, CELO vectors modified at the C terminus were not tested further.
FIG. 4.
Ad-directed EGFP expression in human H1299 and MCF-7 cell lines. (a) Fluorescence microscopy of H1299 and MCF-7 cells following infection with Ad5-EGFP and with CELO-EGFP, CELOEGFP-RGD, and CELOEGFP-HIRGD viruses at an MOI of 10,000 vp/cell for 48 h (Olympus IX-71 microscope; magnification, ×350; Olympus, Germany). Representative fields are presented. (b) Quantitative assessment of Ad vector-mediated EGFP expression. The results of flow cytometric analysis of H1299 and MCF-7 cells infected with the Ad5 and CELO vectors indicated in panel a are shown. The cells were infected for 48 h at an MOI of 10,000 vp/cell. The percentage of EGFP-positive cells as defined by gate M1 is given in each panel.
Surprisingly, while the Ad5-SEAP vector gave approximately fivefold-higher levels of SEAP expression in CAR-positive cells than the CELOSEAP-HIRGD vector (Fig. 3c, HCT116 and H1299), transduction of H1299 with Ad5-EGFP and CELOEGFP-HIRGD vectors (Fig. 4b) resulted in comparable levels of EGFP-expressing cells (Fig. 4b) (94% for Ad5-EGFP versus 83% for CELOEGFP-HIRGD). To explain this difference between the SEAP and EGFP experiments, we estimated the number of Ad intracellular genomes accumulated after infection. H1299 and MCF-7 cells were infected with Ad5-EGFP, CELOEGFP-HIRGD, and CELO-EGFP vectors at an MOI of 104 for 2 h. At 48 h after infection, the number of Ad genomes was measured by real-time PCR using primer and probe sets specifically designed to detect either CELO or Ad5 sequences, as described by Stevenson et al. (34). We have found that, on average, each transduced H1299 cell accumulated about five times more Ad5-EGFP genomes than CELOEGFP-HIRGD genomes (Fig. 5). These data provide a potential explanation for the difference between the levels of SEAP expression and percentages of EGFP-expressing CAR-positive cells for Ad5 and CELO vectors. There were no discrepancies noted in the results of EGFP and SEAP tests for CELO and Ad5 vectors in the context of MCF-7 cells.
FIG. 5.
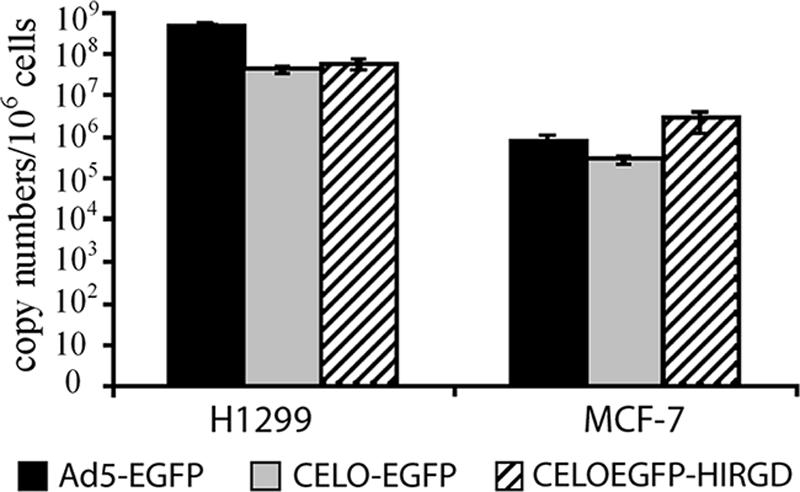
Quantitative analysis of Ad vector DNA accumulated after transduction of CAR-negative and CAR-positive cells. Human H1299 and MCF-7 cells were exposed for 2 h to 10,000 vp/cell of Ad5-EGFP, CELO-EGFP, and CELOEGFP-HIRGD. The virus-containing medium was then removed, and the cells were washed twice with 0.5 ml of ice-cold phosphate-buffered saline and incubated in fresh medium for 48 h. Total DNA was extracted and used in real-time PCR to determine the number of Ad copies present in the cell population. The error bars represent standard deviations from the mean.
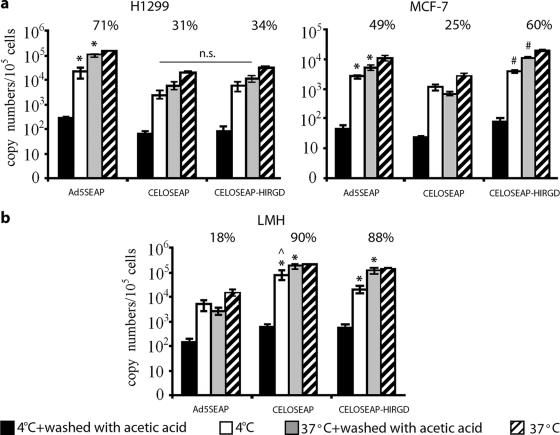
Insertion of an RGD motif into the HI-like loop of CELO fiber 1 affects both attachment and internalization of CELO in CAR-negative cells.
To evaluate whether insertion of the RGD motif in the HI-like loop of CELO fiber 1 influences attachment and/or internalization of the virus, we infected H1299 and MCF-7 cells with wild-type or fiber-modified (with RGD within the HI loop) CELO vectors. The cells were incubated with virus at 4°C (allowing virus binding only) or 37°C (allowing virus binding and internalization) for 90 min, and surface-bound virus was then removed by a mild acetic acid wash. The number of surface-bound or internalized viral particles was determined by measurement of the viral genome copy number by real-time PCR as described above. As predicted, CAR-negative cells bound more fiber-modified virus (CELOSEAP-HIRGD) than wild-type viruses (Ad5-SEAP and CELO-SEAP) (P < 0.05). In contrast, CAR-positive cells accumulated more Ad5-SEAP than CELOSEAP-HIRGD and CELO-SEAP (P < 0.05) (Fig. 6a). Internalization of CELOSEAP-HIRGD in CAR-negative MCF-7 cells was highly efficient, with 60% of the cell-associated virus becoming internalized within 90 min, whereas only 25% of cell-associated CELO-SEAP and 49% of Ad5-SEAP were internalized during the same amount of time. In contrast, there was no difference in the efficiencies of internalization of CELOSEAP-HIRGD and CELO-SEAP in CAR-positive cells. Attachment of CELOSEAP-HIRGD to permissive LMH cells showed a slight (but statistically significant) reduction in comparison to CELO-SEAP (Fig. 6b); however, the two viruses were internalized with similar efficiencies in this cell line. Taken together, these results indicate that insertion of RGD within the HI-like loop improves both attachment and internalization of CELO virus specifically in CAR-negative cells.
FIG. 6.
Analysis of virus internalization. (a) Binding and internalization of Ad5-SEAP and CELO vectors in CAR-positive (H1299) and CAR-negative (MCF-7) cells. *, significant difference between Ad5-SEAP and CELO vectors (P < 0.01); #, significant difference between CELO-SEAP and CELOSEAP-HIRGD (P < 0.05); n.s., statistically nonsignificant. (b) Binding and internalization of Ad5-SEAP and CELO vectors in LMH cells permissive for CELO (but not for Ad5). *, significant difference between CELO vectors and Ad5-SEAP (P < 0.005);, significant difference between CELO-SEAP and CELOSEAP-HIRGD (P < 0.05). H1299, MCF-7, and LMH cells were incubated with the indicated viruses at 1,000 vp/cell at 37°C (allowing internalization) or 4°C (allowing binding but not internalization) for 90 min. The virus was then removed, and the cells were washed with phosphate-buffered saline (PBS). A portion of each sample was then treated with 0.2 M acetic acid to remove virus bound to the cell surface, washed with PBS, and lysed. DNA was extracted, and real-time PCR analysis of Ad5 and CELO genomic sequences was performed. The percentages of total particles that had internalized are indicated. The percentages represent the numbers of vp associated with cells (at 37°C) after the cells were washed with acetic acid divided by the total number of Ad particles that attached to the cells at the same temperature. The error bars represent standard deviations from the mean.
The CELOSEAP-HIRGD virus has greater tolerance to blocking of CAR receptors than the unmodified CELO virus.
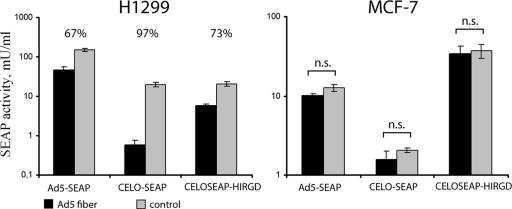
Our next goal was to examine whether blocking of CAR receptors on the cell surface influences the efficiency of gene transfer by the CELOSEAP-HIRGD vector. For this purpose, H1299 and MCF-7 cells were used in an assay based upon inhibition of CAR-mediated gene delivery by soluble (non-virus-associated) Ad5 fiber protein, which efficiently blocks Ad binding to CAR (8).
As shown in Fig. 7, transduction of the SEAP reporter gene by all recombinant CELO viruses and Ad5 was blocked by Ad5 fiber protein in H1299 cells, but not in MCF-7 cells. The same concentration of Ad5 fiber protein caused a threefold decrease in reporter gene expression for Ad5-SEAP and a 3.7-fold decrease for CELOSEAP-HIRGD. Interestingly, CELO-SEAP-mediated gene transfer was reduced 34-fold by Ad5 fiber treatment. It is likely that the difference in gene transfer efficiency by CELO recombinants when CAR is blocked can be explained by the presence of the RGD motif in the structure of CELOSEAP-HIRGD fiber 1. It is known that CELO, like Ad5, utilizes fiber-CAR interaction for attachment of the virus to the host cell surface (35). However, unlike Ad5, the penton base protein of the CELO virus does not contain an RGD motif. Therefore, it seems logical that the CELOSEAP-HIRGD virus bearing RGD possesses more resistance to Ad5 fiber-blocking treatment than the unmodified CELO virus. Our finding that CELOSEAP-HIRGD demonstrates increased resistance to CAR blocking compared to wild-type CELO further validates its ability to infect cells in a CAR-independent manner.
FIG. 7.
SEAP gene transfer by the CELOSEAP-HIRGD vector under CAR-blocking conditions. H1299 cells were preincubated with Ad5 fiber protein (10 μg/ml) for 30 min and then exposed for 2 h to Ad5-SEAP, CELO-SEAP, or CELOSEAP-HIRGD at an MOI of 1,000 vp/cell. SEAP activity was determined 48 h after infection. The percent reduction of SEAP activity in Ad5 fiber-pretreated cells compared to control cells is indicated above each pair of bars. n.s., statistically nonsignificant. The error bars represent standard deviations from the mean.
CELO-HIRGD demonstrates increased efficacy of gene transfer in vivo.
It is known that some types of primary tissues are CAR deficient or CAR negative (22, 26, 28, 30). In particular, samples of mammary gland biopsy specimens showed low levels of CAR expression (23, 32). Since CELOSEAP-HIRGD showed efficient transduction of CAR-deficient cultured cells, we next examined the ability of the modified virus to transduce genes into primary CAR-deficient cells, namely, primary mammary gland cells from rabbits. The cells were transduced with Ad5-SEAP and with CELOSEAP-HIRGD and CELO-SEAP viruses at an MOI of 100, 1,000, or 10,000 vp/cell. As expected based upon our findings in established CAR-deficient cell lines (Fig. 3c), gene transfer into rabbit primary breast cells was improved by using CELOSEAP-HIRGD compared to CELO-SEAP (Fig. 8a). The absolute levels of SEAP expression directed by all vectors were lower in the primary cells than in the established cell lines; however, the profile of SEAP expression was similar to that in CAR-deficient cells (ACHN and H596) (compare Fig. 3c and 8a).
FIG. 8.
SEAP gene transfer by fiber-modified CELO Ad into primary breast cells. (a) SEAP activity in the culture medium from primary rabbit breast cells infected with Ad5-SEAP or with CELO-SEAP or CELOSEAP-HIRGD virus. The cells were infected with the viruses at an MOI of 100, 1,000, or 10,000 vp/cell for 2 h. SEAP activity was determined 48 h postinfection. (b) SEAP activity in milk after rabbit mammary gland transduction with Ad5-SEAP or with CELO-SEAP or CELOSEAP-HIRGD virus. Rabbits (n = 9) were infected with the viruses (1010 vp) on the first day of the lactation period. SEAP activity was determined on the third day of the lactation period. SEAP activity in the milk from control rabbits (injected with phosphate-buffered saline or with the control virus, CELOdlRV (a deletion mutant without the SEAP gene), did not exceed 0.145 mU/ml (data not shown). The error bars represent standard deviations from the mean.
Having demonstrated that CELOSEAP-HIRGD can transduce rabbit primary breast cells in culture, we next tested the fiber-modified CELO virus for gene transfer efficiency in vivo. For this purpose, pregnant rabbits were used as experimental models. On the first day of the lactation period, the rabbits were injected in the mammary gland with Ad5-SEAP and with CELOSEAP-HIRGD and CELO-SEAP viruses at 1010 vp per injection. SEAP activity in milk sera was determined on the third day of the lactation period. In this system, we found that fiber-modified CELO virus was four times more effective in gene delivery than wild-type CELO virus and showed efficiency similar to that of an Ad5-based vector (Fig. 8b). Thus, the modified CELO vector that we have developed illustrates that CELO-mediated gene transfer is a promising technology that is not necessarily limited by CAR expression.
DISCUSSION
Recombinant Ad is a powerful system for mammalian gene transfer that is widely used for gene therapy applications (1, 2, 20, 29, 40). However, two factors can negatively affect the efficacy of Ad-mediated gene transfer. The first is preexisting immunity to human Ads (9, 13, 18, 33, 43), and the second is limited distribution of the Ad-specific receptor CAR on the surfaces of many cell types (22, 23, 28, 30). Use of the avian Ad CELO bypasses the immunity limitation but is still dependent upon CAR expression. It was recently shown that both drawbacks can simultaneously be partially overcome by using an alternative avian Ad CELO vector system modified by polymer coating to retarget the virus to the fibroblast growth factor receptor (34). However, CELO-mediated gene transfer was limited in this case by fibroblast growth factor receptor expression. In the current study, we set out to expand the tropism of CELO through genetic alteration of the fiber1 protein rather than using polymer-based modification. It is known that the HI loop of the fiber knob domain of human Ads is a convenient site for incorporation of different motifs for redirection of virus specificity (1, 8). Another site commonly used for modification of virus tropism is the C terminus of the fiber knob domain (37). Following identification of an HI-like loop in CELO fiber 1, we created and tested CELO viruses modified by RGD motif insertion at both of these sites. By measuring the transduction of SEAP and EGFP reporter genes into cells expressing different levels of CAR, we demonstrated that only the CELO virus bearing the RGD motif at the HI-like loop of the fiber knob showed increased gene transfer efficacy and that the increase was observed only in CAR-negative cells. Notably, following infection with CELOSEAP-HIRGD, the level of SEAP expression and the number of penetrated genomes (data not shown) were lower for CAR-positive and β5 integrin-deficient HCT-116 cells than for H1299 cells expressing high levels of both CAR and β5 integrin. This confirmed successful retargeting of the modified CELO vector to the RGD-binding integrin receptor.
Insertion of the RGD motif at the C terminus of CELO fiber 1 resulted in the opposite effect: a reduction in gene transfer efficiency compared to unmodified CELO. Interestingly, the CELOSEAP-HIRGD vector showed enhanced gene transfer efficiency only in CAR-deficient and CAR-negative cells, but not in CAR-positive cells. One possible explanation for this is that the major interaction of the CELO fiber protein with CAR can partially mask the engagement of integrins with the RGD motif localized in the HI-like loop and, as a consequence, decrease the efficiency of integrin-mediated penetration of the vector into the cell. Therefore, modification of the CELO fiber provides increased gene transfer into CAR-deficient and, especially, CAR-negative cells.
It is well known that a native RGD motif located in the penton base of human Ads does not affect the initial attachment step for CAR-interacting Ads but is crucial for virus internalization (31, 38). To understand whether insertion of an RGD motif into the HI-like loop of CELO fiber 1 affects virus binding and/or internalization, we infected MCF-7, H1299, and LMH cells (permissive for CELO replication). This analysis revealed that insertion of an RGD motif into the CELO fiber significantly increases both binding and internalization of virus into CAR-negative MCF-7 cells. In contrast, we did not observe any differences in the efficiency of interaction of HIRGD-modified CELO virus with H1299 (CAR-positive) cells.
In the next stage of the study, we examined whether blocking of CAR receptors influenced the efficiency of gene transfer by the CELOSEAP-HIRGD vector. We found that gene transfer efficacy by the CELO-SEAP virus was reduced 34-fold under conditions of CAR blocking by Ad5 fiber protein. In contrast, the efficacy of gene transfer provided by CELOSEAP-HIRGD under the same conditions was reduced only 3-fold. The same effect was observed for Ad5 naturally possessing the RGD motif in its penton base. It has been shown that introduction of an RGD motif in the HI loop of Ad5 vectors results in Ad5 tolerance of CAR receptor blocking (8). However, in the case of HI-like loop modification of CELO, total tolerance of CAR blocking has not been achieved.
Ad5-based vectors are widely used for experimental gene therapy of human breast cancer, as well as for production of recombinant proteins in the milk of animals (12). Since expression of CAR on primary breast cells is often low (22, 31), we proposed that the HIRGD-modified CELO vector might be an effective tool for gene transfer into these cells. Our results demonstrate that this fiber-modified CELO virus is capable of providing gene transfer into rabbit mammary gland cells in vitro and in vivo as efficiently as an Ad5-based vector.
In summary, this study describes a novel approach for the generation of new tropism-modified CELO viruses. The identification of an HI-like loop as a potential site for introduction of heterologous receptor binding motifs in the structure of CELO fiber will allow the construction of new genetically modified CELO-based vectors with specified or expanded tropism. This approach can be used for the creation of a new generation of optimized non-human Ad vectors.
Acknowledgments
This work was supported by the following grants: ISTC 3014 and a grant from the Russian Foundation of Basic Research (06-04-48858).
We thank Marieta Cepec and Marina Kasaikina for proofreading of the manuscript.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Bauerschmitz, G. J., J. T. Lam, A. Kanerva, K. Suzuki, D. M. Nettelbeck, I. Dmitriev, V. Krasnykh, G. V. Mikheeva, M. N. Barnes, R. D. Alvarez, P. Dall, R. Alemany, D. T. Curiel, and A. Hemminki. 2002. Tteatment of ovarian cancer with a tropism modified oncolytic adenovirus. Cancer Res. 62:1266-1270. [PubMed] [Google Scholar]
- 2.Bauerschmitz, G. J., S. D. Barker, and A. Hemminki. 2002. Adenoviral gene therapy for cancer: from vectors to target and replication competent agent. Int. J. Oncol. 21:1161-1174. [PubMed] [Google Scholar]
- 3.Cherenova, L. V., D. Y. Logunov, E. V. Shashkova, M. M. Shmarov, L. V. Verkhovskaya, G. L. Neugodova, D. B. Kazansky, K. K. Doronin, and B. S. Naroditsky. 2004. Recombinant avian adenovirus CELO expressing the human interleukin-2: characterization in vitro, in ovo and in vivo. Virus Res. 100:257-261. [DOI] [PubMed] [Google Scholar]
- 4.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. Jpred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 5.Davison, E., I. Kirby, J. Whitehouse, I. Hart, J. F. Marshall, and G. Santis. 2001. Adenovirus type 5 uptake by lung adenocarcinoma cells in culture correlates with Ad5 fibre binding, is mediated by α(v)β1 integrin, and can be modulated by changes in beta1 integrin function. J. Gene Med. 3:550-559. [DOI] [PubMed] [Google Scholar]
- 6.Davison, E., R. M. Diaz, I. R. Hart, G. Santis, and J. F. Marshall. 1997. Integrin α5β1-mediated adenovirus infection is enhanced by the integrin activating antibody TS2/16. J. Virol. 71:6204-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dechecchi, M. C., P. Melotti, A. Bonizzato, M. Santacatterina, M. Chilosi, and G. Cabrini. 2001. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J. Virol. 18:8772-8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flomenberg, P., V. Piaskowski, R. L. Truitt, and J. T. Casper. 1995. Characterization of human proliferative T cells response to adenoviruses. J. Infect Dis. 171:1090-1096. [DOI] [PubMed] [Google Scholar]
- 10.Francois, A., C. Chevalier, B. Delmas, N. Eterradossi, D. Toquin, G. Rivallan, and P. Langlois. 2004. Avian adenovirus CELO recombinants expressing VP2 of infectious bursal disease virus induce protection against bursal disease in chicken. Vaccine 22:2351-2360. [DOI] [PubMed] [Google Scholar]
- 11.Gu, W., A. Ogose, H. Kawashima, M. Ito, T. Ito, A. Matsuba, H. Kitahara, T. Hotta, K. Tokunaga, H. Hatano, T. Morita, S. Urakawa, T. Yoshizawa, H. Kawashima, R. Kuwano, and N. Endo. 2004. High-level expression of the coxsackievirus and adenovirus receptor messenger RNA in osteosarcoma, Ewing's sarcoma, and benign neurogenic tumors among musculoskeletal tumors. Clin. Cancer Res. 10:3831-3838. [DOI] [PubMed] [Google Scholar]
- 12.Han, Z. S., Q. W. Li, Z. Y. Zhang, B. Xiao, D. W. Gao, S. Y. Wu, J. Li, H. W. Zhao, Z. L. Jiang, and J. H. Hu. 2007. High-level expression of human lactoferrin in the milk of goats by using replication-defective adenoviral vectors. Protein Expr. Purif. 1:225-231. [DOI] [PubMed] [Google Scholar]
- 13.Harvey, B. G., N. R. Hackett, T. El-Sawy, T. K. Rosengart, E. A. Hirschowitz, M. D. Lieberman, M. L. Lesser, and R. G. Crystal. 1999. Variability of human systemic humoral immune responses to adenovirus gene transfer vectors administered to different organs. J. Virol. 73:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemminki, A., N. Belousova, K. R. Zinn, B. Liu, M. Wang, T. R. Chaudhuri, B. E. Rogers, D. J. Buchsbaum, G. P. Siegal, M. N. Barnes, J. Gomez-Navarro, D. T. Curiel, and R. D. Alvarez. 2001. An adenovirus with enhanced infectivity mediates molecular chemotherapy of ovarian cancer cells and allows imaging of gene expression. Mol. Ther. 4:223-231. [DOI] [PubMed] [Google Scholar]
- 15.Huang, S., T. Kamata, Y. Takada, Z. M. Ruggeri, and G. R. Nemerow. 1996. Adenovirus interaction with distinct integrins mediates separate events in cell entry and gene delivery to hematopoietic cells. J. Virol. 70:4502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 17.Kasono, K., J. L. Blackwell, J. T. Douglas, I. Dmitriev, T. V. Strong, P. Reynolds, D. A. Kropf, W. R. Carroll, G. E. Peters, R. P. Bucy, D. T. Curiel, and V. Krasnykh. 1999. Selective gene delivery to head and neck cancer cells via an integrin targeted adenoviral vector. Clin. Cancer Res. 5:2571-2579. [PubMed] [Google Scholar]
- 18.Kass-Eisler, A., E. Falck-Pedersen, D. H. Elfenbein, M. Alvira, P. M. Buttrick, and L. A. Leinwand. 1994. The impact of developmental stage, route of administration and the immune system on adenovirus-mediated gene transfer. Gene Ther. 1:395-402. [PubMed] [Google Scholar]
- 19.Krasnykh, V., I. Dmitriev, G. Mikheeva, C. R. Miller, N. Belousova, and D. T. Curiel. 1998. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J. Virol. 72:1844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Gal La Salle, G., J. J. Robert, S. Berrard, V. Ridoux, L. D. Stratford-Perricaudet, M. Perricaudet, and J. Mallet. 1993. An adenovirus vector for gene transfer into neurons and glia in the brain. Science 259:988-990. [DOI] [PubMed] [Google Scholar]
- 21.Li, E., S. L. Brown, D. G. Stupack, X. S. Puente, D. A. Cheresh, and G. R. Nemerow. 2001. Integrin αvβ1 is an adenovirus coreceptor. J. Virol. 75:5405-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., R. C. Pong, J. M. Bergelson, M. C. Hall, A. I. Sagalowsky, C. P. Tseng, Z. Wang, and J. T. Hsieh. 1999. Loss of adenoviral receptor expression in human bladder cancer cells: a potential impact on the efficacy of gene therapy. Cancer Res. 59:325-330. [PubMed] [Google Scholar]
- 23.Liu, Y., T. Ye, J. Maynard, H. Akbulut, and A. Deisseroth. 2006. Engineering conditionally replication-competent adenoviral vectors carrying the cytosine deaminase gene increases the infectivity and therapeutic effect for breast cancer gene therapy. Cancer Gene Ther. 4:346-356. [DOI] [PubMed] [Google Scholar]
- 24.Logunov, D. Y., G. V. Ilyinskaya, L. V. Cherenova, L. V. Verhovskaya, M. M. Shmarov, P. M. Chumakov, B. P. Kopnin, and B. S. Naroditsky. 2004. Restoration of p53 tumor-suppressor activity in human tumor cells in vitro and in their xenografts in vivo by recombinant avian adenovirus CELO-p53. Gene Ther. 1:79-84. [DOI] [PubMed] [Google Scholar]
- 25.Michou, A. L., H. Lehrmann, M. Saltik, and M. Cotten. 1999. Mutational analysis of the avian adenovirus CELO, which provides a basis for gene delivery vectors. J. Virol. 73:1399-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, C. R., D. J. Buchsbaum, P. N. Reynolds, J. T. Douglas, G. Y. Gillespie, M. S. Mayo, D. Raben, and D. T. Curiel. 1998. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res. 58:5738-5748. [PubMed] [Google Scholar]
- 27.Pasch, A., J. Kupper, A. Wolde, R. Kandolf, and H. Selinka. 1999. Comparative analysis of virus-host cell interactions of haemagglutinating and non-haemagglutinating strains of coxsackievirus B3. J. Gen. Virol. 80:3153-3158. [DOI] [PubMed] [Google Scholar]
- 28.Rebel, V. I., S. Hartnett, J. Denham, M. Chan, R. Finberg, and C. A. Sieff. 2000. Maturation and lineage-specific expression of the coxsackie and adenovirus receptor in hematopoietic cells. Stem Cells 18:176-182. [DOI] [PubMed] [Google Scholar]
- 29.Rosenfeld, M. A., K. Yoshimura, B. C. Trapnell, K. Yoneyama, E. R. Rosenthal, W. Dalemans, M. Fukayama, J. Bargon, L. E. Stier, L. Stratford-Perricaudet, et al. 1992. In vivo transfer of the human cystic fibrosis transmembrane conductance regulator gene to the airway epithelium. Cell 68:143-155. [DOI] [PubMed] [Google Scholar]
- 30.Seidman, M. A., S. M. Hogan, R. L. Wendland, S. Worgall, R. G. Crystal, and P. L. Leopold. 2001. Variation in adenovirus receptor expression and adenovirus vector mediated transgene expression at defined stages of the cell cycle. Mol. Ther. 4:13-21. [DOI] [PubMed] [Google Scholar]
- 31.Shayakhmetov, D. M., A. M. Eberly, Z. Y. Li, and A. Lieber. 2005. Deletion of penton RGD motifs affects the efficiency of both the internalization and the endosome escape of viral particles containing adenovirus serotype 5 or 35 fiber knob. J. Virol. 79:1053-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shayakhmetov, D. M., Z. Y. Li, S. Ni, and A. Lieber. 2002. Targeting of adenovirus vectors to tumor cells does not enable efficient transduction of breast cancer metastases. Cancer Res. 62:1063-1068. [PubMed] [Google Scholar]
- 33.Smith, C. A., L. S. Woodruff, C. Rooney, and G. R. Kitchigan. 1998. Extensive cross-reactivitry of adenovirus-specific cytotoxic T cell. Hum. Gene Ther. 9:1419-1427. [DOI] [PubMed] [Google Scholar]
- 34.Stevenson, M., E. Boos, C. Herbert, A. Hale, N. Green, M. Lyons, L. Chandler, K. Ulbrich, N. van Rooijen, V. Mautner, K. Fisher, and L. Seymour. 2006. Chick Lethal Orphan (CELO) virus can be polymer-coated and retargeted to infect mammalian cells. Gene Ther. 4:356-368. [DOI] [PubMed] [Google Scholar]
- 35.Tan, P. K., A. Michou, J. M. Bergelson, and M. Cotten. 2001. Defining CAR as a cellular receptor for the avian adenovirus CELO using a genetic analysis of the two viral fibre proteins. J. Gen. Virol. 82:1465-1472. [DOI] [PubMed] [Google Scholar]
- 36.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 39.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 40.Wickham, T. J., D. M. Segal, P. W. Roelvink, M. E. Carrion, A. Lizonova, G. M. Lee, and I. Kovesdi. 1996. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J. Virol. 70:6831-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickham, T. J., M. E. Carrion, and I. Kovesdi. 1995. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 2:750-756. [PubMed] [Google Scholar]
- 42.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure 12:1259-1270. [DOI] [PubMed] [Google Scholar]
- 43.Yang, Y., Q. Li, H. C. Ertl, and J. M. Wilson. 1995. Cellular and humoral immune responses to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J. Virol. 69:2004-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]