Abstract
H9N2 influenza viruses have become established in terrestrial poultry in different Asian countries over the last 2 decades. Our previous study demonstrated that quail harbor increasingly diverse novel H9N2 reassortants, including both Chicken/Beijing/1/94 (Ck/Bei-like) and Quail/Hong Kong/G1/97 (G1-like) viruses. However, since 1999, the genesis and evolution of H9N2 viruses in different types of poultry have not been investigated systematically. In the present study, H9N2 viruses isolated from chickens, ducks, and other minor poultry species were characterized genetically and antigenically. Our findings demonstrate that Ck/Bei-like H9N2 viruses have been introduced into many different types of poultry in southern China, including quail, partridges, chukar, pheasant, guinea fowl, and domestic ducks, while G1-like viruses were commonly detected in quail, less frequently detected in other minor poultry species, and not detected in chickens and ducks. Genetic analysis revealed 35 genotypes of H9N2 viruses, including 14 novel genotypes that have not been recognized before. Our results also suggested that two-way interspecies transmission exists between different types of poultry. Our study demonstrates that the long-term cocirculation of multiple virus lineages (e.g., H5N1 and H9N2 viruses) in different types of poultry has facilitated the frequent reassortment events that are mostly responsible for the current great genetic diversity in H9N2 and H5N1 influenza viruses in this region. This situation favors the emergence of influenza viruses with pandemic potential.
While highly pathogenic H5N1 influenza viruses have spread widely throughout Eurasia and Africa, another subtype of influenza viruses, H9N2, has also become endemic in different types of terrestrial poultry in multiple countries on the Eurasian continent. Epidemiological and genetic studies revealed that three distinct lineages of H9N2 influenza viruses have been responsible for outbreak events. These included viruses represented by Chicken/Beijing/1/94 (Ck/Bei-like), Quail/Hong Kong/G1/97 (G1-like), and Duck/Hong Kong/Y439/97 (Y439-like or Korean-like). Ck/Bei-like and G1-like viruses have been prevalent mainly in China since the mid-1990s, while the G1-like viruses have also been recorded as causing outbreaks in chickens in the Middle East and Germany (1, 2, 4). While Y439-like viruses were isolated from domestic ducks in Hong Kong in 1997, similar viruses have also been identified from disease outbreaks in chickens in South Korea since 1996 (8, 17).
H9N2 influenza viruses were detected only in domestic ducks during influenza virus surveillance in southern China from 1976 to 1980 (28). Since the late 1990s, Ck/Bei-like and G1-like H9N2 viruses from southern China have become predominant in chickens and quail, respectively (11, 12). In 2000, Ck/Bei-like viruses may have been transmitted reversely back to domestic ducks, wherein multiple reassortant variants of H9N2, or genotypes, were recognized in this region (19). Molecular epidemiological studies of H9N2 viruses from quail isolated from 2000 to 2005 revealed that G1-like viruses are still predominant in quail and are frequently reassorted with either Ck/Bei-like or H5N1/01-like viruses to generate novel reassortants. While only four G1-like reassortants were detected in quail, there were 16 different genotypes of Ck/Bei-like H9N2 viruses. Those H9N2 variants from quail, particularly Ck/Bei-like viruses, contained gene segments from multiple sources, including those closely related to highly pathogenic H5N1 influenza viruses. This updated information suggested that two-way transmission between quail and chickens occurred frequently, as the genotypes of Ck/Bei-like viruses were transient and did not become established in quail (31).
Although the quail is considered a possible intermediate host for the introduction of influenza viruses from aquatic birds to terrestrial poultry species (26), this species is only one type of minor poultry species and chickens still account for approximately 70% of the total poultry population in China. It is noted that in the past 2 decades, a variety of birds, collectively named minor poultry species, including pheasant, chukar, partridges, guinea fowl, and pigeons, have also been farmed. These operations dramatically increased the complexity of the influenza virus ecosystem in this region. Recent studies suggested that most influenza A virus subtypes could replicate asymptomatically in similar kinds of birds under laboratory conditions (16, 21). However, the impact of such changes in the poultry industry on influenza virus ecology has not been investigated.
In addition to the quail isolates, H9N2 influenza viruses have also been isolated regularly from both chickens and other minor poultry species in our surveillance program in southern China, but their genetic diversity and mechanism of genesis have not been determined since 1999. The interrelationships of H9N2 influenza viruses from different types of poultry are still not determined. This situation highlights the necessity of exploring the possible role of different types of poultry in the ecology of influenza virus in southern China, while the broad distribution of H9N2 influenza viruses in those birds provides such an opportunity.
Previous studies revealed that H9N2 influenza viruses from poultry could occasionally be transmitted from poultry to mammalian species, including humans and pigs (3, 20, 24, 25, 30). The Ck/Bei-like and G1-like viruses were initially recognized from both a human and pigs in the late 1990s and were also observed in 2003 and recently in Hong Kong (5). Genetic analyses demonstrated that the human H9N2 influenza virus isolate in 2003 was a novel reassortant and most likely originated directly from local live poultry markets (3). These recent interspecies transmission events suggest that current H9N2 influenza virus variants are still potentially infectious for humans.
Our long-term influenza virus surveillance program in southern China focused mainly on major poultry, including chickens, ducks, and geese. In addition, we also sampled a variety of other minor poultry species. In the present study, H9N2 influenza viruses isolated from chickens, domestic ducks, and other minor poultry species from 2000 to 2005 were characterized genetically and antigenically. The findings of the present study demonstrate that Ck/Bei-like H9N2 viruses have been introduced into many different types of poultry in this region, including quail, partridges, chukar, pheasant, guinea fowl, and domestic ducks, while G1-like viruses were commonly detected in quail, less frequently detected in other minor poultry species, and not detected in chickens and ducks. Genetic studies revealed that two-way interspecies transmission exists between different types of poultry. Phylogenetic analysis suggests that the long-term cocirculation of multiple virus lineages (e.g., H5N1 and H9N2 viruses) in different types of poultry facilitated frequent reassortment events that were mostly responsible for the current great genetic diversity in H9N2 and H5N1 variants in this region (6, 10, 11, 18). The present study provides insight into the genesis and evolution of H9N2 influenza viruses in southern China. The current influenza virus ecosystem in southern China favors the emergence of influenza viruses with pandemic potential.
MATERIALS AND METHODS
Sampling and virus isolation.
Our previous studies characterized H9N2 viruses from quail, which was considered a “minor poultry species.” Therefore, minor poultry species in the present study, namely, chukar, guinea fowl, partridges, and pheasant, are referred to as “other minor poultry species.” A total of 47,255 chickens and 6,925 birds of other minor poultry species were sampled from six provinces in southern China between July 2000 and December 2005. During the same period, a total of 49,150 ducks were also sampled. Of those samples, 20,535 chicken, 3,008 other minor poultry species, and 5,381 duck specimens were paired tracheal and cloacal swabs, while the remaining specimens were either cloacal or fecal swabs. Our influenza virus surveillance program was carried out as previously reported (30). In brief, surveillance was initiated in Guangdong in July 2000. Since 2002, this program was gradually expanded to five other provinces in this region, including Fujian, Guangxi, Guizhou, Hunan, and Yunnan. Viruses were isolated in 9- to 11-day-old embryonated chicken eggs as previously described (18, 19).
Antigenic analysis.
All virus isolates were subtyped by standard hemagglutination inhibition (HI) and neuraminidase (NA) inhibition tests, using a panel of World Health Organization reference antisera as previously described (3). Antigenic analysis was performed using three different panels of monoclonal antibodies (MAbs), against Qa/HK/G1/97, Dk/HK/Y280/97, and Ck/HK/G9/97, as previously described (31). All MAbs were produced at the Department of Infectious Diseases, St. Jude Children's Research Hospital, TN (7). Numerical analysis of HI titers was conducted using PRIMER, version 5.2.9 (PRIMER-E, Plymouth, United Kingdom), also as previously described (31).
Phylogenetic and molecular analyses.
One or two virus isolates from each positive sampling occasion were selected for characterization. Viral RNA extraction, cDNA synthesis, PCR, and sequencing were carried out as previously described (12). All eight gene segments sequenced from these viruses were characterized and phylogenetically analyzed with available virus sequence data from GenBank. The program MrModeltest 2.2 (23) was used to determine the appropriate DNA substitution model and γ-rate heterogeneity. The generated model was used in all subsequent analyses. Neighbor-joining trees were constructed using PAUP* 4.0 (29), and Bayesian analysis was conducted with MrBayes 3.1 (15) by using two replicates of 1 million generations with six chains, sampling every 100 generations. Estimates of the phylogenies were calculated by performing 1,000 neighbor-joining bootstrap replicates, and Bayesian posterior probabilities were calculated from the consensus of 18,000 trees after excluding the first 2,000 trees as burn-in.
Genotype definition.
Virus genotypes were defined by gene phylogeny. A distinct phylogenetic lineage with bootstrap support of ≥80% indicated a common origin. Viruses with G1-like and Ck/Bei-like hemagglutinin (HA) genes were designated genotype A and B series, respectively, as previously described (31).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study are available from GenBank under accession numbers CY023090 to CY024737.
RESULTS
Prevalence of H9N2 influenza viruses in chickens, ducks, and other minor poultry species.
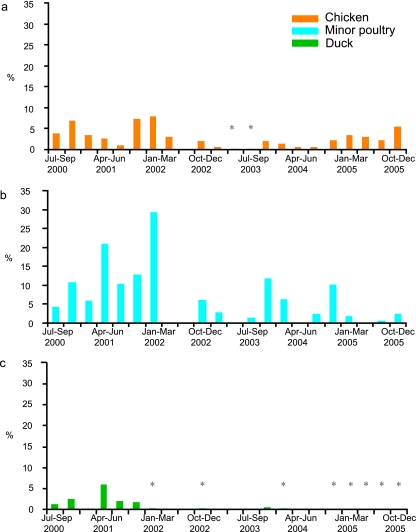
A total of 103,300 samples were collected from chickens, ducks, and other minor poultry species from 2000 to 2005. In general, H9N2 viruses were isolated from apparently healthy birds year-round in live poultry markets in southern China, but the isolation rate was usually higher in the winter season than in the summer season (Fig. 1; Table 1). The majority of viruses were isolated from chickens and other minor poultry species, while ducks tested positive on very few sampling occasions, except for those in 2000 and 2001 (Fig. 1).
FIG. 1.
Comparison of H9N2 influenza virus isolation rates from chickens (a), other minor poultry species (b), and ducks (c) from southern China, July 2000 to December 2005. Surveillance was conducted in live poultry markets in Fujian, Guangdong, Guangxi, Guiyang, Hunan, and Yunnan Provinces. *, positive sampling occasions with low isolation rates of <0.3%.
TABLE 1.
Prevalence of H9N2 viruses from chickens, other minor poultry species, and ducks in southern China during 2000 to 2005
| Month | No. of H9N2 isolates/total sample no.
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chickens
|
Other minor poultry species
|
Ducks
|
||||||||||||||||
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | |
| January | 1/161 | 5/70 | 10/844 | 13/817 | 110/2,056 | 4/43 | 33/79 | 1/42 | 29/224 | 9/254 | 0/502 | 1/200 | 0/769 | 3/587 | 2/1,380 | |||
| February | 15/215 | 4/8 | 2/440 | 10/666 | 46/1,432 | 0/11 | 9/36 | 2/52 | 6/176 | 1/150 | 0/191 | 0/162 | 0/428 | 0/361 | 1/1,035 | |||
| March | 3/182 | 0/35 | 2/689 | 11/834 | 26/1,651 | 0/15 | 3/39 | 1/45 | 10/308 | 0/162 | 2/149 | 0/102 | 0/681 | 0/696 | 0/1,396 | |||
| April | 13/178 | 0/49 | 1/603 | 11/732 | 84/2,204 | 0/5 | 0/47 | 0/47 | 0/206 | 0/138 | 2/118 | 0/108 | 0/561 | 0/695 | 1/1,972 | |||
| May | 0/176 | 0/48 | 2/652 | 1/757 | 67/2,023 | 1/6 | 0/36 | 0/19 | 0/181 | 0/150 | 14/121 | 0/114 | 0/576 | 0/635 | 0/2,153 | |||
| June | 1/162 | 11/256 | 0/604 | 0/733 | 31/1,931 | 3/8 | 0/19 | 0/49 | 0/220 | 0/204 | 5/115 | 0/125 | 0/533 | 0/663 | 1/2,089 | |||
| July | 14/203 | 2/181 | 0/123 | 0/646 | 0/843 | 22/1,881 | 8/60 | 1/11 | 0/18 | 0/40 | 1/207 | 2/174 | 0/119 | 1/135 | 0/341 | 0/567 | 0/818 | 0/2,426 |
| August | 0/115 | 1/154 | 0/115 | 3/584 | 6/1,071 | 27/1,803 | 0/35 | 0/19 | 0/49 | 3/85 | 0/266 | 1/188 | 0/100 | 1/168 | 0/152 | 0/494 | 0/994 | 2/3,034 |
| September | 5/172 | 2/139 | 0/132 | 1/707 | 13/922 | 75/1,725 | 0/89 | 5/28 | 0/43 | 0/99 | 16/241 | 0/160 | 4/131 | 8/211 | 0/190 | 0/605 | 0/952 | 6/2,214 |
| October | 2/242 | 6/177 | 2/537 | 3/725 | 8/994 | 119/1,755 | 0/46 | 5/27 | 0/79 | 4/68 | 39/244 | 3/256 | 0/58 | 3/205 | 1/339 | 6/595 | 0/1,051 | 6/2,574 |
| November | 26/199 | 10/154 | 28/860 | 10/699 | 45/1,360 | 41/1,679 | 8/36 | 6/27 | 6/36 | 14/134 | 21/274 | 0/164 | 2/131 | 3/210 | 4/889 | 2/627 | 1/1,241 | 0/2,670 |
| December | 10/106 | 21/181 | 12/685 | 32/899 | 37/1,738 | 126/1,702 | 5/38 | 5/71 | 3/33 | 33/227 | 15/212 | 12/170 | 4/120 | 4/175 | 1/625 | 0/765 | 1/1,097 | 0/2,910 |
| Total | 57/1,037 | 75/2,060 | 62/2,918 | 66/8,092 | 155/11,467 | 774/21,842 | 21/304 | 30/271 | 54/514 | 58/907 | 137/2,759 | 28/2,170 | 10/659 | 41/2,300 | 6/3,347 | 8/7,201 | 5/9,790 | 19/25,853 |
aThe isolation rates for chickens in 2000 to 2005 were 5.5%, 3.6%, 2.1%, 0.8%, 1.4%, and 3.5%, respectively. The isolation rates for other minor poultry species in 2000 to 2005 were 6.9%, 11.1%, 10.5%, 6.4%, 5.0%, and 1.3%, respectively. The isolation rates for ducks in 2000 to 2005 were 1.8%, 1.8%, 0.17%, 0.12%, 0.06%, and 0.07%, respectively.
There were 1,189 strains of H9N2 influenza viruses isolated from 47,225 chicken samples (overall isolation rate, 2.5%) (Table 1). Of these viruses, 944 were isolated from 20,535 tracheal swabs (isolation rate, 4.6%), while 245 strains were isolated from 26,690 cloacal or fecal swabs (isolation rate, 0.9%) (Table 2). It is noteworthy that the rate of isolation of H9N2 viruses from fecal material and cloacal samples declined from 5.4% in 2000 to 0.5% in 2005, while there was no dramatic change in the isolation rate for tracheal samples from chickens during the study period (Table 2). This suggests that H9N2 influenza viruses have gradually adapted to replicate in the respiratory tract of chickens.
TABLE 2.
Comparison of replication sites of H9N2 viruses from chickens and other minor poultry species
| Year | No. of H9N2 isolates/total sample no. (isolation rate [%])
|
|||
|---|---|---|---|---|
| Chickens
|
Other minor poultry species
|
|||
| Tracheal swabs | Fecal and cloacal swabs | Tracheal swabs | Fecal and cloacal swabs | |
| 2000 | 4/64 (6.3) | 53/973 (5.4) | 20/60 (33.3) | 1/244 (0.4) |
| 2001 | 4/41 (9.8) | 71/2,019 (3.5) | 29/126 (23) | 1/145 (0.7) |
| 2002 | 28/839 (3.3) | 34/2,079 (1.7) | 48/237 (20.3) | 6/277 (2.2) |
| 2003 | 37/3,520 (1.0) | 29/4,572 (0.7) | 34/116 (29.3) | 24/791 (3.0) |
| 2004 | 147/4,985 (3.0) | 8/6,482 (0.1) | 134/1,366 (9.8) | 3/1,393 (0.2) |
| 2005 | 724/11,086 (6.5) | 50/10,756 (0.5) | 27/1,103 (2.4) | 1/1,067 (0.1) |
| Total | 944/20,535 (4.6) | 245/26,690 (0.9) | 292/3,008 (9.7) | 36/3,917 (0.9) |
A total of 89 H9N2 viruses were isolated from 49,150 duck samples (overall isolation rate, 0.18%) (Table 1). Fifty-one of these H9N2 viruses were isolated from 2,959 specimens (isolation rate, 1.7%) during 2000 to 2001, while only 38 viruses were isolated from 46,191 cloacal or fecal swabs (isolation rate, 0.08%) from 2002 to 2005 (Fig. 1; Table 1). While tracheal swabs were collected only in 2005, only a single H9N2 virus was isolated from 5,381 samples, indicating that the major site of H9N2 influenza virus replication in the duck is the intestine. The isolation rates of H9N2 influenza viruses from duck have declined since 2002, with ducks testing positive infrequently (Fig. 1; Table 1).
Three hundred twenty-eight H9N2 influenza viruses were isolated from a total of 6,925 samples collected from other minor poultry species (overall isolation rate, 4.7%) (Table 1). Of these viruses, 292 were isolated from tracheal swabs, while only 36 strains were isolated from cloacal or fecal swabs (Table 2). There is a clear replication pattern where H9N2 viruses replicate mainly in the respiratory tract, not the intestine, in other minor poultry species.
Antigenic analysis.
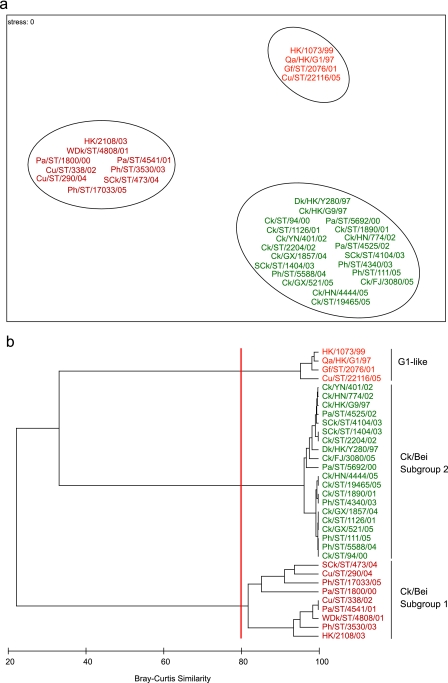
The antigenic properties of representative H9N2 influenza viruses were investigated with a panel of MAbs raised against Qa/HK/G1/97, Dk/HK/Y280/97, and Ck/HK/G9/97 by HI assay (Table 3). Numerical analysis of HI titers was conducted to visualize the antigenic variation and revealed three distinct groups that were in agreement with the results of phylogenetic analyses (Fig. 2; see below). One group included two viruses from other minor poultry species, Gf/ST/2076/01 and Cu/ST/22116/05, which reacted well with two Qa/HK/G1/97 MAbs and both Ck/HK/G9/97 MAbs, a reaction pattern similar to that of Qa/HK/G1/97. The remaining two groups contained Ck/Bei-like viruses (subgroups 1 and 2). Ck/Bei-like subgroup 2 viruses were isolated from both chickens and other minor poultry species and reacted well with all tested MAbs, except G1-9 and 1073-9, a pattern similar to that of the prototype virus, Dk/HK/Y280/97 (Fig. 2; Table 3). Ck/Bei-like subgroup 1 viruses were mostly isolated from other minor poultry species and had high HI titers against MAb Y280-8C4 and only moderate reactivity to Y280-18B10, a reactivity pattern similar to that of the recent human H9N2 isolate (HK/2108/03) and a wild duck isolate (WDk/ST/4108/01) (Fig. 2; Table 3).
TABLE 3.
HI titers from antigenic analysis of influenza A H9N2 viruses
| Virus | Genotype | HI titera
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Qa/HK/G1/97
|
Dk/HK/Y280/97
|
Ck/HK/G9/97
|
||||||||||||||
| 1073-9 | 26 | 29 | 7B10 | 8C4 | 15F1 | 18G4 | 3D11 | 4G3 | 19A10 | 18B10 | 2F4 | 18B1 | G9-6 | G9-25 | ||
| Qa/HK/G1/97 | A0 | 200 | > | > | <100 | <100 | <100 | <100 | <100 | <100 | <100 | 100 | <100 | <100 | > | > |
| HK/1073/99 | A0 | 100 | > | > | <100 | <100 | <100 | 100 | <100 | <100 | <100 | 100 | <100 | <100 | > | > |
| Dk/HK/Y280/97 | B0 | <100 | 400 | > | > | > | > | > | > | > | > | 3,200 | > | > | > | > |
| Ck/HK/G9/97 | Bn | 400 | 800 | > | > | > | > | > | > | > | > | 6,400 | > | > | > | > |
| HK/2108/03 | B7 | <100 | <100 | <100 | 200 | > | <100 | <100 | <100 | <100 | <100 | 1,600 | <100 | <100 | <100 | <100 |
| WDk/ST/4808/01 | B7 | <100 | <100 | <100 | 200 | > | <100 | <100 | <100 | <100 | <100 | 3,200 | <100 | <100 | <100 | <100 |
| Gf/ST/2076/01 | A0 | <100 | > | > | <100 | <100 | <100 | <100 | <100 | <100 | <100 | 200 | <100 | <100 | > | > |
| Cu/ST/22116/05 | A4 | 3,200 | > | > | <100 | <100 | <100 | <100 | <100 | <100 | <100 | 100 | <100 | <100 | > | > |
| Pa/ST/5692/00 | B2 | 800 | 400 | > | > | > | > | > | > | 12,800 | > | 12,800 | > | > | > | > |
| Pa/ST/4525/02 | B3 | <100 | 800 | > | > | > | > | > | > | > | > | 6,400 | > | > | > | > |
| Ph/ST/4340/03 | B23 | 800 | 1,600 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ph/ST/5588/04 | B14 | 200 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ph/ST/111/05 | B3 | 200 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/ST/94/00 | B3 | 200 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/ST/1126/01 | B0 | 100 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/ST/1890/01 | B3 | 800 | 1,600 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/HN/774/02 | B17 | 100 | 800 | > | > | > | > | > | > | > | > | 6,400 | > | > | > | > |
| Ck/YN/401/02 | B18 | 200 | 800 | > | > | > | > | > | > | > | > | 6,400 | > | > | > | > |
| Ck/ST/2204/02 | B3 | 800 | 1,600 | > | > | > | > | > | > | > | > | 6,400 | > | > | > | > |
| SCk/ST/1404/03 | B19 | 800 | 1,600 | > | > | > | > | > | > | > | > | 6,400 | > | > | > | > |
| SCk/ST/4104/03 | B22 | 100 | 1,600 | > | > | > | > | > | > | > | > | 6,400 | > | > | > | > |
| Ck/GX/1857/04 | B25 | 100 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/ST/19465/05 | B29 | 800 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/GX/521/05 | B26 | 100 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/HN/4444/05 | B28 | 800 | 800 | > | > | > | > | > | > | > | > | > | > | > | > | > |
| Ck/FJ/3080/05 | B14 | 1,600 | 1,600 | > | > | > | > | > | > | > | > | 12,800 | > | > | > | > |
| Pa/ST/1800/00 | B1 | 800 | <100 | <100 | 800 | > | <100 | <100 | <100 | <100 | <100 | 12,800 | <100 | <100 | <100 | <100 |
| Pa/ST/4541/01 | B7 | <100 | <100 | <100 | 400 | > | <100 | <100 | <100 | <100 | <100 | 3,200 | <100 | <100 | <100 | <100 |
| Cu/ST/338/02 | B8 | <100 | <100 | <100 | 400 | > | <100 | <100 | <100 | <100 | <100 | 3,200 | <100 | <100 | <100 | <100 |
| Ph/ST/3530/03 | B7 | 100 | <100 | <100 | 400 | > | <100 | 100 | <100 | <100 | <100 | 3,200 | <100 | <100 | <100 | <100 |
| Cu/ST/290/04 | B7 | 3,200 | <100 | <100 | 400 | > | <100 | 100 | 100 | <100 | <100 | 6,400 | <100 | <100 | <100 | <100 |
| Ph/ST/17033/05 | B16 | 200 | <100 | <100 | 400 | > | <100 | 100 | 200 | <100 | <100 | 6,400 | 100 | <100 | <100 | <100 |
| SCk/ST/473/04 | B7 | 1,600 | <100 | <100 | 400 | > | <100 | 100 | 100 | <100 | <100 | 6,400 | 100 | 100 | <100 | <100 |
>, HI titer of >12,800. The HI assay was started at a 1:100 dilution.
FIG. 2.
Numerical analysis of HI titers (Table 3) by nonmetric multidimensional ordination in two dimensions (a) and by using hierarchical agglomerative clustering (b).
Phylogenetic analysis of surface genes.
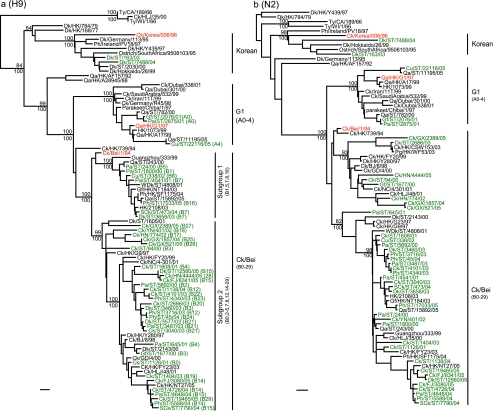
To better understand the evolutionary pathway of H9N2 viruses in southern China, 112 of 1,189 (9.4%) viruses from chickens and 79 of 328 (9.4%) viruses from other minor poultry species were sequenced. A further 15 of 79 (19%) H9N2 viruses from ducks, isolated from 2001 to 2005, were also sequenced. At least one isolate from each positive sampling occasion was sequenced. Those sequence data were analyzed phylogenetically together with data available in public databases. Phylogenetic analysis of the H9 HA gene showed that the majority of isolates tested belonged to the Ck/Bei-like lineage. Three isolates from other minor poultry species (Gf/ST/2076/01, Pa/ST/2875/01, and Cu/ST/22116/05) clustered within the G1-like lineage, and two isolates from ducks (Dk/ST/163/04 and Dk/ST/7448/04) had a Korean-like HA gene (Fig. 3a). The Ck/Bei-like lineage contained two subgroups, including subgroup 1, represented by Qa/ST/243/00, and subgroup 2, represented by Dk/HK/Y280/97, as described in our previous study (31). All H9N2 viruses from chickens belonged to subgroup 2, with the exception of three viruses (SCk/ST/473/04, SCk/ST/999/04, and Ck/ST/6786/04) that clustered in subgroup 1 (Fig. 3a). Similarly, the HA genes of all Ck/Bei-like duck viruses fell into subgroup 2, except for a single virus (Dk/ST/3658/03) that belonged to subgroup 1. In comparison, 47 H9N2 viruses from other minor poultry species clustered into subgroup 1, while 29 viruses belonged to subgroup 2 (Fig. 3a).
FIG. 3.
Phylogenetic relationships of HA (a) and NA (b) genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method in the PAUP* program. Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities, respectively. Analysis was based on nucleotides 129 to 1042 of the HA gene and 231 to 1297 of the NA gene. The HA and NA trees were rooted to Qa/Arkansas/29209-1/93 (H9N2) and Ck/Pennsylvania/8125/83 (H5N2), respectively. Viruses characterized in this study are highlighted in green. Genotypes characterized in this study are shown in parentheses and defined in Table 4. Abbreviations: BJ and Bei, Beijing; Ck, chicken; Dk, duck; FJ, Fujian; GD, Guangdong; Gf, guinea fowl; GX, Guangxi; HK, Hong Kong; HLJ, Heilongjiang; HN, Hunan; NC, Nanchang; Pg, pigeon; Ph, pheasant; Qa, quail; SCk, silky chicken; SD, Shandong; SH, Shanghai; ST, Shantou; Ty, turkey; WDk, wild duck; YN, Yunnan. Bar, 0.01 substitution per site.
Phylogenetic analysis of the NA gene showed a similar evolutionary pattern to that of the HA gene, wherein all viruses clustered within the Ck/Bei-like lineage, except for three viruses from other minor poultry species and two viruses from ducks, which clustered in the G1-like and Korean-like lineages, respectively (Fig. 3b). These results show that Ck/Bei-like viruses are predominant in chickens, ducks, and other minor poultry species, in comparison to previous results that indicated that G1-like viruses were maintained mainly in quail (31).
Phylogenetic analysis of internal genes.
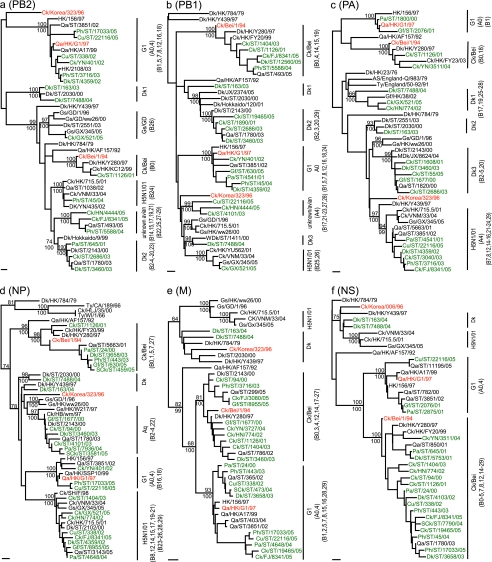
Phylogenetic analysis of the ribonucleoprotein complex genes (PB2, PB1, PA, and NP genes) revealed that these genes had more diversified sources than the surface genes and that H9N2 viruses circulating in chickens, ducks, and other minor poultry species had undergone extensive reassortment to generate multiple novel reassortants or genotypes (Fig. 4a to d; Table 4). Analysis of the PB2 gene revealed seven distinct evolutionary lineages, including G1-like (n = 66), Ck/Bei-like (n = 3), H5N1/01-like (n = 1), and Gs/Gd-like (n = 1) lineages, an unknown avian source (n = 77), and two duck lineages (for Dk1, n = 2; and for Dk2, n = 56) (Fig. 4a).
FIG. 4.
Phylogenetic relationships of the PB2 (a), PB1 (b), PA (c), NP (d), M (e), and NS (f) genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method in PAUP*. Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities, respectively. Analysis was based on the following nucleotides: PB2, 1079 to 2138; PB1, 42 to 1217; PA, 1429 to 2127; NP, 31 to 917; M, 49 to 864; and NS, 88 to 815. The PB2, PA, NP, and M trees were rooted to A/equine/Prague/1/56 (H7N7), the PB1 tree was rooted to Qa/Arkansas/29209-1/93 (H9N2), and the NS tree was rooted to A/swine/Hong Kong/168/93 (H1N1). Viruses characterized in this study are highlighted in green. Aq, aquatic bird. Other virus names and abbreviations can be found in the legend to Fig. 3. Bar, 0.01 substitution per site.
TABLE 4.
Gene constellations of different genotypes of H9N2 influenza viruses and their host distribution in southern China
| Genotype | Host (no. of viruses)a | Lineage of gene segment
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| PB2 | PB1 | PA | HA | NP | NA | M | NS | ||
| A0 | Qa (9), MP (2) | G1 | G1 | G1 | G1 | G1 | G1 | G1 | G1 |
| A1 | Qa (4) | G1 | G1 | G1 | G1 | G1 | G1 | G1 | Ck/Bei |
| A2 | Qa (1) | G1 | G1 | G1 | G1 | G1 | G1 | Ck/Bei | Ck/Bei |
| A3 | Qa (19) | G1 | G1 | H5N1/01 | G1 | G1 | G1 | G1 | G1 |
| A4 | MP (1) | G1 | ? | H5N1/01 | G1 | G1 | G1 | G1 | G1 |
| B0 | Ck (3) | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei |
| B1 | Qa (1), MP (2) | G1 | G1 | G1 | Ck/Bei | Ck/Bei | Ck/Bei | G1 | Ck/Bei |
| B2 | Qa (1), MP (3) | Dk | Dk | Dk | Ck/Bei | Aq | Ck/Bei | G1 | Ck/Bei |
| B3 | Qa (3), Ck (33), MP (8), Dk (7) | Dk | Dk | Dk | Ck/Bei | Aq | Ck/Bei | Ck/Bei | Ck/Bei |
| B4 | Qa (1), Ck (2), MP (1) | Dk | Ck/Bei | Dk | Ck/Bei | Aq | Ck/Bei | Ck/Bei | Ck/Bei |
| B5 | Qa (3), MP (2) | G1 | G1 | Dk | Ck/Bei | Ck/Bei | Ck/Bei | G1 | Ck/Bei |
| B6 | Qa (1) | G1 | G1 | G1 | Ck/Bei | G1 | G1 | G1 | Ck/Bei |
| B7 | Qa (9), Ck (5), MP (36), Dk (1) | G1 | G1 | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei | G1 | Ck/Bei |
| B8 | Qa (6), MP (3) | G1 | G1 | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | G1 | Ck/Bei |
| B9 | Qa (1) | H5N1/01 | G1 | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | G1 | Ck/Bei |
| B10 | Qa (1) | Dk | Dk | H5N1/01 | Ck/Bei | Aq | Ck/Bei | G1 | Ck/Bei |
| B11 | Qa (1) | G1 | G1 | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei |
| B12 | Qa (1), Ck (1), MP (1), Dk (1) | G1 | G1 | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B13 | Qa (1) | G1 | G1 | H5N1/01 | Ck/Bei | G1 | Ck/Bei | Ck/Bei | Ck/Bei |
| B14 | Qa (3), Ck (19), MP (9) | ? | Ck/Bei | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B15 | Qa (5), Ck (22), MP (3), Dk (1) | ? | Ck/Bei | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | G1 | Ck/Bei |
| B16 | Qa (2), Ck (1), MP (3) | G1 | G1 | H5N1/01 | Ck/Bei | G1 | Ck/Bei | G1 | Ck/Bei |
| B17 | Ck (1) | ? | ? | Dk | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B18 | Ck (7) | G1 | G1 | Ck/Bei | Ck/Bei | G1 | Ck/Bei | Ck/Bei | Ck/Bei |
| B19 | Ck (1) | ? | Ck/Bei | Dk | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B20 | Ck (2) | Dk | Dk | Dk | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B21 | Ck (5), MP (3), Dk (3), | ? | ? | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B22 | Ck (1) | ? | ? | H5N1/01 | Ck/Bei | Aq | Ck/Bei | Ck/Bei | Ck/Bei |
| B23 | MP (1) | Dk | ? | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B24 | MP (1) | H5N1/01 | G1 | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B25 | Ck (3) | ? | H5N1/01 | Dk | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B26 | Ck (1) | Gs/GD | H5N1/01 | Dk | Ck/Bei | H5N1/01 | Ck/Bei | Ck/Bei | Ck/Bei |
| B27 | Ck (1) | ? | ? | Dk | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei | Ck/Bei |
| B28 | Ck (3) | ? | ? | Dk | Ck/Bei | H5N1/01 | Ck/Bei | G1 | Ck/Bei |
| B29 | Ck (1) | ? | Dk | H5N1/01 | Ck/Bei | H5N1/01 | Ck/Bei | G1 | Ck/Bei |
Number of viruses characterized in this study and in reference 31. Abbreviations: ?, unknown avian host or lineage; Aq, aquatic bird; Ck, chicken; Ck/Bei, Ck/Beijing/1/94-like; Dk, duck; G1, Qa/HK/G1/97-like; Gs/GD, Gs/GD-like; H5N1/01, H5N1/01-like; MP, minor poultry species, except for quail; Qa, quail.
The PB2 genes of the majority of chicken isolates fell into two groups, namely, unknown avian, for which we could not identify a source, and Dk2 (Fig. 4a). The PB2 genes of the Dk2 lineage are most closely related to an H9N2 virus, Dk/Hokkaido/9/99, an isolate possibly obtained from a migratory duck. Interestingly, one chicken isolate (Ck/GX/521/05) was closely related to an H5N1 genotype G virus (Gs/GX/345/05), with a Gs/GD-like PB2 gene (Fig. 4a). The majority of the H9N2 viruses from other minor poultry species had G1-like PB2 genes, while duck H9N2 isolates had PB2 genes from many different lineages, including G1-like, Dk2, and unknown avian (Fig. 4a; data not shown). However, two of them (Dk/ST/163/03 and Dk/ST/7488/04) contained a PB2 gene from the domestic duck gene pool (Dk1) in this region (Fig. 4a).
Phylogenetic analysis of the PB1 gene showed that the H9N2 viruses formed seven different lineages, including G1-like (n = 66), Ck/Bei-like (n = 61), and H5N1/01-like (n = 4) lineages, three duck lineages (for Dk1, n = 1; for Dk2, n = 53; and for Dk3, n = 1), and an unknown avian lineage (n = 20) (Fig. 4b). The PB1 genes of the majority of chicken isolates fell into the Ck/Bei-like and Dk2 lineages, while those of other minor poultry species isolates were mostly G1-like and those of duck viruses were from diverse sources. The Dk2 PB1 genes were closely related to those of viruses isolated from migratory ducks or sentry ducks in southern China and Japan (e.g., Dk/ST/2030/00 [H9N1] and Dk/Hokkaido/120/01 [H6N2]). It was noted that the PB1 genes of Dk/ST/163/03 and Dk/ST/7488/04, both of which have Korean-like HA and NA genes, clustered into different lineages, namely, Dk1 and Dk3 (Fig. 4b).
Six different PA lineages for the H9N2 influenza viruses tested were recognized, including G1-like (n = 4), Ck/Bei-like (n = 10), and H5N1/01-like (n = 123) lineages and three duck lineages (for Dk1, n = 11; for Dk2, n = 1; and for Dk3, n = 57) (Fig. 4c). Most H9N2 viruses isolated from chickens, ducks, and minor poultry species since 2002 and 2003 had H5N1/01-like PA genes related to that of a contemporary duck virus (Dk/HK/Y439/97 [H9N2]). It is interesting that Gf/HK/38/02 (H5N1; genotype X), together with 10 H9N2 chicken isolates and 1 duck isolate, clustered with the Dk1 lineage (Fig. 4c).
The nucleoprotein (NP) genes of these H9N2 viruses formed five lineages, including G1-like (n = 14), Ck/Bei-like (n = 50), H5N1/01-like (n = 85), duck (n = 2), and aquatic bird (n = 55) lineages (Fig. 4d). It was noted that the NP genes of many H9N2 viruses tested apparently were derived from an H6N9 virus (Gs/HK/W217/97), and therefore, this lineage was assigned as an aquatic bird lineage. The majority of chicken and duck H9N2 isolates had NP genes belonging to either the H5N1/01-like or aquatic bird lineage, while viruses from other minor poultry species were from the Ck/Bei-like or H5N1/01-like lineage. It is interesting that one H5N1 virus isolated in 1997 from central China (Ck/Hubei/wm/97) also grouped with the aquatic bird lineage.
The matrix (M) and nonstructural (NS) protein genes showed much less diversity than the other genes and belonged to either the G1-like or Ck/Bei-like lineage (Fig. 4e and f). Thirty-two chicken, 2 duck, and 55 minor poultry species isolates had G1-like M genes, while 80 chicken, 11 duck, and 24 minor poultry species isolates contained Ck/Bei-like M genes. For the NS gene, all of these H9N2 viruses had a Ck/Bei-like gene segment, except for three viruses (Gf/ST/2076/01, Pa/ST/2875/01, and Cu/ST/22116/05) whose genes belonged to the G1-like lineage, similar to the results for the HA gene. It is interesting that two duck H9N2 viruses (Dk/ST/163/04 and Dk/ST/7488/04) had both M and NS genes that always clustered with those of viruses from the gene pool in this region, including the Korean-like H9N2 virus.
Genotyping.
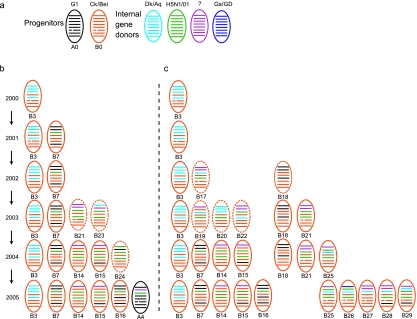
All H9N2 viruses from chickens and 76 of 79 viruses from minor poultry species belonged to the Ck/Bei-like lineage (genotype B series), while only 3 viruses from other minor poultry species belonged to the G1-like lineage (genotype A series). Except for two duck isolates that were closely related to the Korean-like H9N2 virus and were of pure duck origin for each gene segment, the remaining duck H9N2 isolates were all genotype B series (Table 4). Thus, a total of 35 genotypes of H9N2 influenza viruses were identified from different types of poultry under our surveillance, including 14 novel genotypes that were not recognized in our previous study (31) and were designated genotypes B17 to B29 and genotype A4 (Fig. 5; Table 4). Nineteen genotypes were identified from chickens, including the progenitor genotype B0 in 2001, and 16 additional genotypes were detected from other minor poultry species (Table 4). These novel genotypes were all triple or even quadruple reassortants, with gene segments from Ck/Bei-like, G1-like, aquatic bird, duck, and H5N1/01-like viruses (Fig. 4 and 5; Table 4).
FIG. 5.
Genotypes of H9N2 influenza viruses of chickens and other minor poultry species in southern China. The figure shows progenitors of H9N2 influenza virus genotype A and B series and internal gene donors (a) and genotypes from other minor poultry species (b) and chickens (c) in southern China. Dashed lines represent transient and short-lived genotypes. Details of transient genotypes are given in Table 6. The eight gene segments (horizontal bars starting from the top) are PB2, PB1, PA, HA, NP, NA, M, and NS. Each color represents a virus lineage. Genotype definitions are described in Materials and Methods. Abbreviations are listed in Table 4.
It is noted that genotype B3 viruses have been detected in chickens and other minor poultry species every year since they appeared in 2000, suggesting that this subtype of viruses has become established in these birds. Genotype B7 viruses were initially detected in other minor poultry species and quail from 2001 onwards and failed to be detected in quail in 2005 (31), being detected only in chickens in 2004-2005, suggesting that the interspecies transmission direction of this genotype was from quail or other minor poultry species to chickens. The remaining genotypes, genotype B17 to B29, were only occasionally and transiently detected in chickens or minor poultry species. It has been noted that the genotype number being recognized in both chickens and other minor poultry species has increased since 2003, suggesting that H9N2 influenza viruses have become more and more diversified in this region (see Table 6). However, the number of genotypes detected in ducks has decreased since 2003, which correlates with the decreased number of isolates detected from ducks, suggesting that the Ck/Bei-like variants are not adaptive in domestic ducks.
TABLE 6.
Distribution of different H9N2 virus genotypes among different types of poultry from southern China
| Year | Genotype (no. of isolates)a
|
|||
|---|---|---|---|---|
| Chickens | Other minor poultry species | Quail | Ducksb | |
| 2000 | B3 (8) | B1 (2), B2 (1), B3 (1), B5 (2) | A0 (3), A1 (1), B1 (1), B2 (1), B3 (1) | B1 (2), B3 (5), B4 (1), B5 (1), B-n8 (1) |
| 2001 | B0 (3), B3 (8), B4 (2) | A0 (2), B3 (2), B4 (1), B7 (2) | A0 (4), A1 (3), A2 (1), B3 (1), B4 (1), B5 (3), B6 (1) | B3 (4), B7 (1), B-n6 (1) |
| 2002 | B3 (7), B17 (1), B18 (3) | B3 (2), B7 (5), B8 (2) | A0 (2), A3 (3), B3 (1), B7 (2), B8 (3) | B3 (2), B12 (1) |
| 2003 | B3 (3), B18 (2),B19 (1), B20 (1), B21 (5), B22 (1) | B2 (2), B3 (1), B7 (11), B12 (1), B21 (3), B23 (1) | A3 (7), B7 (6), B9 (1), B10 (1), B11 (1), B12 (1), B13 (1) | B3 (1), B7 (1), B21 (2) |
| 2004 | B3 (5), B18 (2), B21 (1), B7 (4), B12 (1), B14 (5), B15 (1), B25 (2) | B3 (1), B7 (16), B8 (1), B14 (8), B15 (2), B24 (1) | A3 (5), B7 (1), B14 (2), B15 (3) | B21 (1) |
| 2005 | B3 (2), B7 (1), B14 (14), B15 (21), B25 (1), B16 (1), B26 (1), B27 (1), B28 (3), B29 (1) | A4 (1), B3 (1), B7 (2), B14 (1), B15 (1), B16 (3) | A3(4), B8 (3), B14 (1), B15 (2), B16 (2) | B15 (1) |
Number of isolates recognized among representative strains tested.
Viruses characterized in reference 19 were also included.
Of the three G1-like viruses, two were nonreassortant viruses (genotype A0), while the other was a novel reassortant with a PB1 gene of unknown source that was designated genotype A4, in contrast to genotype A3 viruses. This novel PB1 gene had also been detected in Ck/Bei-like H9N2 influenza viruses from minor poultry species but not from quail, suggesting that genotype A4 viruses might be generated directly within other minor poultry species (Table 4).
Molecular characterization.
The deduced amino acid sequences of the viruses were aligned and compared with those of other representative H9N2 viruses in this region. Except for six viruses from other minor poultry species, the HAs of all viruses tested had 226Leu at receptor binding sites (H3 numbering), as recognized in our previous studies, while those of the other six viruses had Glu at position 226 (Table 5) (22). Other substitutions related to receptor binding sites have not been recognized. Most H9N2 viruses analyzed maintained an Arg-Ser-Ser-Arg (R-S-S-R) motif at the connecting peptide of their HA, but a few substitutions were observed at each site of the connecting peptide (Table 5). However, no additional basic amino acids were found in all tested viruses. Thirty-one of the Ck/Bei-like H9N2 viruses had the same three-amino-acid deletion (positions 62 to 64) at the NA stalk region, as previously recognized (11, 13), while one G1-like virus had a two-amino-acid deletion (positions 38 and 39) which was also previously recognized in G1-like viruses from quail (11, 12, 31).
TABLE 5.
Comparison of amino acid sequences of HA, NA, and M2 genes of representative viruses from southern China
| Virus | Genotype | Residue at RBSa
|
NA deletion (aa) | Connecting peptidec | M2 residue at amantadine resistance mutation position
|
||
|---|---|---|---|---|---|---|---|
| 226 | 228 | 27 | 31 | ||||
| Qa/HK/G1/97 | A0 | L | G | 38-39 | R-S-S-R | V | S |
| Dk/HK/Y280/97 | B0 | L | G | 62-64 | R-S-S-R | V | S |
| Dk/HK/Y439/97 | Korean | Q | G | A-S-N-R | V | S | |
| Dk/HK/289/78 | Q | G | A-S-N-R | ||||
| Dk/ST/163/04 | Korean | Q | G | A-S-D-R | V | S | |
| Dk/ST/7448/04 | Korean | Q | G | A-S-G-R | V | S | |
| Cu/ST/22116/05 | A4 | Q | G | 38-39 | R-S-S-R | V | S |
| Pa/ST/2063/00 | B1 | Qb | G | R-S-S-R | V | N | |
| Pa/ST/24/00 | B5 | L | G | R-S-S-R | V | S | |
| Gf/ST/1677/00 | B3 | L | G | 62-64 | R-S-S-R | V | S |
| Cu/ST/338/02 | B8 | L | G | R-S-S-R | V | S | |
| Pa/ST/4525/02 | B3 | L | G | R-S-S-R | V | S | |
| Ph/ST/443/03 | B7 | L | G | K-S-S-R | V | S | |
| Ck/ST/94/00 | B3 | L | G | 62-64 | R-S-S-R | V | S |
| Ck/ST/1579/00 | B3 | L | G | 62-64 | R-S-S-R | V | N |
| Ck/ST/4608/02 | B3 | L | G | R-S-S-R | A | S | |
| Ck/HN/774/02 | B17 | L | G | 62-64 | R-L-S-R | V | G |
| Ck/ST/3040/03 | B21 | L | G | R-S-I-R | V | S | |
| Ck/GX/1857/04 | B25 | L | G | 62-64 | R-A-S-R | V | S |
| Ck/GX/187/05 | B25 | L | G | 62-64 | R-A-S-K | V | S |
| Ck/ST/22504/05 | B14 | L | G | R-S-S-R | V | N | |
RBS, receptor binding site.
Representative of six viruses from other minor poultry species.
Connecting peptide from positions −4 to −1 of HA1. Italics represent newly identified motifs.
An Arg292Lys mutation in the NA gene that has been associated with oseltamivir resistance was not detected in any of the H9N2 viruses tested (14). However, 11 viruses had Asn at residue 31 of the M2 protein, which is responsible for amantadine resistance of influenza viruses (1). Four of 112 H9N2 influenza viruses from chickens had a Val27Ala mutation, which is also associated with amantadine resistance of influenza virus (1). One virus, Ck/HN/774/02, had a Ser31Gly mutation which had not been observed before. Three G1-like viruses from minor poultry species had Glu at position 92 of the NS1 protein, a mutation related to the pathogenicity of H5N1 influenza virus in pigs, but the remaining 203 viruses had Asp, a residue typically observed in avian influenza viruses, at this position (27).
DISCUSSION
Since the late 1990s, two distinct H9N2 virus lineages have become established in chickens and quail in southern China (11-13). In the present study, we genetically and antigenically characterized H9N2 viruses isolated from chickens, ducks, and other minor poultry species in our surveillance from 2000 to 2005. All H9N2 viruses from chickens and most of the viruses from ducks and other minor poultry species belonged to the Ck/Bei-like lineage, while G1-like viruses prevailed mainly in quail and were rarely detected in other minor poultry species. Phylogenetic studies revealed that two-way interspecies transmission occurred between different types of poultry and that reassortment events among established virus lineages (e.g., H5N1 and H9N2 viruses) were mostly responsible for the current great genetic diversity in H9N2 and H5N1 variants in this region.
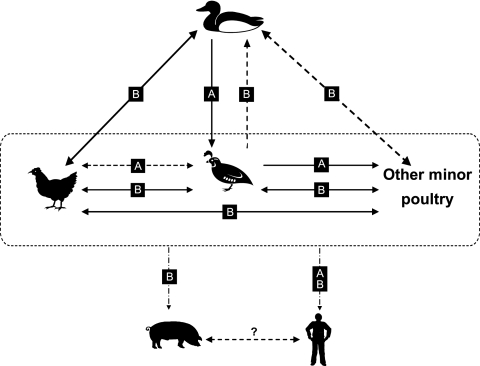
Even though many novel reassortants have been generated, relative host restriction is present for H9N2 viruses. Quail are able to harbor both Ck/Bei-like and G1-like viruses and may play a central role in the current ecosystem of southern China (Fig. 6), while chickens could support only Ck/Bei-like viruses, but other minor poultry species seem to be in between quail and chickens, as most of their isolates were Ck/Bei-like, with a few G1-like viruses. It was also noted that only a few Ck/Bei-like viruses have been recognized in domestic ducks since 2002 (Fig. 1 and Table 6). These findings suggest that the current direction of gene flow of Ck/Bei-like H9N2 viruses is from chickens to quail, other minor poultry species, and ducks. G1-like viruses and those Ck/Bei-like viruses with G1-like gene segments could be introduced from quail to other minor poultry species.
FIG. 6.
Ecology of H9N2 influenza viruses in southern China. “A” and “B” shown in black boxes indicate Qa/HK/G1/97-like and Ck/Bei/1/94-like virus lineages, respectively. Solid lines represent confirmed gene flow directions. Dashed lines indicate indirect evidence of gene flow.
It is noted that G1-like viruses were not detected in chickens in the present study but did cause outbreaks in chickens in Middle Eastern and European countries, including Iran, Saudi Arabia, United Arab Emirates, and Germany (1, 2, 4). Even though those G1-like viruses underwent further reassortment with “local” influenza viruses and generated novel reassortants (1), the reasons that G1-like viruses have not been detected in other types of terrestrial poultry in our surveillance are still unknown. Obviously, the G1-like viruses detected in southern China and Middle Eastern countries shared the same progenitor and evolutionary pathway. Thus, current G1-like viruses in southern China might have originally been introduced from Middle Eastern countries, or it is also likely that the virus spread the other way around, similar to the transmission of Qinghai-like H5N1 virus from the east transmitting in a western or northwestern direction (6). It has been understood that there is influenza virus gene exchange between the extremities of Europe and Asia (9).
The present study revealed that prototypes of Ck/Bei-like and G1-like viruses (genotypes B0 and A0) have been replaced by their descendant reassortants since 2002 and 2003, respectively (Table 6). Genotypes B3 and B7 were persistent in either chickens or other minor poultry species from 2001 to 2005, but these two genotypes still have not become predominant in their host, as novel reassortants continued to emerge every year (Fig. 5 and Table 6). This suggests that the Ck/Bei-like viruses are of genetically unstable and transient gene constellations. This situation could give rise to great uncertainty for the current ecosystem in southern China. First, novel reassortants could continue to have further interspecies transmission and further reassort with other viruses to cause new outbreaks; and second, novel reassortants keep challenging the species barrier between birds and mammals.
The findings of the present investigation show a dynamic ecosystem with multiple interspecies transmissions of H9N2 influenza virus (Fig. 6). Two-way transmissions of H9N2 between different types of poultry in southern China promote the development of various genotypes, not only for H9N2 but also for H5N1 and other influenza viruses. The combination of a dynamic H9N2 ecosystem and the presence of multiple novel genotypes increases the risk of H9N2 viruses entering the human population themselves or, like the Hong Kong H5N1 bird flu incident (11), indirectly contributing their internal gene complex to promote the introduction of other subtypes to humans. This situation has posed a persistent and significant pandemic threat in the past 10 years. However, to reduce this kind of risk, these interspecies transmissions must be disrupted. This study provides clear clues about how to interrupt these two-way transmissions between different poultry species, e.g., with modified market systems and farming practices in the affected regions.
H5N1 viruses have usually caused infections with high mortality in humans, which resulted in early detection and prevention of further adaptation and reassortment to develop human-to-human transmission; however, H9N2 human infections manifest with a typical human flu-like illness that can easily be overlooked (3, 5). At this point, H9N2 viruses have a greater chance and time to develop the ability of human-to-human transmission. Continuing influenza virus surveillance of both animal and human aspects seems to be the best option for detecting and interrupting this kind of development.
Acknowledgments
This study was supported by the Li Ka Shing Foundation, the Research Fund for Control of Infectious Diseases and Research Grants Council (HKU1/05C) of the Hong Kong SAR Government, and the National Institutes of Health (NIAID contract HHSN266200700005C).
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Aamir, U. B., U. Wernery, N. Ilyushina, and R. G. Webster. 2007. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology 361:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks, J., E. C. Speidel, P. A. Harris, and D. J. Alexander. 2000. Phylogenetic analysis of influenza A viruses of H9 haemagglutinin subtype. Avian Pathol. 29:353-360. [DOI] [PubMed] [Google Scholar]
- 3.Butt, K. M., G. J. D. Smith, H. Chen, L. J. Zhang, Y. H. Leung, K. M. Xu, W. Lim, R. G. Webster, K. Y. Yuen, J. S. M. Peiris, and Y. Guan. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 43:5760-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron, K. R., V. Gregory, J. Banks, I. H. Brown, D. J. Alexander, A. J. Hay, and Y. P. Lin. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:36-41. [DOI] [PubMed] [Google Scholar]
- 5.Centre for Health Protection. 20 March 2007. Girl recovered from influenza A virus (H9N2). Centre for Health Protection, Department of Health, Hong Kong, SAR, China. http://www.chp.gov.hk/content.asp?lang=en&info_id=9158&id=116.
- 6.Chen, H., G. J. D. Smith, K. S. Li, J. Wang, X. H. Fan, J. M. Rayner, D. Vijaykrishna, J. X. Zhang, L. J. Zhang, C. T. Guo, C. L. Cheung, K. M. Xu, L. Duan, K. Huang, K. Qin, Y. H. Leung, W. L. Wu, H. R. Lu, Y. Chen, N. S. Xia, T. S. Naipospos, K. Y. Yuen, S. S. Hassan, S. Bahri, T. D. Nguyen, R. G. Webster, J. S. Peiris, and Y. Guan. 2006. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl. Acad. Sci. USA 103:2845-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, Y. K., H. Ozaki, R. J. Webby, R. G. Webster, J. S. Peiris, L. Poon, C. Butt, Y. H. Leung, and Y. Guan. 2004. Continuing evolution of H9N2 influenza viruses in southeastern China. J. Virol. 78:8609-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, Y. K., S. H. Seo, J. A. Kim, R. J. Webby, and R. G. Webster. 2005. Avian influenza viruses in Korean live poultry markets and their pathogenic potential. Virology 332:529-537. [DOI] [PubMed] [Google Scholar]
- 9.Duan, L., L. Campitelli, X. H. Fan, C. Y. H. Leung, D. Vijaykrishna, J. X. Zhang, I. Donatelli, M. Delogu, K. S. Li, E. Foni, C. Chiapponi, W. L. Wu, H. Kai, R. G. Webster, K. F. Shortridge, J. S. M. Peiris, G. J. D. Smith, H. Chen, and Y. Guan. 2007. Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 81:7529-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan, Y., J. S. Peiris, A. S. Lipatov, T. M. Ellis, K. C. Dyrting, S. Krauss, L. J. Zhang, R. G. Webster, and K. F. Shortridge. 2002. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. USA 99:8950-8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 16:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, Y. J., S. Krauss, D. A. Senne, I. P. Mo, K. S. Lo, X. P. Xiong, M. Norwood, K. F. Shortridge, R. G. Webster, and Y. Guan. 2000. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267:279-288. [DOI] [PubMed] [Google Scholar]
- 14.Herlocher, M. L., R. Truscon, S. Elias, H. L. Yen, N. A. Roberts, S. E. Ohmit, and A. S. Monto. 2004. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J. Infect. Dis. 190:1627-1630. [DOI] [PubMed] [Google Scholar]
- 15.Huelsenbeck, J. P., and F. R. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Humberd, J., Y. Guan, and R. G. Webster. 2006. Comparison of the replication of influenza A viruses in Chinese ring-necked pheasants and chukar partridges. J. Virol. 80:2151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, Y. J., J. Y. Shin, M. S. Song, Y. M. Lee, J. G. Choi, E. K. Lee, O. M. Jeong, H. W. Sung, J. H. Kim, Y. K. Kwon, J. H. Kwon, C. J. Kim, R. J. Webby, R. G. Webster, and Y. K. Choi. 2007. Continuing evolution of H9 influenza viruses in Korean poultry. Virology 359:313-323. [DOI] [PubMed] [Google Scholar]
- 18.Li, K. S., Y. Guan, J. Wang, G. J. D. Smith, K. M. Xu, L. Duan, A. P. Rahardjo, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. T. Estoepangestie, A. Chaisingh, P. Auewarakul, H. T. Long, N. T. Hanh, R. J. Webby, L. L. Poon, H. Chen, K. F. Shortridge, K. Y. Yuen, R. G. Webster, and J. S. M. Peiris. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209-213. [DOI] [PubMed] [Google Scholar]
- 19.Li, K. S., K. M. Xu, J. S. Peiris, L. L. Poon, K. Z. Yu, K. Y. Yuen, K. F. Shortridge, R. G. Webster, and Y. Guan. 2003. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans? J. Virol. 77:6988-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova, N. V., H. Ozaki, H. Kida, R. G. Webster, and D. R. Perez. 2003. Replication and transmission of influenza viruses in Japanese quail. Virology 310:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 23.Nylander, J. A. A. 2004. MRMODELTEST 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 24.Peiris, J. S., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. Ip, R. W. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 26.Perez, D. R., W. Lim, J. P. Seiler, G. Yi, M. Peiris, K. F. Shortridge, and R. G. Webster. 2003. Role of quail in the interspecies transmission of H9 influenza A viruses: molecular changes on HA that correspond to adaptation from ducks to chickens. J. Virol. 77:3148-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 28.Shortridge, K. F. 1992. Pandemic influenza: a zoonosis? Semin. Respir. Infect. 7:11-25. [PubMed] [Google Scholar]
- 29.Swofford, D. L. 2001. PAUP*: phylogenetic analysis using parsimony (and other methods) 4.0 beta. Sinauer Associates, Sunderland, MA.
- 30.Xu, C., W. Fan, R. Wei, and H. Zhao. 2004. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003 (H9N2) virus. Microbes Infect. 10:919-925. [DOI] [PubMed] [Google Scholar]
- 31.Xu, K. M., K. S. Li, G. J. Smith, J. W. Li, H. Tai, J. X. Zhang, R. G. Webster, J. S. Peiris, H. Chen, and Y. Guan. 2007. Evolution and molecular epidemiology of H9N2 influenza A viruses from quail in southern China, 2000 to 2005. J. Virol. 81:2635-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]