Abstract
Several different herpes simplex viruses (HSVs) and vectors are being explored as therapeutic products for use in the treatment of cancer and neurological disorders. The viral strain and the combination of mutant viral genes that ultimately may serve as a safe and optimal backbone for such products are still being explored. The large genome size and complexity of the viral life cycle make such determinations difficult, because the significance of differences between proposed products is difficult to evaluate. For example, we previously reported that two lineages of γ34.5-deleted HSVs used in clinical studies differ from each other in the size of the UL3 protein expressed (M. J. Dambach et al., Mol. Ther. 13:891-898, 2006). Because the function of UL3 is not known and UL3 gene expression is poorly understood, the significance of such a difference cannot be predicted. Here, I begin to address the function of UL3 by investigating UL3 gene expression. I report that the transcript start site of UL3 mRNA isolated from HSV type 1 (HSV-1)-infected cells maps to a position downstream of the predicted translation start site. By constructing and characterizing the recombinant virus CB8116, which has a mutation in the first in-frame start codon of this UL3 transcript, I demonstrated that UL3 protein translation initiates at the second in-frame start codon of the UL3 open reading frame. This information adds to the body of basic knowledge of HSV-1 biology that forms the foundation for our current understanding of HSV-based products. Future research on HSV-1 biology will facilitate the rational design and evaluation of future generations of therapeutic viruses.
Herpes simplex virus type 1 (HSV-1) is commonly associated with infections of the mucocutaneous membranes of the mouth and eyes, of the brain, and of internal organs of infected neonates (35). Recently, modified forms of HSV-1 have gained attention for their use in cancer and gene therapy clinical studies (2, 21, 36). HSVs proposed for use in cancer therapy studies are replication competent but are highly attenuated in the mouse nervous system. For some HSVs, this attenuation is due to the absence of the γ34.5 gene that encodes the neurovirulence factor infected cell protein 34.5. Recently, we reported that one lineage of γ34.5-deleted HSVs proposed for use in clinical studies expresses a truncated UL3 protein (3). This UL3 truncation appears to be limited to HSVs derived from the recombinant virus backbone R3617 (e.g., G207, MGH1, 3616UB, and G47Δ) (13, 14, 18, 27). In contrast, viruses derived from other HSV strains or lineages (e.g., R3659, HSV1716, and OncovexGM-CSF) are expected to express an intact UL3 protein (3, 5-7, 33). Because the function of UL3 is unknown, it is unclear whether the expression of a truncated or intact UL3 protein is optimal for viruses and vectors used in clinical trials. One focus of studies in my laboratory is to determine the function of UL3 in the viral life cycle.
UL3 belongs to a group of 36 HSV-1 accessory proteins that are not required for productive viral replication in cell culture (1, 19). Like many of these proteins, the role played by the HSV-1 UL3 protein in cell culture, animal models, and natural infection has not been reported. However, several studies on the UL3 protein and UL3 gene expression have provided valuable information about this accessory protein. Based on HSV-1 DNA sequence information, the UL3 open reading frame (ORF) is predicted to encode a 235-amino-acid (aa) protein (17). UL3 commonly is described as a nuclear phosphoprotein (4, 11, 28, 38). However, UL3 is found in both nuclear and cytoplasmic fractions of infected cells (4). Immunofluorescence microscopy has shown that UL3 relocates from the cytoplasm (38, 39) to the nucleus (16, 38, 39) as the viral replication cycle progresses. At late times postinfection, UL3 colocalizes in small intranuclear structures (SINs) with other viral proteins (e.g., ICP22 and UL4) (10, 16). Cells infected with viruses such as R325, lacking infected cell protein number 22 (ICP22), have abnormally shaped SINs (16, 30). In addition to their colocalization in the nucleus, UL3 and ICP22 both are present in multiple isoforms and are posttranslationally modified in the presence of the HSV-1 protein kinase UL13 (4, 15, 24, 38).
The size of the UL3 protein and the transcript from which it is translated remain undetermined. HSV-1 is reported to express two transcripts capable of encoding UL3: a short transcript of 0.6 to 1 kb in size and a longer transcript, 2.6 kb in size. Singh and Wagner (31) reported that the 5′ end of the shorter transcript maps to a site internal to the putative translation initiation site of UL3 (Fig. 1A). This report prompted several groups to suggest that UL3 is translated from the 2.6-kb polycistronic transcript that also contains the coding sequences of the viral genes UL1 and UL2 (15, 38). Because protein expression from a dicistronic or polycistronic transcript is rare in viruses and eukaryotes (reviewed in reference 12), I was particularly interested in investigating UL3 protein expression.
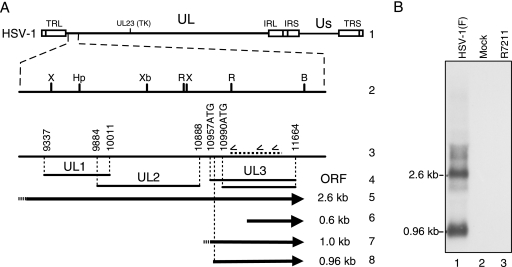
FIG. 1.
(A) Schematic diagram of the HSV-1 genome illustrating the relative sizes and locations of HSV-1 UL3-specific transcripts in the UL1-UL3 region. The HSV-1 genome is shown in line 1 in the prototype orientation. The left end of the unique long region (UL) of HSV-1 is shown expanded in lines 2 and 3. The relevant restriction endonuclease cleavage sites of HSV-1 are marked (line 2) relative to the coding regions of the UL1, UL2, and UL3 ORFs indicated in line 4. The nucleotide positions that form the boundaries of UL1, UL2, and UL3 ORFs as described by McGeoch et al. (17) are shown in lines 3 and 4. The 5′ position of the first and second UL3 ORF start codons, 10957ATG and 10990ATG, and the location of the antisense UL3 probe (horizontal dashed line) used in these experiments are indicated in line 3. Nucleotide positions are based on the HSV-1 strain 17+ sequence (GenBank accession no. X14112). The 2.6- and 0.6-kb transcripts reported by Singh and Wagner (31) are indicated as arrows in lines 5 and 6. The ∼1-kb transcript identified for HSV-1 (26) and HSV-2 (38) is shown in line 7. The 0.96-kb transcript reported here is shown in line 8. (B) Northern blot analysis of total RNA from uninfected (Mock) RS cells or RS cells infected with HSV-1(F) or R7211. RS cells were infected at an MOI of 10 with HSV-1(F) or R7211 and were harvested at 19 hpi. Total RNA from mock-infected cells or cells infected with HSV-1(F) or R7211 were loaded in lanes 1 to 3. Transcripts of 2.6 and 0.96 kb were identified only in HSV-1(F)-infected cell total RNA. TRL, terminal repeat long; IRL, inverted repeat long; IRS, inverted repeat short; TRS, terminal repeat short; US, unique short region; B, BamHI; Hp, HpaI; R, EcoRV; Xb, XbaI; X, XhoI.
Here I report the existence of a monocistronic UL3 transcript that has the capacity to encode a 224-aa UL3 protein. By constructing and characterizing recombinant virus CB8116, in which the first in-frame methionine codon of this transcript was mutated selectively, I show that this methionine codon is used exclusively to initiate UL3 protein translation in HSV-1 strain F [HSV-1(F)]-infected cells. Based on these studies, I suggest that UL3 is translated from a unique monocistronic transcript by the ribosomal scanning mechanism of translation initiation. I also show that virus CB8116 is a minimal deletion virus that is the only reported HSV-1(F) UL3 deletion virus described to date that retains the capacity to express neighboring proteins UL1, UL2, and UL4.
MATERIALS AND METHODS
Cells.
Virus stocks were grown in Vero cells (ATCC), and their titers were determined on Vero cells. Rabbit skin (RS) cells and human 143 thymidine kinase-deficient (143tk−) cells were obtained from Bernard Roizman and were used for selection of recombinant viruses. Cell lines (Vero, RS, and 143tk−) were grown in Dulbecco's modified Eagle medium (DMEM) (Biowhittaker) supplemented with 5% heat-inactivated bovine calf serum. Medium for 143tk− cells was supplemented either with bromodeoxyuridine or with hypoxanthine, aminopterin, and thymidine (HAT). Infected cells were maintained in medium 199V, which consists of medium 199 (Biowhittaker) supplemented with 1% heat-inactivated bovine calf serum, 50 U of penicillin/ml, and 50 μg of streptomycin/ml.
Viruses.
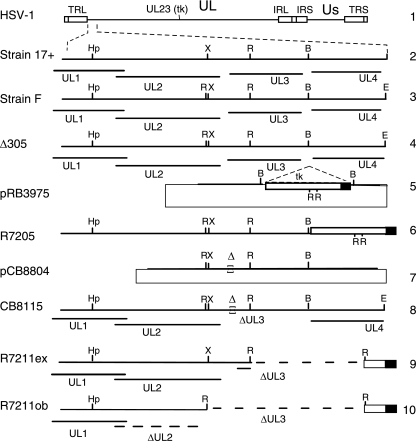
As described in more detail in Results, viruses CB8115 and CB8116 were derived from recombinant virus R7205. As described previously (1), R7205 contains the coding sequence of the thymidine kinase (tk) gene that is fused to the α27 promoter and inserted into the BamHI site between the UL3 and UL4 ORFs. Virus R7205 was derived from HSV-1(F) recombinant virus Δ305, which lacks the 500-bp SacI-BglII fragment of the tk gene, and is therefore tk− (23, 24). Recombinant virus R7211 also was derived from R7205. This recombinant virus was designed to be deleted in the UL3 coding sequence downstream of the EcoRV site (Fig. 2, line 9), which is located approximately 43 codons from the start of the UL3 ORF (1, 17). As described in Discussion, an unexpected EcoRV site 5′ to UL3 resulted in a complete deletion of the UL3 ORF as well as the 3′ terminus of the UL2 ORF (Fig. 2, line 10).
FIG. 2.
Schematic diagrams of the HSV-1 genome and viruses used in this study. The HSV-1 genome in the prototype orientation is shown in line 1. The left ends of the unique long region (UL) of HSV-1 strains 17+ and F and recombinant viruses derived from strain F are shown expanded in lines 2 to 10. The relevant restriction endonuclease sites of HSV-1 strains 17+ (line 2) and F (line 3) are indicated in relation to the coding regions of the UL1, UL2, and UL3 ORFs. The recombinant virus R7205 (line 6) was derived from cotransfection of viral DNA from Δ305 (line 4) with plasmid pRB3975 (line 5), containing the α27 promoter-driven tk gene cassette inserted at the BamHI site located between UL3 and UL4. Recombinant virus CB8115 (line 8) was derived from homologous recombination of R7205 viral DNA (line 6) with plasmid pCB8804 (line 7). The expected and observed deletions of the UL3 ORF of recombinant virus R7211 (R7211ex and R7211ob, respectively) are shown in lines 9 and 10, respectively. TRL, terminal repeat long; IRL, inverted repeat long; IRS, inverted repeat short; TRS, terminal repeat short; US, unique short region; B, BamHi; E, EcoRI; Hp, HpaI; R, EcoRV; X, XhoI.
Virus infections.
RS cells were exposed to virus at a multiplicity of infection (MOI) of 10 in medium 199V at 37°C with gentle rotation. After a 1- to 2-h adsorption period at 37°C, the inoculum was removed and replaced with fresh medium 199V. The incubation of infected cells was continued for the desired time period at 37°C, and the cells were harvested as described below.
Kits and reagents.
Restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs. Calf intestinal alkaline phosphatase and shrimp alkaline phosphatase were obtained from Boehringer Mannheim and Pharmacia, respectively.
Antisera.
The production of rabbit polyclonal antiserum to a glutathione S-transferase (GST)-UL3 fusion protein or a GST-UL4 fusion protein has been described previously (10, 15). The UL3-B and UL4-A antisera were used at dilutions of 1:1,000 and 1:500, respectively. The UL1-2 rabbit polyclonal antiserum was raised against a peptide corresponding to the C-terminal aa 200 to 224 of HSV-1 UL1. The UL2.1 rabbit polyclonal antiserum was raised against a peptide corresponding to HSV-2 UL2 aa 90 to 211. The UL1 and UL2 antisera were kind gifts from David Johnson and Sal Caradonna, respectively (8, 38).
Polyacrylamide gel electrophoresis and immunoblotting.
Lysates of virally infected RS cells were separated on a sodium dodecyl sulfate (SDS)-polyacrylamide gel with 10% (UL1, UL2, and UL3) or 15% (UL4) polyacrylamide and were transferred to a nitrocellulose membrane for immunoblotting. Ponceau S was used to verify equivalent transfer and loading and to identify the locations of the broad-range molecular weight standards (Bio-Rad). Immunoblotting was performed as described previously (15). Briefly, the protein-antibody complex was detected by alkaline phosphatase-conjugated goat anti-rabbit antiserum (A3687; Sigma) using an alkaline phosphatase conjugate substrate kit (Bio-Rad) for protein visualization. Digital photographs of immunoblots were imported into Adobe Photoshop and were contrast enhanced to ensure visibility of detected bands through photoreproduction.
RNA isolation and Northern blotting.
Total RNA was isolated (TRIzol reagent; GIBCO-BRL) from uninfected RS cells or RS cells infected with the indicated virus at an MOI of 10. Ten micrograms of RNA was separated on a 1% agarose gel and was subjected to Northern blotting (NorthernMax kit; Ambion) according to the manufacturer's instructions. Blots were incubated with radioactively labeled probe antisense to UL3, described below. Kodak Biomax film was exposed to the radiolabeled blot. Images of the resulting autoradiogram were imported into Adobe Photoshop and were adjusted for contrast.
Radiolabeled probe.
An antisense UL3 probe was generated in a Taq-based PCR using 250 nM of primer 11556R, 2.5 ng of UL3 PCR amplification product, [32P]dCTP, Redivue deoxycytidine 5′-[α-32P]triphosphate, triethylammonium salt (code no. AA0005; 500 μCi; Amersham), 3 U Taq (Promega), and 150 μM dATP, dGTP, and dTTP, all in a 1× PCR buffer (Promega). Cycling conditions were 35 cycles of 94°C for 15 s, 55°C for 20 s, and 72°C for 30 s. The probe was purified with ProbeQuant G50 MicroSpin columns (Amersham). The resulting probe contained 466 nucleotides of the UL3 coding sequence (nucleotides 11090 to 11556 of GenBank accession no. X14112). The sequence of the primers used were the following: 11090F, 5′-AGATACGACTCCCGCAGATTC-3′; and 11556R, 5′-GTGTAATACTTGCGCGGCTTGC-3′.
RACE. (i) SMART RACE and subcloning.
Total RNA was isolated (TRIzol reagent; GIBCO-BRL) from RS cells infected with HSV-1(F) at an MOI of 10. The BD SMART RACE cDNA amplification kit (BD Clontech) was used to amplify the 5′ end of the UL3 transcript by following the manufacturer's instructions. Briefly, 1 μg of RNA was used for first-strand DNA synthesis using PowerScript reverse transcriptase (BD Clontech). The first-strand cDNA was subjected to PCR amplification (Advantage 2 PCR enzyme system; Clontech) using gene-specific oligonucleotide primers and primers specific for the adapter ligated to the double-stranded DNA. Two independent PCR products were amplified, each obtained with different specific primers for the 5′ rapid amplification of cDNA ends (RACE) (11534R, 5′-GAAGCTGCTCTCGCACTCGAGCGGCGTG-3′; 11340R, 5′-CTTGGCCAGCGACAACCGCAGGTCCTTG-3′). Digital imaging of electrophoretically separated PCR products was performed with an Eagle Eye documentation system (Strategene). 5′ RACE PCR products were subjected to ligation with the pCR4-TOPO vector (Invitrogen). Ligation products were used to transform electrocompetent XL1-BLUE cells. Transformants were selected on Luria-Bertani-kanamycin plates. Colonies were screened for inserts by restriction digestion with EcoRI.
(ii) RLM-RACE and subcloning.
Vero cells were infected with HSV-1 strains F, 17+, and KOS and recombinant virus CB8116 at an MOI of 10. Total RNA was isolated from infected cells at 19 h postinfection (hpi) using an RNeasy mini kit (QIAGEN). 5′ RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) was performed using a FirstChoice RLM-RACE kit (Ambion) according to the manufacturer's instructions. Nested PCR was performed using the primer 11114R (5′-ATGGAATCTGCGGGAGTCGTATCT-3′) as the outer primer and 11077R (5′-ACTCGTGGTTTGGCGTGTCGAA-3′) as the inner primer. The UL3 forward primer 10959F (5′-GGTTAAACCTCTGGTCTCATACGGGT-3′) was used as an internal control. PCRs were performed in a PTC-100 programmable thermal controller (MJ Research, Inc.) using Titanium Taq DNA polymerase (BD Biosciences).
Cycling conditions for the outer 5′ RACE were 94°C for 3 min followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Cycling conditions for the inner 5′ RACE were 94°C for 3 min followed by 35 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s, and then a final incubation at 72°C for 7 min. RLM-RACE PCR products were ligated into the pCR4-TOPO vector (Invitrogen). Electrocompetent TOP10 cells were electroporated with the ligation product and transformants selected on Luria-Bertani-kanamycin plates.
Construction of plasmids.
Plasmid pCB8804 was constructed from the plasmid pRB5453 by using a QuikChange site-directed mutagenesis kit (Stratagene) as recommended by the manufacturer, along with the primer 5′-GGTCTCATACGGGTCGGTTAACTCGGGCGTCGGGGG-3′ and its complementary strand. Deviations of the oligonucleotide sequence from that of the HSV-1(F) sequence are underlined. In this plasmid, the second ATG of the UL3 ORF is mutated to AAC, which creates an HpaI restriction site that is used for diagnostic purposes.
Plasmid pRB5453 contains the entire UL3 ORF and an ∼1-kbp flanking region, including most of UL2 and the entire UL4 ORF in pGem3Z. It was created from the ∼2.7-kbp PstI-EcoRI (nucleotides 99000 to 12593) fragment of HSV-1(F) cosmid AX3-3 ligated into the PstI-EcoRI-digested vector pGem3Z.
Virus construction.
Recombinant virus CB8115 (tk−) was constructed in Vero cells by cotransfection of 1 μg of R7205 DNA and between 0.1 and 1 μg of plasmid pCB8804 DNA (Fig. 2, line 7) using Lipofectamine Plus reagent (Invitrogen). R7205 DNA was prepared from potassium acetate gradients as described elsewhere (9, 15). tk− progeny were selected in 143tk− cells in the presence of bromodeoxyuridine. Plaque isolates were plaque purified twice on Vero cells and were screened for the desired UL3 mutation by PCR using the primers 10588F and 11715R, which anneal to sites flanking the first ATG of the UL3 ORF. Amplification products were digested with HpaI restriction endonuclease and were separated by agarose gel electrophoresis. The desired isolates were further plaque purified on Vero cells.
CB8116, a tk+ version of recombinant virus CB8115, was constructed by cotransfection of CB8115 DNA with plasmid pRB165, containing the native tk sequence. tk+ viruses were selected on 143tk− cells overlaid with DMEM containing HAT and 5% calf serum (Sigma), as described previously (22, 23). Isolates were plaque purified in Vero cells and were screened for an intact tk gene by PCR using primers tk forward (5′-CAGCGTCTTGTCATTGGCGA-3′) and tk reverse (5′-TGGCAAGCCCATAAACG-3′) as described by O'Toole et al. (20).
Recombinant virus R8109 was made in RS cells by cotransfection of R4660 DNA with plasmid pRB165, containing the native tk sequence. tk+ viruses were selected and plaque purified as described above. Recombinant virus R4660 contains DNA encoding a 20-aa tag in the correct orientation and reading frame with the UL4 ORF, as described previously (10).
PCR analysis of viral DNA.
Viral DNA for PCR analysis was prepared from infected Vero cells using the Wizard genomic DNA purification kit (Promega). The resulting DNA was PCR amplified using 100 ng of infected cell DNA as the template and 100 ng of primers in a 50-μl PCR mixture containing Pfu DNA polymerase (Stratagene) and 5% dimethylsulfoxide. To verify that the viral DNA contained the desired mutation in the UL3 region, the following primers were used for amplification: UL2Xba3′For, 5′-TACTAAACACGACCCTGACC-3′; and UL4-3′For 5′-GCAGACAAACTTTTGGGGTG-3′, as described by Dambach et al. (3). Amplification products were digested with the restriction endonuclease HpaI (for viruses CB8115 and CB8116). The PCR product size was estimated by comparison with a 100-bp molecular size standard (NEB), as visualized by agarose gel electrophoresis.
Sequencing and analysis.
5′ RACE-RLM samples were sequenced by the CBER Facility for Biotechnology Resources. All other sequencing reactions were performed using a Big Dye terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). Automated sequence determination was performed with the ABI Prism 310 genetic analyzer, as described previously (3). Sequence analysis was performed using Vector NTI Advance 9.0 (Invitrogen) and nucleotide-nucleotide BLAST searches (BLASTn; NCBI).
RESULTS
UL3 transcripts.
The two 3′ coterminal UL3-specific transcripts identified in RS cells infected with HSV-1 strains 17+ and KOS that were identified by Singh and Wagner (31) are illustrated in Fig. 1 (Fig. 1A, lines 5 and 6). The first two ATG codons of the UL3 ORF are indicated at positions 10957 and 10990 of the viral genome (Fig. 1A, lines 3 and 4). Because the corresponding AUGs would be absent from the 0.6-kb transcript, we and others (15, 38) hypothesized that UL3 is translated from the 2.6-kb transcript by using alternative mechanisms of translation initiation. We first investigated the possibility that UL3 was translated from an internal ribosome entry site (IRES) in the 2.6-kb transcript. Despite significant experimental effort using standard techniques for IRES identification, we were unable to identify an IRES responsible for UL3 translation initiation (N. Martin and N. Markovitz, unpublished data).
HSV-1(F) was used in our previous studies on UL3. As the sequence of strain F differs from that of strains 17+ and KOS, which were used by Singh and Wagner (31), I examined the possibility that, at least for HSV-1(F), a unique transcript did exist from which a full-length UL3 protein was translated. To determine the size and, thus, potential coding capacity of HSV-1(F)-infected cell UL3 transcripts, Northern blotting was performed using a 32P-labeled probe antisense to UL3 (Fig. 1A, line 3, dashed bar) and total RNA from RS cells infected with either HSV-1(F) or the UL3 deletion virus R7211 (1) or from uninfected RS cells (Fig. 1B). Two transcripts from the HSV-1(F)-infected cells, 2.6 and 0.96 kb in size, hybridized to the UL3-specific probe (Fig. 1B, lane 1). No UL3-specific transcripts were identified in samples from R7211-infected or uninfected cells (Fig. 1B, lanes 2 and 3). The inability to detect the UL3-specific transcripts was not unexpected for R7211 infected cells, because the expected genome of R7211 (R7211ex) lacks the sequence complementary to the UL3 probe (Fig. 2, line 9) (1). Based on these findings, I concluded that a full-length UL3 protein may be translated from the 0.96-kb transcript by using a classical ribosomal scanning model of translation initiation.
5′ RACE analysis.
To accurately map the start site of this UL3 transcript, RACE analysis was performed on total RNA isolated from HSV-1(F)-infected RS cells using one of two UL3-specific primers, 11534R or 11340R (second and third short arrows in line 3 of Fig. 1A), and the universal primer provided with the SMART RACE kit (Clontech). Using a UL3-specific primer paired with the universal primer, two 5′ RACE PCR products were amplified that contained the 5′ end of the UL3 transcript (Fig. 3A). The size of the resulting amplification products, approximately 400 and 600 bp, was consistent with a transcript start site near the beginning of the UL3 ORF (Fig. 1A, line 7). The amplification products were subcloned into the pCR4-TOPO vector (Invitrogen). One subclone from each amplification product was sequenced. Sequence analysis revealed that the 5′ terminus of the UL3 transcript was located at position 10974 by using primer 11534R and at position 10975 by using primer 11340R (Fig. 3B, lines 2 and 3).
FIG. 3.
(A) HSV-1(F) 5′ RACE PCR products separated by agarose gel electrophoresis. 5′ RACE UL3 PCR products, amplified using a universal primer and UL3 sequence-specific primers 11534R and 11340R, are shown in lanes 2 and 3, respectively. (B) Alignment of the HSV-1 DNA nucleotide sequence with the sequence of the 5′ end of the UL3 transcripts reported here. The DNA sequence of the HSV-1 genome near the 5′ end of the UL3 ORF is shown in line 1. The sequence corresponding to the 5′ end of the UL3 transcripts derived from the amplification products depicted in panel A are shown in lines 2 and 3. The sequence corresponding to the 5′ end of UL3 transcripts from HSV-1 strains F, 17+, and KOS using the RLM-RACE method are shown in lines 4 to 6, respectively. The relevant portion of the DNA sequence of plasmid pCB8804 is shown in line 7. The 5′ end of the UL3 transcript from CB8116 is shown in line 8. The mutation of the six-nucleotide sequence of UL3 containing the second ATG codon, GTGATG to GTTAAC (boldface and italics), in plasmid pCB8804 and virus CB8116 was verified by sequence analysis, as shown in lines 7 and 8.
To rule out the possibility that the 5′ terminus I identified at position 10975 resulted from a degraded transcript, a second set of experiments was performed with HSV-1(F)-infected cells using a 5′ RACE method that selected for only 5′-capped mRNA (RLM-RACE; Ambion) (29). Briefly, Vero cells were infected with HSV-1(F) at an MOI of 10 and were harvested at 19 hpi. Total RNA was isolated and processed for RLM-RACE as described in Materials and Methods. A 5′ RACE PCR product of ∼150 bp was amplified using the gene-specific primer 11114R (Fig. 1A, line 3, first short arrow). The amplification product was subcloned into the pCR4-TOPO vector, and one isolate was sequenced. Sequence analysis identified the 5′ initiation site of the HSV-1(F) UL3 transcript to be at position 10974 (Fig. 3B, line 4).
To determine whether the UL3 transcript start site of HSV-1(F) differs from that of strains KOS and 17+, I included total RNA from cells infected with HSV-1 strains KOS and 17+ in parallel studies with the HSV-1(F) sample from the RLM-RACE experiment. The results of these studies revealed that the UL3 transcript initiation site of HSV-1 strains 17+ and KOS was located at position 10974 (Fig. 3B, lines 5 and 6). I thus concluded that HSV-1 strains F, KOS, and 17+ shared the same UL3 transcript start site (Fig. 3B, lines 4, 5, and 6) and thus encoded a 224-aa UL3 protein.
I next examined the possibility that a larger (235-aa) UL3 protein is translated from the larger 2.6-kb transcript using the first ATG codon of the UL3 ORF (10957ATG) (Fig. 1A, line 3), which is absent from the 0.96-kb transcript (Fig. 1A, line 8). To address this possibility, I constructed recombinant virus CB8115 and its tk repair virus CB8116, described below. In both viruses, the second ATG codon of the UL3 ORF was mutated to AAC (Asp) (Fig. 3B, line 8). I predicted that if the entire UL3 ORF, which encoded a 235-aa UL3 protein, were translated from the 2.6-kb transcript, then these recombinant viruses should express detectable levels of UL3 protein.
Construction and characterization of recombinant viruses CB8115 and CB8116, in which the predicted start codon is mutated.
The construction strategies for viruses CB8115 (tk−) and CB8116 (tk+) are illustrated in Fig. 2 and are described in Materials and Methods. Briefly, CB8115 was constructed in Vero cells by cotransfection of R7205 DNA with plasmid pCB8804 DNA (Fig. 2, lines 6, 7, and 8), in which the native sequence GTGATG containing the putative UL3 start codon at position 10990 was mutated to GTTAAC, encoding the HpaI restriction endonuclease site (Fig. 3B, line 7). Subsequently, a tk+ virus, CB8116, was constructed by cotransfection of CB8115 DNA with plasmid pRB165, containing the native tk sequence. Mutagenesis of the start codon was confirmed for viruses CB8115 and CB8116 by HpaI restriction endonuclease cleavage of PCR amplification products that spanned the mutation. The sequences of the UL3 ORF and flanking regions for CB8116 were further characterized by sequence analysis of PCR amplification products from viral DNA, as described in Materials and Methods. The presence of the intact UL23 and tk genes was verified by the absence of the syn phenotype in infected cells and by PCR analysis of the viral genome.
Recombinant viruses CB8115 and CB8116 fail to express UL3.
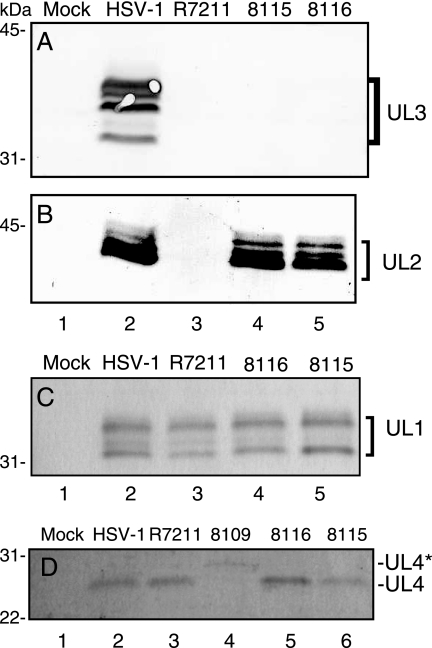
To test for the expression of UL3 from the 2.6-kb transcript, I performed an immunoblot of uninfected and infected RS cell lysates using rabbit polyclonal antiserum to a GST-UL3 fusion protein (7). RS cells were infected at an MOI of 10 with the following virus: HSV-1(F), R7211, CB8115, or CB8116. Multiple isoforms of the UL3 protein were readily detected in lysates of HSV-1(F)-infected RS cells (Fig. 4A, lane 2). In contrast, UL3 was undetectable in lysates of uninfected RS cells or RS cells infected with R7211, CB8115, or CB8116 (Fig. 4A, lanes 1 and 3 to 5). These data demonstrate that viruses mutated in the second ATG codon of the UL3 ORF fail to express UL3. I further show that the absence of UL3 protein was not due to the lack of a UL3 transcript, because results of 5′ RACE analysis of RNA isolated from CB8116-infected cells demonstrated that the UL3 transcript was present (Fig. 3B, line 8). I concluded from these studies that the UL3 protein is not translated from the first ATG codon of the UL3 ORF, as was initially predicted from the analysis of the DNA sequence (17). Instead, it is translated exclusively from the second ATG codon of the UL3 ORF. It is therefore appropriate to refer to the second ATG codon of the UL3 ORF as the UL3 start codon.
FIG. 4.
Immunoblot detection of the HSV-1 protein UL1, UL2, UL3, or UL4 in electrophoretically separated RS cell lysates. Lysates of uninfected RS cells (Mock; lane 1) or RS cells, infected at an MOI of 10 with the indicated virus, were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with antiserum to UL3 (A), UL2 (B), UL1 (C), or UL4 (D). RS cells were harvested 18 to 24 hpi. Approximately 2 × 105 (A and B) and 7 × 104 (C and D) infected cells were loaded per lane.
Cells infected with recombinant viruses CB8115 and CB8116 express viral proteins UL1, UL2, and UL4.
I next asked whether mutation of the UL3 start codon in viruses CB8115 and CB8116 affected protein expression from the neighboring genes UL1, UL2, and UL4. To address this question, I performed immunoblot analysis of electrophoretically separated lysates of uninfected RS cells or RS cells infected with HSV-1(F) or recombinant viruses. As expected, viral proteins were not detected in uninfected cells (Fig. 4, lanes 1). UL1 and UL2 proteins are easily detected in lysates of HSV-1(F)-infected cells (Fig. 4B and C, lanes 2) using antiserum to UL1 or UL2 protein (8, 37). UL4 was also detected in lysates of HSV-1(F)-infected cells, but it was detected with more difficulty (Fig. 4D, lane 2). I included lysates from R8109-infected cells in our immunoblots in order to verify the identity of the UL4 band detected by the UL4 antiserum. The UL4 protein detected from lysates of R8109-infected cells migrated more slowly in SDS-polyacrylamide gels (Fig. 4D, lane 4). This was expected, because the size of the R8109 UL4 protein is 20 aa larger than that of the native UL4 protein due to the insertion of an epitope-encoding sequence within the R8109 UL4 gene. As described in Materials and Methods, R8109 was derived from the tk− virus R4660, which also expresses an epitope-tagged UL4 protein (10).
Viral proteins UL1, UL2, and UL4 were also detected in lysates of cells infected with CB8115 and CB8116 (Fig. 4B and C, lanes 2, 4, and 5, and D, lanes 2, 5, and 6). UL1 and UL4 were also detected in the lysates of RS cells infected with R7211 (Fig. 4C and D, lanes 3). Surprisingly, the UL2 protein was not detected in the lysates of RS cells infected with R7211 (Fig. 4B, lane 3). I conclude from these data that UL3 deletion virus CB8116 is the only reported HSV-1(F) UL3 deletion virus described to date that retains the capacity to express neighboring proteins UL1, UL2, and UL4.
DISCUSSION
The rational design of therapeutic viruses and viral vectors requires an intimate knowledge of the structure and function of the viral components and an understanding of how they work together as a whole biological system during interaction with their host. Such an understanding will require more detailed knowledge of the functions of many HSV-1 proteins. This report has contributed to the understanding of UL3 gene expression. I have shown that the UL3 coding sequence is contained within an ∼1-kb transcript that initiates at position 10974, which is within the UL3 ORF. I further showed that the ATG codon used exclusively for UL3 translation is located at position 10990 of this UL3 transcript and that these features are conserved across HSV-1 strains F, 17+, and KOS. Finally, I report that the UL3 deletion virus CB8116 expresses viral proteins UL1, UL2, and UL4 and is thus well suited for future studies on the function of UL3. I also conclude that the UL3 gene is translated from a unique monocistronic transcript and encodes a 224-aa UL3 protein and that the UL3 leader sequence of only 16 nucleotides is sufficient for gene expression. These data provide a new understanding of UL3 gene expression and allowed us to make available a recombinant HSV-1 lacking UL3 protein expression.
Results reported here differ from those of initial studies on UL3 transcription. Singh and Wagner (31) identified two UL3-specific transcripts, 2.6 and 0.6 kb in size. In those studies, the 5′ transcript start site of the 0.6-kb UL3-specific transcript was mapped to a position 342 bases internal to the UL3 ORF start site for HSV-1 strains KOS and 17+, as well as for HSV-2 strain 333 (31). The results of these studies suggested that the entire UL3 coding sequence could be translated only from the 2.6-kb transcript.
In the studies reported here, no evidence of a 0.6-kb transcript was observed. Rather the results reported here are in closer agreement with results reported by Pyles and Thompson (25) and by Worrad and Caradonna (37). These groups also reported the presence of a 2.6-kb transcript and identified an approximately 1-kb UL3-specific transcript in cells infected with HSV-1 strain 17+ or HSV-2 strain 333. While these studies did not map the 5′ terminus of the shorter UL3-specific transcript (38), they also did not exclude the possibility that UL3 could be encoded by the shorter, 1-kb transcript.
Here, I suggest that UL3 is not translated by an alternate mechanism of translation initiation, as I and others had originally proposed. A simpler explanation consistent with existing observations is that UL3 is translated by using a standard scanning mechanism of translation initiation, in which the first in-frame AUG codon of an approximately 1-kb UL3-specific transcript serves as the UL3 translation initiation site. The first AUG codon of this transcript corresponds to the second ATG codon of the UL3 ORF, it is present in the transcript I have identified, and it has the proper Kozak context.
There are two additional observations that are noteworthy. One intriguing observation is the identification of a leader sequence for the UL3 transcript that is only 16 nucleotides long. Such an unusually short leader sequence is not unprecedented. I identified one report indicating that the K8.1 gene of Kaposi's sarcoma-associated herpesvirus has a 14-nucleotide 5′ untranslated region (32).
Another interesting observation is the absence of UL2 protein in lysates of RS cells infected with R7211. In hindsight, the lack of UL2 protein expression can be explained by the combination of the subcloning strategy used to construct R7211 (1) and a single-nucleotide difference between strains F and 17+ (accession nos. X14112 and AY730706, respectively). As reported by Dambach et al. (3), this single-nucleotide difference, located within the C terminus of the UL2 protein of strain F, created a novel EcoRV site that does not exist in strain 17+ (Fig. 2, compare line 2 to line 3). Because the subcloning strategy for the plasmid used in the construction of R7211 involved an EcoRV collapse (Fig. 2, compare line 6 to line 9), the presence of an unexpected EcoRV site in the UL2 ORF of strain F predicts the deletion of the C terminus of UL2, the entire UL3 coding region, and the UL3 5′ untranslated region (Fig. 2, line 10). Sequence analysis of DNA from R7211-infected cells confirmed this prediction (Martin and Markovitz, unpublished). This C-terminal deletion may destabilize UL2 at the protein or transcript level and explains the observed absence of UL2 protein in R7211-infected cells.
HSV-based viruses proposed for use in clinical trials are large, complex biological systems that still are poorly understood. Sequence analysis recently has revealed the presence of unexpected genomic mutations predicted to alter the structure of viral proteins encoded by HSV-based therapeutic viruses slated for, or already used in, clinical trials (3, 34). The UL3 protein is one example. C-terminally truncated UL3 protein is expressed by HSV-based viruses derived from the R3617 virus lineage (3). R3617-derived viruses are currently in or are proposed for use in clinical trials for cancer therapy (e.g., G207, MGH1, 3616UB, and G47Δ) (13, 14, 18, 27). In contrast, an intact UL3 protein most likely is expressed by other HSV-based viruses currently in or proposed for use in clinical trials (e.g., HSV1716, OncovexGM-CSF, HF10, and R3659); the backbones of these viruses are derived from different HSV-1 strains or lineages that express intact UL3 proteins (3, 5-7, 33, 34). Although the effect of an intact or truncated UL3 protein currently is not understood, the results of the studies presented here and the availability of a minimal UL3 deletion virus, CB8116, provide a solid foundation for future studies on the function of UL3. This knowledge and that gained from continued research on other poorly understood HSV-1 proteins may allow us to design future generations of HSV-based products that are safer and more effective than those currently available.
Acknowledgments
I thank David C. Johnson (Oregon Health and Science University) and Sal Caradonna (University of Medicine and Dentistry of New Jersey) for the gifts of UL1-2 antiserum and UL2.1 antiserum, respectively, and Bernard Roizman (The University of Chicago) for RS and 143 cells, viruses HSV-1(F), R7211, and R7205, cosmid AX3-3, and plasmid pRB165. I thank Sandra K. Weller (University of Connecticut Health Center) for HSV-1 strain KOS. HSV-1 strain 17+ originally was obtained from Duncan McGeoch. HSV-1(F) used in the RLM-RACE assay was obtained from the ATCC. I thank Natalia Martin, Ahmad Akhgar, Eli E. Bar, Prashanth Chandramani, Laura Corvette, Megan J. Dambach, Stephen J. Daniell, and Nidhi Rumpal for technical assistance. I thank Jerry Weir, Malcolm Moos, and Shuang Tang for their critical reviews of earlier drafts of the manuscript.
Fellowships awarded to Natalia Martin, Eli E. Bar, Prashanth Chandramani, Megan J. Dambach, Stephen J. Daniell, and Nidhi Rumpal were administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the FDA. This work was supported by intramural funding from the U.S. Food and Drug Administration allocated through the Division of Cellular and Gene Therapies.
The findings and conclusions in this report are those of the author and do not necessarily represent the views of the Food and Drug Administration.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bharatan, N. S., M. A. Currier, and T. P. Cripe. 2002. Differential susceptibility of pediatric sarcoma cells to oncolysis by conditionally replication-competent herpes simplex viruses. J. Pediatr. Hematol. Oncol. 24:447-453. [DOI] [PubMed] [Google Scholar]
- 3.Dambach, M. J., J. Trecki, N. Martin, and N. S. Markovitz. 2006. Oncolytic viruses derived from the γ34.5-deleted herpes simplex virus recombinant R3616 encode a truncated UL3 protein. Mol. Ther. 13:891-898. [DOI] [PubMed] [Google Scholar]
- 4.Ghiasi, H., G. C. Perng, S. Cai, A. B. Nesburn, and S. L. Wechsler. 1996. The UL3 open reading frame of herpes simplex virus type 1 codes for a phosphoprotein. Virus Res. 44:137-142. [DOI] [PubMed] [Google Scholar]
- 5.Harrow, S., J. Harland, R. Mabbs, R. Petty, M. Fraser, D. Hadley, J. Patterson, S. M. Brown, and R. Rampling. 2004. HSV1716 injection into the brain adjacent to tumour following surgical resection of high-grade glioma: safety data and long-term survival. Gene Ther. 11:1648-1658. [DOI] [PubMed] [Google Scholar]
- 6.Hu, J. C. C., Z. Han, B. Liu, M. Robinson, R. Branston, R. C. Coombes, and R. S. Coffin. 2003. Combination of OncoVEX with chemotherapy for cancer treatment. Cancer Gene Ther. 10:67. [Google Scholar]
- 7.Hu, J. C., M. J. Booth, G. Tripuraneni, D. Davies, S. A. A. Zaidi, M. Tamburo de Bella, M. J. Slade, S. B. Marley, M. Y. A. Gordon, R. S. Coffin, R. C. Coombes, and T. Kamalati. 2006. A novel HSV-1 virus, JS1/34.5-/47-, purges contaminating breast cancer cells from bone marrow. Clin. Cancer Res. 12:6853-6862. [DOI] [PubMed] [Google Scholar]
- 8.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igarashi, K., R. Fawl, R. J. Roller, and B. Roizman. 1993. Construction and properties of a recombinant herpes simplex virus 1 lacking both S-component origins of DNA synthesis. J. Virol. 67:2123-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jahedi, S., N. S. Markovitz, F. Filatov, and B. Roizman. 1999. Colocalization of the herpes simplex virus 1 UL4 protein with infected cell protein 22 in small, dense nuclear structures formed prior to onset of DNA synthesis. J. Virol. 73:5132-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klupp, B. G., H. Granzow, W. Fuchs, E. Mundt, and T. C. Mettenleiter. 2004. Pseudorabies virus UL3 gene codes for a nuclear protein which is dispensable for viral replication. J. Virol. 78:464-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak, M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361:13-37. [DOI] [PubMed] [Google Scholar]
- 13.Kramm, C. M., M. Chase, U. Herrlinger, A. Jacobs, P. A. Pechan, N. G. Rainov, M. SenaEsteves, M. Aghi, F. H. Barnett, E. A. Chiocca, and X. O. Breakefield. 1997. Therapeutic efficiency and safety of a second-generation replication-conditional HSV1 vector for brain tumor gene therapy. Hum. Gene Ther. 8:2057-2068. [DOI] [PubMed] [Google Scholar]
- 14.Liu, R., R. L. Martuza, and S. D. Rabkin. 2005. Intracarotid delivery of oncolytic HSV vector G47Δ to metastatic breast cancer in the brain. Gene Ther. 12:647-654. [DOI] [PubMed] [Google Scholar]
- 15.Markovitz, N. S., F. Filatov, and B. Roizman. 1999. The UL3 protein of herpes simplex virus 1 is translated predominantly from the second in-frame methionine codon and is subject to at least two posttranslational modifications. J. Virol. 73:8010-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markovitz, N. S., and B. Roizman. 2000. Small dense nuclear bodies are the site of localization of herpes simplex virus 1 UL3 and UL4 proteins and of ICP22 only when the latter protein is present. J. Virol. 74:523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA-sequence of the long unique region in the genome of herpes-simplex virus type-1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 18.Mineta, T., S. D. Rabkin, T. Yazaki, W. D. Hunter, and R. L. Martuza. 1995. Attenuated multi-mutated herpes-simples virus-1 for the treatment of malignant gliomas. Nat. Med. 1:938-943. [DOI] [PubMed] [Google Scholar]
- 19.Mori, I., and Y. Nishiyama. 2006. Accessory genes define the relationship between the herpes simplex virus and its host. Microbes Infect. 8:2556-2562. [DOI] [PubMed] [Google Scholar]
- 20.O'Toole, J. M., M. Aubert, A. Kotsakis, and J. A. Blaho. 2003. Mutation of the protein tyrosine kinase consensus site in the herpes simplex virus 1 alpha 22 gene alters ICP22 posttranslational modification. Virology 305:153-167. [DOI] [PubMed] [Google Scholar]
- 21.Parikh, N. S., M. A. Currier, Y. Y. Mahller, L. C. Adams, B. Di Pasquale, M. H. Collins, and T. P. Cripe. 2005. Oncolytic herpes simplex virus mutants are more efficacious than wild-type adenovirus type 5 for the treatment of high-risk neuroblastomas in preclinical models. Pediatr. Blood Cancer 44:469-478. [DOI] [PubMed] [Google Scholar]
- 22.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24:555-565. [DOI] [PubMed] [Google Scholar]
- 23.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25:227-232. [DOI] [PubMed] [Google Scholar]
- 24.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pyles, R. B., and R. L. Thompson. 1994. Evidence that the herpes-simplex virus type-1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 68:4963-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyles, R. B., and R. L. Thompson. 1994. Mutations in accessory DNA replicating functions alter the relative mutation frequency of herpes simplex virus type 1 strains in cultured murine cells. J. Virol. 68:4514-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyles, R. B., R. E. Warnick, C. L. Chalk, B. E. Szanti, and L. M. Parysek. 1997. A novel multiply-mutated HSV-1 strain for the treatment of human brain tumors. Hum. Gene Ther. 8:533-544. [DOI] [PubMed] [Google Scholar]
- 28.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2457. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 29.Schaefer, B. C. 1995. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem. 227:255-273. [DOI] [PubMed] [Google Scholar]
- 30.Sears, A. E., I. W. Halliburton, B. Meignier, S. Silver, and B. Roizman. 1985. Herpes simplex virus 1 mutant deleted in the alpha 22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh, J., and E. K. Wagner. 1993. Transcriptional analysis of the herpes-simplex virus type 1 region containing the TR(L)-U(L) junction. Virology 196:220-231. [DOI] [PubMed] [Google Scholar]
- 32.Tang, S., and Z. M. Zheng. 2002. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J. Biol. Chem. 277:14547-14556. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, S. K., Y. McGrath, M. J. Robinson, P. A. Reay, and R. S. Coffin. 2003. Herpes simplex virus as a vaccine delivery platform. Cancer Gene Ther. 10:95. [DOI] [PubMed] [Google Scholar]
- 34.Ushijima, Y., C. Luo, F. Goshima, Y. Yamauchi, H. Kimura, and Y. Nishiyama. 2007. Determination and analysis of the DNA sequence of highly attenuated herpes simplex virus type 1 mutant HF10, a potential oncolytic virus. Microbes Infect. 9:142-149. [DOI] [PubMed] [Google Scholar]
- 35.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 36.Williams, D. A., and T. P. Cripe. 2006. Adventitious mutations in clinical grade vectors: an issue to consider? Mol. Ther. 13:831-832. [DOI] [PubMed] [Google Scholar]
- 37.Worrad, D. M., and S. Caradonna. 1988. Identification of the coding sequence for herpes simplex virus uracil-DNA glycosylase. J. Virol. 62:4774-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Worrad, D. M., and S. Caradonna. 1993. The herpes simplex virus type 2 UL3 open reading frame encodes a nuclear localizing phosphoprotein. Virology 195:364-376. [DOI] [PubMed] [Google Scholar]
- 39.Yamada, H., Y. M. Jiang, H. Y. Zhu, K. Inagaki-Ohara, and Y. Nishiyama. 1999. Nucleolar localization of the UL3 protein of herpes simplex virus type 2. J. Gen. Virol. 80:2157-2164. [DOI] [PubMed] [Google Scholar]