Abstract
Coronaviruses express open reading frame 1a (ORF1a) and ORF1b polyproteins from which 16 nonstructural proteins (nsp) are derived. The highly conserved region at the carboxy terminus of ORF1a is processed by the nsp5 proteinase (Mpro) into mature products, including nsp7, nsp8, nsp9, and nsp10, proteins with predicted or identified activities involved in RNA synthesis. Although continuous translation and proteolytic processing of ORF1ab by Mpro is required for replication, it is unknown whether specific cleavage events within the polyprotein are dispensable. We determined the requirement for the nsp7 to nsp10 proteins and their processing during murine hepatitis virus (MHV) replication. Through use of an MHV reverse genetics system, in-frame deletions of the coding sequences for nsp7 to nsp10, or ablation of their flanking Mpro cleavage sites, were made and the effects upon replication were determined. Viable viruses were characterized by analysis of Mpro processing, RNA transcription, and growth fitness. Deletion of any of the regions encoding nsp7 to nsp10 was lethal. Disruption of the cleavage sites was lethal with the exception of that of the nsp9-nsp10 site, which resulted in a mutant virus with attenuated replication. Passage of the attenuated nsp9-nsp10 cleavage mutant increased fitness to near-wild-type kinetics without reversion to a virus capable of processing nsp9-nsp10. We also confirmed the presence of a second cleavage site between nsp7 and nsp8. In order to determine whether a distinct function could be attributed to preprocessed forms of the polyprotein, including nsp7 to nsp10, the genes encoding nsp7 and nsp8 were rearranged. The mutant virus was not viable, suggesting that the uncleaved protein may be essential for replication or proteolytic processing.
Coronavirus infections were described as early as the 1930s and were classified as a distinct family in the 1960s. Since then, coronaviruses have been isolated from almost all species of animals tested, including humans. Disease in animals is often severe and contributes to significant agricultural and economic loss. Until an emergent coronavirus was identified as the etiological agent of the severe acute respiratory syndrome (SARS) outbreak in 2003 (14, 30), human coronaviruses were typically associated with mild upper respiratory tract infections in winter (23, 37). In addition to the more-severe SARS coronavirus (SARS-CoV) (14, 30), two new human coronaviruses were found to be associated with lower respiratory disease, NL-63 and HKU-1 (16, 17, 53, 55). Since the SARS epidemic, new research into coronaviruses has been driven by the desire to better understand coronavirus replication, pathology, and mechanisms of host range expansion and has spurred the development of anticoronavirus therapies in the event of another deadly emergence.
Coronavirus replication is mediated by a complex of cellular and virus-encoded proteins of open reading frame 1 (ORF1), which comprises the first two-thirds of the viral 27- to 32-kb positive-strand RNA genome (Fig. 1A). Translation of ORF1 produces two polyproteins designated ORF1a and ORF1ab (Fig. 1B). The larger of the two polyproteins, pp1ab, is a carboxyl extension of pp1a that depends upon a −1 ribosomal frameshift at a pseudoknot structure to read through the translational stop at the end of ORF1a and translate through ORF1b (6, 31). For murine hepatitis virus (MHV), the polyproteins are processed into intermediate and mature nonstructural proteins (nsp) by two papain-like proteinase activities in nsp3 (PLP1 and PLP2) and by the nsp5 proteinase (3CLpro or Mpro) (18, 19, 33, 35, 52). Continuous translation and proteolytic processing of the polyproteins is required for productive infection, making the proteases attractive targets for antiviral therapies and suggesting that distinct roles may exist for both the mature and intermediate precursor forms of the proteins (5, 11, 29, 32, 45, 56).
FIG. 1.
MHV genome organization, proteolytic processing of the replicase polyproteins, and putative cleavage sites of nsp7 to nsp10. (A) The 5′ two-thirds of the MHV genome encode the ORF1 replicase proteins pp1a and pp1ab. ORF2 to ORF7 encode the major structural proteins S, E, M, and N, along with several accessory proteins. (B) The replicase polyproteins are processed by three proteases to produce 16 mature proteins. PLP1 is responsible for cleaving between nsp1-nsp2 and nsp2-nsp3 (black arrows), while PLP2 cleaves between nsp3-nsp4 (open circle). Mpro processes the remainder of the polyproteins (open triangles). The replicase proteins include a number of functionally conserved domains, including the two PLP proteases, an ADP-ribose-1‴-monophosphate-processing enzyme (X), three hydrophobic transmembrane domains (TM, MP1, and MP2), the nsp5 protease (Mpro), RdRp, a putative zinc-binding domain (Z) and helicase (Hel), an exonuclease (ExoN), an endoribonuclease (EU), and an S-adenosylmethionine-dependent ribose 2′-O-methyltransferase (OMT). (C) The amino acid sequences of the Mpro cleavage sites falling within the nsp7 to nsp10 region of the replicase polyproteins are shown (open arrows denote points of cleavage) along with their P1 Gln amino acid positions. A second putative cleavage site falling between nsp7-nsp8 is identified with a question mark and a vertical dotted line illustrating its proposed cleavage site.
The components of the coronavirus replication complex and their individual roles have been only partially defined. Processing of pp1a and pp1ab by nsp3 and nsp5 of most coronaviruses, MHV, and SARS-CoV yields 16 nsp proteins that are associated with the replication complex (Fig. 1B) (19, 40, 47, 48, 59, 60). Although functions have been assigned to many of these proteins by comparative sequence or biochemical analysis (2, 18, 19, 31), several are either poorly understood or unknown. In addition to the proteinases, putative or known functions of ORF1a-derived proteins include hydrophobic transmembrane domains in nsp3 (TM) (24), nsp4 (MP1), and nsp6 (MP2) that likely anchor the replication complex to cellular membranes. An ADP-ribose-1‴-phosphatase domain has also been identified in nsp3 (41-43). The proteins encoded by ORF1b have been associated directly with transcription and replication of viral genes. These include the RNA-dependent RNA polymerase (RdRp) in nsp12 (7, 13, 19), a putative zinc-binding domain (46), a nucleoside triphosphatase and superfamily 1 helicase (Hel) in nsp13 (26, 27, 46), a 3′-to-5′ exonuclease (ExoN) in nsp14 (38), an endoribonuclease in nsp15 (1), and an S-adenosylmethionine-dependent ribose 2′-O-methyltransferase in nsp16 (49).
Among the poorly understood replicase proteins are the “cassette” of nsp7, nsp8, nsp9, and nsp10 near the 3′ end of ORF1a (Fig. 1B). These proteins are present in all coronaviruses with significant identity and similarity and are processed by Mpro into mature products of 10, 22, 12.7, and 15 kDa, respectively. The nsp7 to nsp10 proteins colocalize with the replication complex and are presumably involved in viral RNA synthesis (3, 4, 20, 34, 54). Although the exact function of these proteins is unknown, recent work has provided some insight into their role in replication. Structural analyses of SARS-CoV nsp7 and nsp8 demonstrated that the two proteins form a hexadecameric supercomplex with electrostatic properties favorable for nucleic acid binding that may function as a processivity factor for the RdRp (58). nsp8 has also been shown to possess a low-fidelity primase activity and was proposed to provide the RNA primers required by the nsp12 RdRp during replication or transcription (10, 25). The SARS-CoV nsp9 crystal structure has also been resolved and has been shown to form homodimers possessing single-stranded RNA-binding properties; it has been suggested that the protein may serve to stabilize nascent and template RNA during replication, transcription, and processing (9, 15, 51). Putative temperature-sensitive mutations localized within nsp10 suggest that the protein may be involved with negative-strand synthesis (45). Recent reports describing the refinement of the nsp10 structure have revealed that the protein includes two Zn fingers (28, 36), exhibits nucleic acid-binding affinity (28, 36), and can crystallize to form a spherical dodecameric structure made up of 12 nsp10-nsp11 subunits (50) or nsp10 monomers and homodimers (28). Collectively, these data imply that nsp7 to nsp10 are important in—if not critical to—coronavirus replication.
In this report we describe the requirements for nsp7 to nsp10 and their proteolytic processing in MHV replication. Using the established MHV reverse genetics system (57), we individually deleted the genes encoding each protein, disrupted each of the cleavage sites associated with nsp7 to nsp10, and evaluated whether the protein domain was essential for productive virus infection. For all viable mutants, we characterized virus growth and RNA synthesis in cultured cells, transcription function, and in vitro growth fitness. A potential second cleavage site at the interface of nsp7 and nsp8 in class 2 coronaviruses was previously identified (Fig. 1C) (34), and we determined whether ablation of either or both of the putative cleavage sites affects replication. We also determined whether the precursor protein containing nsp7 to nsp10 performs a distinct role in viral replication by attempting to rescue viable virus when rearranging the nsp7 and nsp8 genes within the infectious clone. This work contributes to the growing body of data indicating that these small proteins are intimately involved with and critical to viral replication and as such are attractive targets for research aimed at understanding the intricacies of the coronavirus replication complex and as the focus of antiviral therapies.
MATERIALS AND METHODS
Cells.
Viruses were generated from infectious clones with delayed brain tumor (DBT) cells and baby hamster kidney cells stably expressing the MHV receptor (BHK-MHVR), as previously described (57). DBT cells were maintained in Eagle's minimal essential medium (MEM) supplemented with 10% fetal clone II, 5% tryptose phosphate broth, 0.05 μg of gentamicin/ml, and 0.25 μg of kanamycin/ml. BHK-MHVR cells were maintained in alpha MEM supplemented with 10% fetal calf serum (FCS), 10% tryptose phosphate broth, 0.05 μg of gentamicin/ml, 0.25 μg of kanamycin/ml, and 800 μg of G418 sulfate (Geneticin; Sigma)/ml.
Assembly of full-length MHV strain A59 (MHV-A59) and mutant infectious cDNA templates.
Viruses were produced from an MHV infectious clone, as previously described (57). Plasmids containing the viral genome were grown to a high concentration, isolated, and digested with Esp3I, BglI, or NotI, according to the manufacturer's directions (New England Biolabs). Viral cDNA inserts were visualized in 1% agarose gels in Tris-acetate-EDTA buffer on a Darkreader (Claire Chemical Research, Denver, CO) and isolated with the QIAquick Gel Extraction Kit (QIAGEN Inc., Valencia, CA). The concentrations of the individual MHV A-G DNA fragments were measured, pooled in stoichiometrically equivalent amounts to 1 μg of total DNA, and ligated with T4 DNA ligase (15 U/100 μl) at room temperature overnight in 30 mM Tris-HCl (pH 7.8)-10 mM MgCl2-10 mM dithiothreitol-1 mM ATP. The ligated products were purified by phenol-chloroform-isoamyl alcohol (1:1:24) and chloroform extraction, ethanol precipitated, and resuspended in H2O. Efficient ligation was confirmed by gel electrophoresis prior to in vitro transcription reactions.
Transcripts of the MHV N gene were coelectroporated with full-length transcripts of the genome. The N gene transcripts were driven from a T7 promoter at the 5′ end of a DNA template generated by PCR from the MHV-G plasmid with a T7-bearing 5′ primer (5′-ATGCATTAATACGACTCACTATAGGGAGAATGTCTTTTGTTCCTGGGCAAG-3′) (5′T7MHV-N) and a poly(A)-containing 3′ primer [5′-TCCGGA(TTT)8TTACACATTAGAGTCATCTTCTAACC-3′] [A59Ng3′(−)].
RNA transfection.
Full-length transcripts of MHV-A59 constructs were generated in vitro with some modification to the manufacturer's instructions (mMessage mMachine; Ambion, Austin, TX). Reactions were performed for 3 h at 37°C with 20-μl reaction mixtures supplemented with 3 μl of a 30 mM GTP stock for a 1:1 ratio of GTP to cap analog. The transcripts were treated with DNase I, precipitated in the provided LiCl solution, and resuspended in H2O. Full-length transcripts were verified by electrophoresis in 0.5% agarose gels in Tris-acetate-EDTA buffer containing 0.1% sodium dodecyl sulfate (SDS). Subconfluent cultures of BHK-R and DBT cells were trypsinized, washed twice with ice-cold phosphate-buffered saline (PBS), and resuspended in PBS at 107 cells/ml. Full-length RNA transcripts were mixed with N gene transcripts and electroporated into 800 μl of the BHK-R cell suspension with three pulses at 850 V and 25 μF in a Bio-Rad Gene Pulser II electroporator. The transfected BHK-MHVR cells were diluted 1:10, 1:100, and 1:1,000 in 5-ml volumes of fresh medium, mixed with 105 DBT cells, seeded in 60-mm-diameter cell culture dishes, and incubated at 37°C. Three hours later, the medium was removed from each dish and replaced with 5 ml of 1% agarose melted in 10% FCS-MEM. The overlaid plates were then incubated at 37°C in 5% CO2 and checked for plaques the next day. Plaques were isolated at 24 or 48 h postelectroporation and amplified on DBT monolayers in 60-mm-diameter cell culture dishes. The virus-containing medium was removed 18 to 24 h postinfection (hpi), aliquoted, and frozen at −70°C until being titered. RNA was isolated from the monolayers for sequence analysis of the plaque-purified viruses by reverse transcription (RT)-PCR. If plaques were not visible by 72 h postelectroporation, the transfection was repeated and the cells were transferred to a single 75-cm2 flask with 105 DBT cells and passaged 1:10 (cells and medium) every 2 to 3 days. RNA was extracted from cells during each passage and analyzed for leader-containing transcript.
Cloning of MHV deletion and cleavage mutants.
Overlapping PCR was employed for construction of mutant cDNA templates. Mutations were introduced with two rounds of PCR. The first round generated two amplicons, which were then fused by a second round of PCR using the 5′ primer for amplicon 1, the 3′ primer of amplicon 2, and the two overlapping amplicons for a template. The first round of PCR consisted of 35 cycles of 94°C annealing for 30 s, 55°C annealing for 30 s, and 68°C extension for 30 s. The second round of PCR increased the extension time to 1 min. The fused, overlapped PCR products were purified (PCR Purification Kit; QIAGEN), digested at two unique restriction sites, purified a second time (PCR Purification Kit), and ligated into a similarly digested MHV plasmid. Primers, template DNAs, restriction sites, and the plasmid backbone into which the PCR product was ligated are provided in Table 1. Mutations inserted into the MHV infectious clone were verified by sequence analysis.
TABLE 1.
Primers, template DNAs, and restriction sites used in the generation of deletion and cleavage mutants
| Mutant | Amplicon | Primer | Primer name | Sequence (5′→3′) | Template DNA | Restriction sites |
|---|---|---|---|---|---|---|
| Δnsp7 | 1 | 5′ | D 3500 | CGGAGGCTTTTGACTTTCTG | MHVD | PstI+NdeI |
| 3′ | D 470c | AATTTGAGATACTTCAATGACTGG | ||||
| 2 | 5′ | 5′Δnsp7 | GTATCTCAAATTCAAAGTGAATTTGTTAATATGGC | MHVD | ||
| 3′ | D 1001c | GCAGACACTACCTTACTCTTC | ||||
| Δnsp8 | 1 | 5′ | D 3500 | CGGAGGCTTTTGACTTTCTG | MHVD | PstI+HindIII |
| 3′ | D743c | TAAGGCTTGCAAGACAGTATTGTC | ||||
| 2 | 5′ | 5′Δnsp8 | GTCTTGCAAGCCTTACAGAACAATGAGTTGATGCCTCAG | MHVD | ||
| 3′ | D 1570c | CAGTTACGCTGGAGTCTG | ||||
| Δnsp9-D | 1 | 5′ | D 880 | CAGCAGATTAAGCAGCTAG | MHVD | HindIII+NdeI |
| 3′ | D 1328c | CAAAACAACAGTAGACACTTC | ||||
| 2 | 5′ | 5′Δnsp9-D | CTACTGTTGTTTTGCAGCCTAAGAGACGAAGGGCG | MHVD | ||
| 3′ | D 1769c | GCGCTCTGCTGAAGCCAG | ||||
| Δnsp9-E | 1 | 5′ | E 4031 | CCACGCTGATGAGCTTTACC | MHVE | ClaI+MscI |
| 3′ | E 1c | TAGGAGAGACGAAGGGC | ||||
| 2 | 5′ | 5′Δnsp9-E | CCTTCGTCTCTCCTAGCGGGTACGGCAACTGAG | MHVE | ||
| 3′ | E 462c | CCATCAACATCTGGATGTTC | ||||
| Δnsp10 | 1 | 5′ | E 4031 | CCACGCTGATGAGCTTTACC | MHVE | ClaI+KpnI |
| 3′ | E 199c | CAATCTCACTGTCGAGG | ||||
| 2 | 5′ | 5′Δnsp10 | CGACAGTGAGATTGCAGTCAAAAGACACGAACTTTTTAAACG | MHVE | ||
| 3′ | E 1021c | CATTGCGGTCAAAATGACGC | ||||
| nsp6*7 | 1 | 5′ | D 3500 | CGGAGGCTTTTGACTTTCTG | MHVD | PstI+NdeI |
| 3′ | D 470c | AATTTGAGATACTTCAATGACTGG | ||||
| 2 | 5′ | QAnsp6/7 | GTATCTCAAATTGCATCAAGATTGACG | MHVD | ||
| 3′ | D 1001c | GCAGACACTACCTTACTCTTC | ||||
| nsp7*8 | 1 | 5′ | D 3500 | CGGAGGCTTTTGACTTTCTG | MHVD | PstI+NdeI |
| 3′ | D 1328c | CAAAACAACAGTAGACACTTC | ||||
| 2 | 5′ | QAnsp7/8 | CAAGCCTTAGCGAGTGAATTTGTTAATATG | MHVD | ||
| 3′ | D 1001c | GCAGACACTACCTTACTCTTC | ||||
| nsp8*9 | 1 | 5′ | D 880 | CAGCAGATTAAGCAGCTAG | MHVD | NdeI+HindIII |
| 3′ | D 1328c | CAAAACAACAGTAGACACTTC | ||||
| 2 | 5′ | QAnsp8/9 | CTACTGTTGTTTTGGCGAACAATGAGTTGATGC | MHVD | ||
| 3′ | D 1570c | CAGTTACGCTGGAGTCTG | ||||
| nsp9*10 | 1 | 5′ | E 4373 | GTTCGGTGTAGGTCGTTC | MHVE | ClaI+MscI |
| 3′ | E 199c | CAATCTCACTGTCGAGG | ||||
| 2 | 5′ | QAnsp9/10 | GTGAGATTGGCGGCGGGTACGG | MHVE | ||
| 3′ | E 462c | CCATCAACATCTGGATGTTC | ||||
| nsp10*11 | 1 | 5′ | E 4031 | CCACGCTGATGAGCTTTACC | MHVE | ClaI+KpnI |
| 3′ | E 606c+ | GCAAACTGGGAGCCTGTGCCTAC | ||||
| 2 | 5′ | QAnsp10/nsp11 | CCCAGTTTGCGTCAAAAGACACG | MHVE | ||
| 3′ | E 1021c | CATTGCGGTCAAAATGACGC |
Rearrangement of the nsp7 and nsp8 genes.
DNAs encoding rearranged nsp7 and nsp8 genes were synthesized (Bio Basic Inc., Ontario, Canada), digested with PstI and HindIII according to the manufacturer's instructions (New England Biolabs), and ligated into the similarly digested MHV-D plasmid. The mutated template was verified by sequence analysis and assembled as part of the infectious clone.
Plaque assay titration of virus titer.
DBT cells in 60-mm-diameter cell culture dishes (∼106 cells) were infected with 200 μl of serially diluted virus in PBS. After 1 h of incubation at 25°C, cells were washed three times with PBS, overlaid with 5 ml of 1% agarose melted in 10% FCS-MEM, and incubated at 37°C in 5% CO2. The next day, plates were stained with neutral red and plaques were counted.
RT-PCR verification of nonviable mutants.
Mutants failing to generate plaque-forming viruses were tested for the ability to generate leader-containing transcripts, whose presence would indicate at least low levels of replication even in the absence of cytopathic effects. RNA harvested from passaged cells was used as a template for generating cDNA by reverse transcription using Superscript II (Invitrogen) and random hexamers (Invitrogen), according to the manufacturer's instructions. Following cDNA synthesis, PCR was completed with primers MHV-4 (5′-AAGAGTGATTGGCGTCCGTA-3′), which anneals to the leader sequence of MHV, and 30019c (5′-GCAGTAATTGCTTCTGCTG-3′), which is complementary to the N gene. Simultaneous PCRs for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were also run as RNA quality and RT controls using primers GAPDHF (5′-CATGGGGAAGGTGAAGGTCG-3′) and GAPDHR (5′-TTGATGGTACATGACAAGGTGC-3′).
Northern blot analysis.
DBT cells in 60-mm-diameter cell culture dishes (106 cells) were infected with MHV-A59 or one of the recombinant viruses at a multiplicity of infection (MOI) of 0.05. At 12 hpi, intracellular RNA was isolated with TRIzol reagent (Invitrogen), as directed by the manufacturer, and 0.05 μg of total mRNA was treated with glyoxal and separated on agarose gels using NorthernMax-Gly according to the manufacturer's directions (Ambion). The RNA was transferred to BrightStar-Plus membrane (Ambion) for 3.5 h and then cross-linked to the membrane by UV light. The blot was prehybridized and probed with a labeled RNA complementary to ∼200 bp of the 5′ portion of the N gene. The RNA probe was generated by PCR using primers MHV-5′N (5′-ATGTCTTTTGTTCCTGGGCAAG-3′) and T7-30019c (5′ATATATTAATACGACTCACTATAGGGAGACCAGAAAACCAGGAGTAATGG-3′). RNA transcripts were driven from the T7 promoter included in the 3′ primer. RNA was biotinylated with the BrightStar Psoralen-Biotin Nonisotopic Labeling Kit, as directed by the manufacturer (Ambion). Following prehybridization, blots were hybridized overnight and washed with low- and high-stringency buffers, as recommended by the manufacturer (Ambion). Filters were incubated with the chemiluminescent substrate CDP-STAR (Ambion). The blots were overlaid with film and developed.
Radiolabeling of MHV proteins and immunoprecipitation of cell lysates.
DBT cells in 60-mm-diameter cell culture dishes (3 × 106 cells) were infected with MHV or one of its mutants or were mock infected with PBS. At 4.5 hpi, the medium was replaced with fresh 5% FCS-DMEM lacking methionine and cysteine and containing actinomycin D (20 μg/ml). At 6 hpi, [35S]methionine-cysteine (100 μCi/ml) was added and the mixture was incubated at 37°C for 3 h. Cells were washed with 1 M Tris and then lysed in 1 ml of lysis buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate [DOC], 50 mM Tris [pH 8.0]). Immunoprecipitations were performed in a final volume of 1 ml with protein A-Sepharose beads (Sigma), 100 μl of radiolabeled lysate, and 2 to 10 μl of polyclonal antiserum specific for one of the nsp7 to nsp10 proteins (4) after the lysate was boiled for 5 min in 1% SDS in buffer C (150 mM NaCl, 1% NP-40, 1% DOC, 1% SDS, 10 mM Tris [pH 7.4]). Protein-bead conjugates were washed three times in the buffer used for immunoprecipitations, and the proteins were eluted from the beads, followed by boiling for 5 min in 2× protein loading buffer (200 mM dithiothreitol, 100 mM Tris [pH 6.8], 0.04% bromophenol blue, 20% glycerol). The proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) in 5 to 18% polyacrylamide gradient gels and analyzed by fluorography. The 14C high-molecular-weight standard (Gibco) and full-range rainbow marker (Invitrogen) were used as molecular weight standards.
Statistical analysis.
Statistical differences between the titers of viruses, expressed as P values, were calculated with Student's t test.
Immunofluorescence assays and confocal microscopy.
DBT cells grown on glass coverslips were infected with MHV or mock infected with PBS and then rocked at 25°C for 30 min. Following virus adsorption, the infected medium was replaced with fresh, prewarmed 10% FCS-MEM and the cells were incubated at 37°C. At 8 hpi, the cells were fixed and permeabilized with 100% methanol cooled to −20°C. Indirect immunofluorescence assays were performed as previously described (4). Secondary antibodies conjugated to fluorophores were used at a 1:1,000 dilution and included anti-guinea pig Alexa 546, anti-rabbit Alexa 488, and anti-mouse Alexa 633. Immunofluorescence was detected with a Zeiss LSM 510 laser scanning confocal microscope with a 40× oil immersion objective. Image analysis and merging were performed with Adobe Photoshop, version 7.0.
RESULTS
Viability of nsp7 to nsp10 deletion mutants.
Mpro targets amino acid sequences (L, F, I)Q↓(S, N/A) and cleaves after the essential Gln at position 1 (P1) (31). The nsp7 to nsp10 protein domains were individually deleted from the MHV genome while preserving functional cleavage sites by fusing the N-terminal P1 amino acid to the carboxyl P1′ residues of the flanking proteins (Table 2). Virus was produced from cDNA templates as previously described (57), and viability was determined by syncytium formation in cells electroporated with in vitro-transcribed mutant genome RNA. All of the deletion mutants failed to yield virus or produce viral cytopathic effect. A series of MHV temperature-sensitive mutants which grow only at a lower permissive temperature have been described (45). With this in mind, a second attempt to generate nonviable viruses was made, and electroporated cells were incubated at 32°C instead of 37°C. However, none of the deletion mutants demonstrated the temperature-sensitive phenotype; no viable viruses were detected at 32°C even after several passages spanning at least a week, and no leader-containing transcripts indicative of subgenomic mRNA synthesis were detected in transfected cultures by RT-PCR, indicating that these deletions were truly lethal for replication.
TABLE 2.
nsp7 to nsp10 cleavage sites for wild-type MHV and deletion mutants
| Protein | Amino acid sequence of indicated cleavage site
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | P5′ | |
| Wild type | ||||||||||
| nsp6-nsp7 | V | S | Q | I | Q | S | R | L | T | D |
| nsp7-nsp8 | L | Q | A | L | Q | S | E | F | V | N |
| nsp8-nsp9 | T | V | V | L | Q | N | N | E | L | M |
| nsp9-nsp10 | T | V | R | L | Q | A | G | T | A | T |
| nsp10-nsp11 | G | S | Q | F | Q | S | K | D | T | N |
| Mutant | ||||||||||
| Δnsp7 | V | S | Q | I | Q | S | E | F | V | N |
| Δnsp8 | L | Q | A | L | Q | N | N | E | L | M |
| Δnsp9 | T | V | V | L | Q | A | G | T | A | T |
| Δsnp10 | T | V | R | L | Q | S | K | D | T | N |
Viability of cleavage site-disrupted mutants.
To evaluate the requirement of nsp7 to nsp10 proteolytic processing for virus replication, cleavage sites flanking nsp7, nsp8, nsp9, and nsp10 were individually disrupted by replacing the P1 Gln with Ala (Table 3). There are two potential cleavage sites at the nsp7-nsp8 interface, an LQ↓A and an LQ↓S (present at positions P5-P3 and P2-P1′, respectively, in Table 1). Although the LQ↓S site has been shown to be cleaved during nsp7-nsp8 processing (34), it is possible that the upstream LQ↓A site is also functional. To address this possibility, both sites were substituted, either individually (MHV7/8A and MHV7/8B) or in combination (MHV7/8AB).
TABLE 3.
Mutagenesis of the nsp7 to nsp10 cleavage sites
| Virusa | Mutation of P1 (Q to A) at cleavage sitesb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | P5′ | |
| MHV6/7 | V | S | Q | I | A | S | R | L | T | D |
| MHV7/8A* | L | A | A | L | Q | S | E | F | V | N |
| MHV7/B† | L | Q | A | L | A | S | E | F | V | N |
| MHV7/8AB‡ | L | A | A | L | A | S | E | F | V | N |
| MHV8/9 | T | V | V | L | A | N | N | E | L | M |
| MHV9/10 | T | V | R | L | A | A | G | T | A | T |
| MHV10/11 | G | S | Q | F | A | S | K | D | T | N |
Symbols: *, the upstream LQ↓A site is substituted; †, the downstream LQ↓S site is substituted; ‡, both sites are substituted.
Boldface type, introduced Ala.
Full-length cDNAs of each of the seven cleavage site mutants were assembled by standard techniques, and transcripts were electroporated into cells. Viability was determined by syncytium formation. For viruses that failed to produce syncytia, the possibility of low-level replication in the absence of syncytium formation was tested by RT-PCR specific for subgenomic mRNAs produced during coronavirus replication. This test was performed on cells collected during three sequential passages of the electroporated cells (Fig. 2). Following electroporation of mutant genome RNA into cells, no syncytia or subgenomic mRNA was detected for mutants MHV6/7, MHV7/8AB, MHV8/9, and MHV10/11. A second attempt to generate nonviable viruses was made with incubations at 32°C, but none of the cleavage mutants failing to grow at 37°C were rescued by growth at the lower temperature. These data indicated that disruption of the cleavage sites at nsp6-nsp7 and both sites at nsp7-nsp8, nsp8-nsp9, and nsp10-nsp11 were lethal for viral replication. In contrast, disruptions of either the nsp7-nsp8 or nsp9-nsp10 cleavage sites were not lethal, and recombinant viruses of each were plaque purified for future use. Plaque morphologies of MHV7/8A and MHV7/8B were similar to those of the wild type, but MHV9/10 displayed a small-plaque phenotype (data not shown). Sequence analysis of RNA from virus-infected cells demonstrated the presence of the appropriate mutations in each of the viable viruses. Growth kinetics of viable viruses were compared to those of wild-type MHV in DBT cells at an MOI of 0.05 PFU/cell (Fig. 3A). MHV, MHV7/8A, and MHV7/8B reached comparable peak titers at 18 hpi of 7.4 ± 0.1 (mean ± standard deviation), 7.3 ± 0.2 (P = 0.2), and 7.4 ± 0.1 (P = 0.8) log10 PFU/ml, respectively. The MHV9/10 mutant displayed an attenuated growth phenotype, with peak titers almost 2 logs lower than those of MHV (P < 0.0001), of 5.5 ± 0.1 log10 PFU/ml at the 18-h point.
FIG. 2.
RT-PCR verification of the replication deficiency of non-syncytium-forming cleavage mutant viruses. RNA was extracted from cells over three passages of electroporated DBT cells and supernatants. PCR was completed with primers specific for leader-containing N gene transcripts and GAPDH as a control for RNA quality and successful RT reactions. RNA from cells infected with wild-type MHV was used as a positive control.
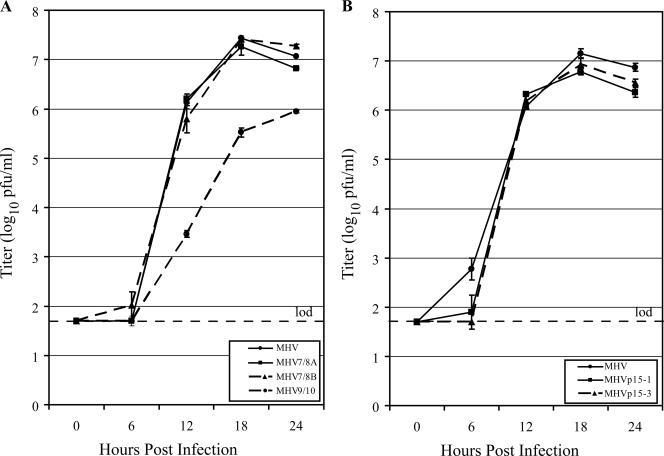
FIG. 3.
Replication kinetics of viable cleavage mutants and MHV9/10 revertants. (A) Comparison of growth curves for MHV (black circle and solid line), MHV7/8A (black square and solid line), MHV7/8B (black triangle and dashed line), and MHV9/10 (black circle and dashed line). (B) Replication fitness of revertant MHV9/10 passage 15 viruses. Growth curves compare MHV (black circle and solid line) to the passage 15 isolates MHVp15-1 (black square and solid line) and MHVp15-3 (black triangle and dashed line). Growth curves were performed on DBT cell monolayers infected at an MOI of 0.05 PFU/cell. Supernatants were sampled for replicating virus at 0, 6, 12, 18, and 24 hpi, and titers were determined by plaque assay. Data points represent the average of three replicate experiments, and error bars show the standard deviation. The limit of detection (lod) is represented by a horizontal dashed line at 1.7 log10 PFU/ml.
Stability of the replication-attenuated MHV9/10 mutation was tested by successive blind serial passages on DBT cells for 15 passages, followed by 3× plaque purification of the resulting virus. The growth kinetics of two passage 15 isolates, MHVp15-1 and MHVp15-3, were compared to those of MHV (Fig. 3B). In contrast to the parent mutant virus, the passage 15 isolates displayed near-wild-type growth kinetics in DBT cells: MHVp15-1 and MHVp15-3 reached peak titers of 6.8 ± 0.1 and 6.9 ± 0.1 log10 PFU/ml, respectively, which approached that of the 7.2 ± 0.1 log10 PFU/ml for MHV.
Sequence analysis of the nsp7 to nsp10 region of the revertant viruses illustrated that the inserted mutation did not revert to wild-type sequence at the nsp9-nsp10 cleavage site. Two patterns of mutations were found. Three of the six plaque isolates sequenced, including MHVp15-1, had mutated the guanosine at nucleotide 13164 to an adenosine, changing the introduced alanine at P1 of the nsp9-nsp10 cleavage site to a threonine (Table 4). The other mutants, including MHVp15-3, maintained the alanine at the P1 position but had a guanosine instead of an adenosine at nucleotide 13102, producing an arginine instead of a lysine 21 amino acids upstream of the nsp9-nsp10 P1 position.
TABLE 4.
Genomic variations in nsp9 among MHV, MHV9/10, and the passage 15 mutants
| Virus | Amino acid at indicated position
|
|
|---|---|---|
| 4298 | 4319 | |
| MHV | Lys | Gln |
| MHV9/10 | Lys | Ala |
| MHVp15-1 | Lys | Thr |
| MHVp15-3 | Arg | Ala |
| MHVQ4319T | Lys | Thr |
| MHVK4298R | Arg | Gln |
| MHV9/10K4298R | Arg | Ala |
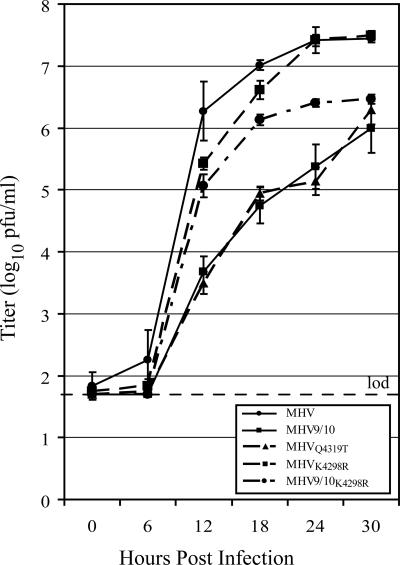
Three mutants were constructed to determine whether the changes found in nsp9 of the passage 15 isolates could be attributed to improved fitness of the viruses (Table 4). The engineered mutant MHVQ4319T contained the P1 Gln4319Thr substitution, reproducing the nsp9 sequence found in MHVp15-1. In order to determine whether the Lys4298Arg substitution in nsp9 identified in MHVp15-3 affects viral replication in DBT cells, the single mutation was introduced in the wild-type infectious clone and used to produce the mutant virus MHVK4298R. The final mutant, MHV9/10K4298R, possessed the nsp9 Lys4298Arg substitution in addition to the P1 Gln4319Ala substitution introduced to disrupt nsp9-nsp10 cleavage, giving it an nsp9 sequence identical to that of MHVp15-3. Viable viruses were recovered for all three, and the growth curves of the plaque-purified viruses indicated that the none of the changes in nsp9 of MHVp15-1 or MHVp15-3 were solely responsible for the improved growth kinetics of the passage 15 revertants (Fig. 4). MHVQ4319T reached a titer of 6.3 ± 0.2 log10 PFU/ml at 30 hpi, comparable to that of MHV9/10 at 6.0 ± 0.4 (P = 0.3) and indicating that the nsp9 mutation found in MHVp15-1 was not responsible for its improved fitness relative to the attenuated parent. Introduction of Lys4298Arg into the MHV backbone did not reduce the peak titer of the virus; MHVK4298R attained a titer of 7.5 ± 0.1 log10 PFU/ml at 30 hpi, which was comparable to the wild-type titer of 7.4 ± 0.1 (P = 0.4). The combination of K4298R and Q2319T found in the nsp9 protein of MHVp15-3 was not sufficient to return the mutant MHV9/10K4298R to a wild-type growth level (P < 0.0001 [at 30 hpi]). Although attaining a peak titer only at 30 hpi comparable to that of MHV9/10 (P = 0.1), the MHV9/10K4298R virus maintained better growth than the parent mutant from 12 to 24 hpi. MHV9/10K4298R displayed higher average titers than did MHV9/10: 5.1 ± 0.2 (P = 0.0007), 6.1 ± 0.1 (P = 0.0007), and 6.4 ± 0.1 (P = 0.004) log10 PFU/ml at 12, 18, and 24 hpi, respectively. In comparison, MHV9/10 had titers of 3.7 ± 0.2, 4.7 ± 0.3, and 5.4 ± 0.3 log10 PFU/ml at the same time points. The mutation(s) responsible for the increased in vitro fitness of the serially passaged virus at least requires the contribution of more-distal mutations within the genome that remain to be identified.
FIG. 4.
Characterization of the nsp9 genetic components of MHVp15-1 and MHVp15-3. Shown is a comparison of MHV (black circle and solid line), the attenuated parent virus MHV9/10 (black square and solid line), MHVQ4319T (black triangle and dotted line), MHVK4298R (black square and dotted line), and MHV9/10K4298R (black circle and dotted line). Growth curves were performed on DBT cell monolayers infected at an MOI of 0.05 PFU/cell. Supernatants were sampled for replicating virus at 0, 6, 12, 18, and 24 hpi, and titers were determined by plaque assay. Data points represent the average of three replicate experiments, and error bars show the standard deviation. The limit of detection (lod) is represented by a horizontal dashed line at 1.7 log10 PFU/ml.
Rearrangement of nsp7 and nsp8.
The domain order of coronavirus ORF1ab nsp proteins is conserved. However, it has been possible to recover mutant viruses with deletions and rearrangements of nsp2 (21; also unpublished data). To test whether the nsp7 to nsp10 order must be maintained for efficient replication, the nsp7 and nsp8 genes were rearranged. Transcripts were driven from the cDNA templates and electroporated into cells. Viability was determined by syncytium formation and detection of leader-containing transcripts by RT-PCR of RNA harvested at each of three passages. Following electroporation of cells with mutant genome RNA containing the nsp8-nsp7 rearrangement, no virus was obtained and no leader-containing transcripts were detected, suggesting that the domain orders for nsp7, nsp8, and perhaps the other components of the precursor polyprotein were essential for replication.
Verification of ablated cleavage sites in viable mutant viruses.
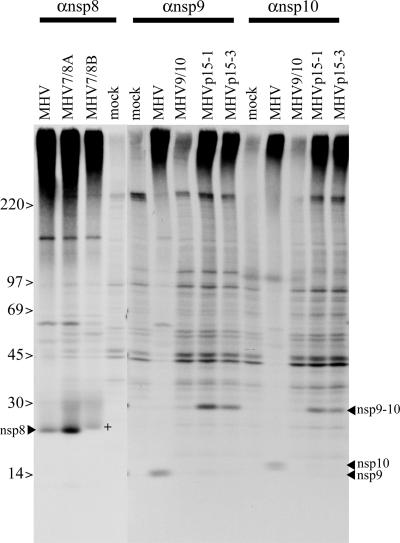
Processing of the ORF1a nsp8, nsp9, and nsp10 replicase proteins was evaluated in DBT cells infected at an MOI of 1, treated with actinomycin D at 4.5 hpi, and radiolabeled from 6 to 9 hpi. Immunoprecipitation using either anti-nsp8 antibody (for the MHV7/8 mutants) or anti-nsp9 and anti-nsp10 antibody (for the MHV9/10 and passage 15 mutants) (3) was used to evaluate the impact of the specific cleavage site mutations on ORF1a polyprotein processing. Substitution of the P1 Gln to Ala prevented processing at the cleavage sites for the MHV9/10, MHVp15-1, and MHVp15-3 viruses (Fig. 5). Normal processing of nsp8 (22 kDa), nsp9 (12 kDa), and nsp10 (15 kDa) was detected during wild-type MHV infection but not in mock-infected cells. In contrast, nsp10 was absent following MHV9/10 and MHVp15 infection. However, a slower-migrating band of approximately 27 kDa, which is the predicted size for an nsp9-nsp10 fusion protein, was present in the mutants but absent in the wild-type controls. These data indicate that neither the nsp9 P1 Ala110Thr substitution in MHVp15-1 nor the nsp9 Lys89Arg change in MHVp15-3 restored cleavage at the nsp9-nsp10 junction.
FIG. 5.
ORF1a polyprotein processing in recombinant viruses. Cultures of cells were infected with MHV, MHV7/8A, MHV7/8B, MHV9/10, MHVp15-1, or MHVp15-3 for 4.5 h. The cultures were radiolabeled for 3 h, and antisera against nsp8 (22 kDa), nsp9 (12 kDa), or nsp10 (15 kDa) was used for immunoprecipitation. DBT cells were infected at an MOI of 1 PFU/cell and labeled with [35S]Met/Cys-containing medium from 6 to 9 hpi in the presence of actinomycin D. At approximately 9 hpi, cells were lysed and proteins were immunoprecipitated with polyclonal sera against nsp8, nsp9, or nsp10 and then resolved by SDS-PAGE and identified by fluorography. Bands corresponding to nsp8, nsp9, nsp10, and the fused nsp9-nsp10 (nsp9-10) are indicated. The protein corresponding to nsp8 isolated from MHV7/8B-infected cells (+) migrated slower than those of MHV or MHV7/8A.
Analysis of MHV7/8A- and MHV7/8B-infected cultures indicated that both the LQ↓A and LQ↓S sites bordering the nsp7-nsp8 junction are functional cleavage sites. Following immunoprecipitation, 22-kDa proteins were precipitated with anti-nsp8 antibody for the wild-type control, MHV7/8A, and MHV7/8B. Notably, the immunoprecipitated protein from MHV7/8B had slightly slower migration than that of the control or the MHV7/8A mutant, consistent with the prediction that the LQ↓A site was cleaved and yielded an nsp8 protein that was 3 amino acids larger (∼366 Da) than that of the LQ↓S cleaved protein.
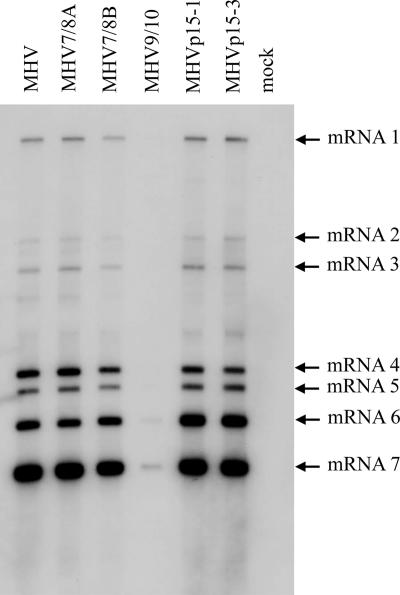
Transcriptional profile of viable mutant viruses.
With the exception of the attenuated MHV9/10 mutant, viable mutants were found to be similar to MHV in their transcriptional activities and generation of subgenomic RNA (Fig. 6). To determine whether differences in RNA synthesis were associated with the different growth phenotypes, cultures of DBT cells were infected with the mutant panel at an MOI of 0.05 and total intracellular RNA was harvested at 12 hpi. Northern blots hybridized with an RNA probe complementing the 5′ end of the N gene showed no differences in either the pattern or relative amounts of subgenomic to genomic RNA in most mutants compared to control virus. Consistent with the reduced growth of the mutant virus in vitro relative to that of MHV, MHV9/10 had significantly reduced amounts of RNA, with only the mRNA 6 and 7 bands clearly resolved. Importantly, revertant viruses had restored efficient growth kinetics and transcription of full-length and subgenomic mRNAs. We did not identify any significant differences in the relative molar ratios of the viral-plus-sensed RNAs (data not shown).
FIG. 6.
RNA synthesis in recombinant and wild-type viruses. Cultures of cells were infected with wild-type MHV or recombinant viruses, and intracellular RNA was harvested from DBT cells at 12 hpi. RNA was separated on a 1% agarose gel, transferred to nylon filters, and hybridized with a biotinylated RNA probe specific for N protein mRNA. The filters were incubated with a chemiluminescent substrate and exposed on film.
Association of mutant proteins with replication complexes.
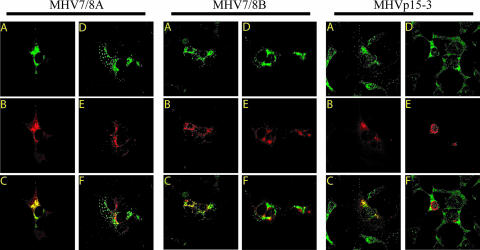
The distribution of the mutant proteins within cells was compared to their wild-type counterparts. The nsp7 to nsp10 proteins are known to colocalize with sites of viral replication while being excluded from regions of virion assembly (3, 4). In order to determine whether abolition of processing in the mutant viruses affected the distribution, subcellular localization, or ability of the protein to traffic into the replication complex, we used confocal microscopy to compare the colocalization of wild-type and mutated proteins to sites of replication and assembly. DBT cultures were infected with either MHV-A59, mock virus, or mutant virus and at 8 hpi were methanol fixed and dually stained for either nsp8 or nsp10 (depending on the mutant, as above) and nucleocapsid (N), which colocalizes with sites of active viral replication, or membrane (M), which is targeted to regions of virus assembly. Regardless of the construct, the nsp8 and nsp10 proteins colocalized with N in subcellular compartments that were separate from M (Fig. 7). These results are identical to those for wild-type MHV (not shown), indicating that incorporation of the mutated proteins into the replication complex is not disrupted (3).
FIG. 7.
Immunofluorescence of MHV in cells infected with cleavage mutant virus. Cells were infected with either MHV7/8A, MHV7/8B, or MHVp15-3, fixed and permeabilized with MeOH, and then dually stained with antibody specific for nsp8 (MHV7/8A and MHV7/8B) or nsp10 (MHVp15-3) and nucleocapsid or membrane protein. The green fluorescent anti-nsp image (A and D) was overlaid with the corresponding red fluorescent anti-N (B) or anti-M image (E) to determine points of colocalization (yellow) between the nsp and either N, representing localization with the replication complexes (C), or M, which is excluded from sites of replication (F).
DISCUSSION
The nsp7 to nsp10 proteins are highly conserved among, and perhaps unique to, the family Coronaviridae. Indeed, even the arteriviruses, another family member of the order Nidovirales, do not appear to possess homologs to the coronavirus nsp7 to nsp10 proteins (39). The roles of these proteins in coronavirus replication are only just beginning to be studied, with the existing body of data suggesting that they are components of the replication complex. However, the details of their involvement in replication and RNA synthesis remain to be determined. This study used an infectious clone of MHV to define fundamental features of the nsp7 to nsp10 proteins during viral replication in culture. Each of the four proteins appears to be critical for viral replication, since deletion of any of the four protein domains was lethal for RNA synthesis and productive virus infection. Furthermore, the results indicate that processing of the proteins from each other is necessary for replication, with the one exception of the nsp9-nsp10 cleavage site. Finally, we determined that rearrangement of two of the replicase proteins, nsp7 and nsp8, was not permissive for virus replication.
To date, only the nsp2 coronavirus replicase protein has been shown to be dispensable for replication in both MHV-A59 and SARS-CoV, albeit attenuating in vitro and in vivo (22). Portions the carboxy-terminal half of MHV nsp1 have also been deleted in viable mutants (8), but otherwise no full or partial deletions of replicase protein domains in viable mutants of any coronavirus have been reported. In contrast, deletion of each of the nsp7 to nsp10 proteins resulted in a lethal phenotype, as evidenced by the lack of recoverable viruses and an inability to detect subgenomic mRNAs by RT-PCR. These data suggest that each of the nsp7 to nsp10 proteins may be an indispensable component of the replication complex, which is consistent with the highly conserved nature of these proteins. Alternatively, deletion of nsp coding sequences may sufficiently alter the structure of the polyprotein template to interfere with Mpro accessibility to its cleavage sites. Interestingly, an MHV temperature-sensitive mutant, LA6 (45), contains a mutation in nsp10 that blocks processing of nsp4 to nsp10 at the nonpermissive temperature (13a), suggesting that mutations or deletions at the C terminus of ORF1a may disrupt Mpro activity.
Although it is known that global inhibition of coronavirus proteinases that process the replicase polyproteins prevents replication (29), the requirements for each of the 15 cleavage sites in the ORF1ab polyprotein are not completely determined. Cleavage of nsp1, nsp2, and nsp3 has been abolished in viable MHV mutants (12, 21). Otherwise, little is known of the requirements for processing, including those for nsp7 to nsp10. Our results show that changes at cleavage sites between nsp6 and nsp7, nsp7 and nsp8, nsp8 and nsp9, and nsp10 and nsp11 are not replication viable. Lethality could be due to the disruption of nsp7 to nsp10 proteolytic processing, causing a failure of precursor, intermediate, or mature protein function within the replication complex. However, not all of the cleavage site mutants were nonviable. Based on genetic analysis, MHV has two functional nsp7-nsp8 cleavage sites, LQ↓A and LQ↓S, and disruption of either of these potential sites failed to affect replication competence, cleavage patterns, or cellular localization in vitro. Interestingly, the LQ↓A site is conserved across all coronavirus families, while the LQ↓S site is limited to group II coronaviruses, including MHV, BCoV, HKU1, and OC43 but not SARS-CoV. Wild-type replication efficiency when either one or the other site was knocked out suggests that either or both sites are cleaved during replication. Although we cannot detect any significant impact on in vitro replication, variations in N- or C-terminal processing of nsp7-nsp8 may influence in vivo pathogenesis or affect cell signaling pathways. However, simultaneous mutation of both sites was lethal, indicating that nsp7 and nsp8 must be fully separated to function in mRNA synthesis.
The only cleavage site that tolerated inactivation was the nsp9-nsp10 cleavage site. The mutant MHV9/10 virus produced an nsp9-nsp10 fusion protein and was highly attenuated in its replication efficiency. Serial passage of this virus restored near-wild-type replication fitness but did so without reverting at the mutated cleavage site or regaining the ability to process nsp9-nsp10, demonstrating that efficient replication can be achieved without nsp9-nsp10 proteolytic processing. The data demonstrate that with the exception of cleavage between the nsp9 and nsp10 proteins, Mpro processing of the nsp7 to nsp10 proteins is essential in coronavirus RNA transcription and replication.
Previous work has indicated that nsp7 and nsp8 in solution form a complex hexadecameric structure that is proposed to function in processivity and generation of RNA primers for the RNA replicase (25, 58). If these structures represent those formed during infection, then they would require cleavage of nsp7 from nsp8. Similarly, the virus could not replicate when the relative positions of the genes encoding nsp7 and nsp8 were switched. This loss of viability could be due to an alteration of the precursor polyprotein that interfered with processing or prevented a distinct function associated with the noncleaved precursor. It is unclear why only the MHV9/10 mutant was viable. nsp9 associates with the replication complex, interacting at least with nsp8 (51), and has been shown to possess single-stranded RNA-binding affinity (3, 15, 51). Nsp10 is known to associate with several proteins of the replication complex, including nsp1, nsp5, nsp7, nsp8, and nsp12 (7, 8). nsp10 has been shown to be critical for the formation of functional replication complexes (45) and has recently been shown to crystallize to form monomers and homodimers as well as a complex dodecameric structure when expressed as an nsp10-nsp11 fusion (28, 50). It is puzzling that this critical protein with broad interactions with other replicase proteins would retain its function without full separation from nsp9. Interestingly, mutation of the nsp10-nsp11 cleavage site was nonviable despite a report that the spherical structure formed by 12 units of nsp10 was crystallized as an nsp10-nsp11 construct (50). Collectively, our data indicate that the C-terminal cleavage site for the nsp10 protein is essential for infectivity, raising doubts about the biological relevance of the reported nsp10-nsp11 dodecameric crystal structure (50).
Prior to this study, two viable cleavage mutants of coronaviruses had been reported, as the PLP1-mediated cleavage sites between nsp1-nsp2 and nsp2-nsp3 were removed in MHV (12, 21). Loss of cleavage site function resulted in attenuated replication and suggested that efficient cleavage of nsp1-nsp2 and nsp2-nsp3 was important, but not required, for replication in tissue culture (12, 21). Indeed, viable mutant virus could be generated even when PLP1, which solely mediates nsp1-nsp2 and nsp2-nsp3 processing in MHV, was inactivated (21). With this report, three cleavage sites in the MHV ORF1a polyprotein have been shown to be dispensable for replication: nsp1-nsp2, nsp2-nsp3, and nsp9-nsp10 (12, 21). It is possible that this reflects the use of these proteins in natural precursors, such as has been reported for nsp2-nsp3 and nsp4 to nsp10. Thus, the engineered changes may reproduce some component of the normal life cycle and at least residual function of these proteins. Interestingly, rearrangement of the nsp7 and nsp8 coding sequences was lethal, a result that lends support to the idea that there may be an independent function in replication associated with the nsp4 to nsp10 precursor (45).
There are still many aspects of coronavirus replication that are not clearly understood. Several conserved components of the coronavirus replicase have no known homologs and have unknown or poorly defined functions. The proteolytic processing of the replicase polyproteins is a critical step in replication of these viruses, and such processing my provide a level of regulation over replication in general, such as providing the molecular switch for altering the output of the replication complex from negative-strand RNA to that of positive-strand RNA (5, 44). This work will progress by experimentally pursuing new questions defined by these results. Although processing of either of the sites at the nsp7-nsp8 boundary had no impact on virus growth in cell culture, work will be done to determine whether both sites are required for efficient replication in animals. Some insight may be gained into nsp7 to nsp10 function, such as cis versus trans activity, by determining whether lethal cleavage mutants can complement each other to form viable replication complexes to rescue virus. The distal mutations that arose during serial passaging to allow the attenuated MHV9/10 mutant to recover have been identified for both MHV9/10p15 revertants. MHVp15-1 contained a 5-nucleotide insertion of CTAAT at nucleotide position 65 with the 5′ untranslated region (UTR), a Met-to-Leu mutation at amino acid 225 of nsp2, a His-to-Leu mutation at amino acid 112 of nsp4, a Phe-to-Val mutation in nsp13 at amino acid 145, a Phe-to-Tyr mutation in the Spike glycoprotein at position 937, and a Ser-to-Ile mutation in ORF6 at amino acid 78. Mutations found in MHVp15-3 included the same 5-nucleotide insertion within the 5′ UTR seen in MHV9/10p15-1, the His-to-Leu change at amino acid position 112 of nsp4, a Ser-to-Gly change at amino acid 74 of nsp5, and a Phe-to-Tyr change at amino acid 937 of S. The changes found for MHVp15-3 were cloned into the infectious clone of the parent MHV9/10, and replication efficiency was restored (data not shown). Additional work to determine whether the mutations shared between the two MHV9/10p15 revertants at the 5′ UTR, nsp4, and S are sufficient to restore efficient growth is also planned. The data in this report support existing information that the proteins of the 3′ C terminus of ORF1a are critical for replication, establish the importance of processing in their function, and lay the foundation for future studies aimed at expanding our understanding of the interactions that occur between proteins of the coronavirus replication complex.
Acknowledgments
This work was supported by NIH/NIAID grant AI023946 to R.S.B.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonilla, P. J., S. A. Hughes, J. D. Pinon, and S. R. Weiss. 1995. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology 209:489-497. [DOI] [PubMed] [Google Scholar]
- 3.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brayton, P. R., M. M. Lai, C. D. Patton, and S. A. Stohlman. 1982. Characterization of two RNA polymerase activities induced by mouse hepatitis virus. J. Virol. 42:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bredenbeek, P. J., C. J. Pachuk, A. F. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59: a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockway, S. M., X. T. Lu, T. R. Peters, T. S. Dermody, and M. R. Denison. 2004. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 78:11551-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campanacci, V., M. P. Egloff, S. Longhi, F. Ferron, C. Rancurel, A. Salomoni, C. Durousseau, F. Tocque, N. Bremond, J. C. Dobbe, E. J. Snijder, B. Canard, and C. Cambillau. 2003. Structural genomics of the SARS coronavirus: cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein. Acta Crystallogr. D 59:1628-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, A., W. Zhang, Y. Xie, W. Jiang, E. Arnold, S. G. Sarafianos, and J. Ding. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denison, M. R., J. C. Kim, and T. Ross. 1995. Inhibition of coronavirus MHV-A59 replication by proteinase inhibitors. Adv. Exp. Med. Biol. 380:391-397. [DOI] [PubMed] [Google Scholar]
- 12.Denison, M. R., B. Yount, S. M. Brockway, R. L. Graham, A. C. Sims, X. Lu, and R. S. Baric. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis, D. E., and D. A. Brian. 1982. RNA-dependent RNA polymerase activity in coronavirus-infected cells. J. Virol. 42:153-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Donaldson, E. F., R. L. Graham, A. C. Sims, M. R. Denison, and R. S. Baric. 2007. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 81:7086-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 15.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esper, F., C. Weibel, D. Ferguson, M. L. Landry, and J. S. Kahn. 2005. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J. Infect. Dis. 191:492-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya, A. E., A. P. Donchenko, V. M. Blinov, and E. V. Koonin. 1989. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 243:103-114. [DOI] [PubMed] [Google Scholar]
- 19.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, R. L., and M. R. Denison. 2006. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J. Virol. 80:11610-11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 79:13399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamre, D. P. J. 1966. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121:190-193. [DOI] [PubMed] [Google Scholar]
- 24.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbert, I., J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25:4933-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov, K. A., and J. Ziebuhr. 2004. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 78:7833-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseph, J. S., K. S. Saikatendu, V. Subramanian, B. W. Neuman, A. Brooun, M. Griffith, K. Moy, M. K. Yadav, J. Velasquez, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2006. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 80:7894-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J. C., R. A. Spence, P. F. Currier, X. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 31.Lee, H. J., C. K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. La Monica, J. Tuler, A. Bagdzhadzhyan, and M. M. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang, P. H. 2006. Characterization and inhibition of SARS-coronavirus main protease. Curr. Top. Med. Chem. 6:361-376. [DOI] [PubMed] [Google Scholar]
- 33.Lu, X., Y. Lu, and M. R. Denison. 1996. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology 222:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, Y., X. Lu, and M. R. Denison. 1995. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 69:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthes, N., J. R. Mesters, B. Coutard, B. Canard, E. J. Snijder, R. Moll, and R. Hilgenfeld. 2006. The non-structural protein Nsp10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 580:4143-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McIntosh, K., J. H. Dees, W. B. Becker, A. Z. Kapikian, and R. M. Chanock. 1967. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA 57:933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 40.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78:9977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putics, A., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1‴-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79:12721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putics, A., A. E. Gorbalenya, and J. Ziebuhr. 2006. Identification of protease and ADP-ribose-1‴-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 87:651-656. [DOI] [PubMed] [Google Scholar]
- 43.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1‴-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawicki, S. G., and D. L. Sawicki. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seybert, A., C. C. Posthuma, L. C. van Dinten, E. J. Snijder, A. E. Gorbalenya, and J. Ziebuhr. 2005. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 79:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su, D., Z. Lou, F. Sun, Y. Zhai, H. Yang, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 80:7902-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus: structure and functional insights. Structure 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tibbles, K. W., I. Brierley, D. Cavanagh, and T. D. Brown. 1996. Characterization in vitro of an autocatalytic processing activity associated with the predicted 3C-like proteinase domain of the coronavirus avian infectious bronchitis virus. J. Virol. 70:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, Z. Y., H. C. Werner, W. P. Kong, K. Leung, E. Traggiai, A. Lanzavecchia, and G. J. Nabel. 2005. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. USA 102:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]