Abstract
Avian influenza A H7 subtype viruses pose a significant threat to human health because of their ability to transmit directly from domestic poultry to humans and to cause disease and, sometimes, death. Although it is important to develop vaccines against viruses of this subtype, very limited information is available on the immune response and pathogenesis of H7 viruses in animal models such as mice and ferrets. Ten H7 viruses were selected for possible vaccine development on the basis of their phylogenetic relationships and geographical locations. The virulence of the 10 viruses for mice and the immunogenicity of the viruses in mice and ferrets were evaluated to study the extent of antigenic relatedness and the level of cross-reactivity of antibodies. Most of the viruses showed similar patterns of cross-reactivity with mouse and ferret antisera. The Eurasian viruses elicited broadly cross-reactive antibodies that neutralized viruses from both Eurasian and North American lineages, but the converse was not true. A subset of the viruses was also evaluated for the ability to replicate and cause disease in BALB/c mice following intranasal administration. H7 subtype viruses were able to infect mice without adaptation and manifested different levels of lethality and kinetics of replication. On the basis of phylogenetic data, induction of broadly cross-neutralizing antibodies in mouse and ferret antisera, and their ability to replicate in mice, we have selected A/Netherlands/219/03 (subtype H7N7) and A/chicken/BC/CN-7/04 (subtype H7N3) viruses for vaccine development. The mouse model can be used for the preclinical evaluation of these vaccines against H7 subtype viruses.
Influenza A viruses are divided into subtypes on the basis of serological and genetic differences in their major surface glycoproteins, the hemagglutinin (HA) and neuraminidase (NA). Sixteen different HA (H1 to H16) and 9 NA (N1 to N9) subtypes have been identified among influenza A viruses (7, 31). Viruses of all 16 HA and 9 NA subtypes infect aquatic birds, and these birds serve as the reservoir from which novel subtypes of influenza viruses are introduced into domestic poultry and the human population. On the basis of their ability to cause disease in chickens, avian influenza viruses are divided into two groups, highly pathogenic (HP) and low-pathogenicity (LP) viruses. HP avian influenza viruses are restricted to H5 and H7 HA subtypes and cause lethal systemic infection that may result in 100% mortality within a flock, whereas LP avian influenza viruses include viruses of all subtypes and cause milder infections, with a lower rate of morbidity and no mortality (29).
Occasionally, avian influenza A viruses are transmitted directly from birds to humans, with variable consequences. Introduction of a new influenza A virus subtype into a susceptible human population could result in a pandemic if the virus causes disease and spreads efficiently from person to person. Although H5N1 viruses currently are the focus of concern, the next pandemic of influenza could be caused by a virus of another subtype.
Avian influenza A H7 subtype viruses have caused large outbreaks of disease in domestic poultry in Asia, Europe, North America, and South America in recent years, resulting in severe economic losses to the poultry industry (5). Because of their ability to transmit directly from domestic poultry to humans and to cause disease and, sometimes, death, H7 viruses also have been recognized as a concern for human health. Although isolated cases of human infections with HP or LP avian influenza H7 viruses have occurred (2, 4, 14, 27, 30), H7 viruses became a major concern with the direct transmission of H7N7 viruses to humans in The Netherlands in 2003. An HP avian influenza H7N7 virus caused severe outbreaks of disease in domestic poultry in The Netherlands in March 2003. Culling of 30 million chickens, i.e., about 28% of the total chicken population in The Netherlands, controlled further spread of the infection (13). This outbreak in poultry also resulted in the direct transmission of the virus to at least 86 people who were involved in the culling of infected poultry. There also was evidence of limited human-to-human transmission from an infected family member in three cases (8, 13). Of these 89 human infections, most of the patients developed conjunctivitis, and a few others developed mild influenza-like illness (8). There was one fatal case of pneumonia and acute respiratory distress syndrome in a veterinarian who visited farms with HP avian influenza virus-infected poultry flocks (8, 13). In 2004, an HP avian influenza H7N3 virus emerged in domestic poultry in British Columbia, Canada. This outbreak resulted in the infection of two workers on a poultry farm, causing mild respiratory disease and conjunctivitis (9, 18, 28). A serological survey in Italy detected anti-H7 antibodies in 7 out of 185 poultry workers who were exposed to an LP avian influenza H7N3 virus during the 2002 to 2003 avian influenza outbreaks in that country (20). Thus, direct transmission of H7 subtype viruses to humans occurs, and this highlights the threat posed by both HP and LP avian influenza viruses of this subtype to human health in terms of pandemic potential.
Phylogenetic analysis of the H7 HA gene reveals a separation into two lineages, Eurasian and North American, that correspond to the geographic separation of the birds that they infect, and they generally correspond to the Eurasian and North American flyways of migratory birds (3). Antigenic relatedness among viruses from the two lineages and the range of cross-protective immune response induced by viruses from the two lineages currently are not known. Such information is vital for the development of efficacious vaccines.
Very limited information is available on the immune response and pathogenesis of H7 viruses in mammalian species, such as ferrets and mice, that currently are used for preclinical studies of pandemic influenza vaccines. Ferrets are considered the best available model for human influenza research, and sera from ferrets postinfection is the WHO standard for antigenic analysis of human influenza viruses. Human influenza viruses replicate in the respiratory tract of ferrets, resulting in acute respiratory illness that is characterized by clinical signs such as fever, rhinorrhea, and sneezing, and ferrets develop antibodies to the infecting strain. In contrast, human influenza viruses rarely cause symptoms of respiratory tract disease in mice and require adaptation to replicate to high titers in mice. Because of these reasons, the use of mouse models for the study of influenza is limited. However, the advantage of a mouse model for avian influenza virus infection is that reagents for immunologic studies of mice are widely available. It is important to assess the immune response and kinetics of replication of H7 viruses in different animal models in order to establish appropriate models for the preclinical evaluation of vaccines.
In this study, we selected 10 H7 viruses on the basis of their phylogenetic relationships and geographical locations and evaluated their ability to induce broadly cross-neutralizing antibodies in mice and ferrets. We also evaluated the ability of a subset of these viruses to replicate and cause disease in mice following intranasal administration.
MATERIALS AND METHODS
Selection of viruses.
Nucleotide sequences of the HA1 domain of the HA gene of 24 avian influenza A H7 subtype viruses were obtained either from GenBank or by sequencing. The virus strains and GenBank accession numbers used for phylogenetic analysis are listed in Table 1. Phylogenetic analysis was performed by the neighbor-joining method, using the MacVector 9.0 software package. The following 10 avian influenza A H7 subtype viruses were selected for further study: A/chicken/BC/CN-7/04 (subtype H7N3) (designated BC/04), A/Netherlands/219/03 (H7N7) (NL/03), A/chicken/Chile/4322/02 (H7N3) (CH/02), A/chicken/Queensland/95 (H7N3) (QL/95), A/turkey/England/63 (H7N3) (EG/63), A/FPV/Rostock/34 (H7N1) (RK/34), A/turkey/VA/55/02 (H7N2) (VA/02), A/turkey/UT/24721-10/95 (H7N3) (UT/95), A/rhea/NC/39482/93 (H7N1) (NC/93), and A/turkey/OR/71 (H7N3) (OR/71). Viruses used in this study were kindly provided by Robert G. Webster, St. Jude Children's Research Hospital, Memphis, TN; David Swayne, Southeast Poultry Research Laboratory, USDA, Athens, GA; Nancy Cox, Influenza Division, Centers for Disease Control and Prevention, Atlanta, GA; and John Pasick, Canadian Food Inspection Agency, National Centre for Foreign Animal Disease, Winnipeg, Canada.
TABLE 1.
Viruses used for phylogenetic analysis
| Virus | Subtype | GenBank accession no. of HA gene sequence |
|---|---|---|
| A/FPV/Dutch/27 | H7N7 | Z12617 |
| A/FPV/Rostock/34 | H7N1 | M24457 |
| A/turkey/England/63 | H7N3 | CY015065 |
| A/turkey/Oregon/71 | H7N3 | M31689 |
| A/chicken/Victoria/75 | H7N7 | Z47199 |
| A/turkey/Minnesota/1200/80 | H7N3 | CY014778 |
| A/chicken/Victoria/85 | H7N7 | M17735 |
| A/chicken/Jena/87 | H7N7 | U20469 |
| A/rhea/North Carolina/39482/93 | H7N1 | EF470586 |
| A/quail/Arkansas/16309-7/94 | H7N3 | AF072401 |
| A/chicken/New Jersey/15086-3/94 | H7N3 | AF072383 |
| A/turkey/Utah/24721-10/95 | H7N3 | EF470585 |
| A/chicken/Pakistan/447/95 | H7N3 | AF202226 |
| A/common iora/Singapore/95 | H7N1 | AF202228 |
| A/chicken/Queensland/667/95 | H7N3 | AF202231 |
| A/mallard/Ohio/322/98 | H7N3 | CY016188 |
| A/chicken/New York/12273-11/99 | H7N3 | AY240892 |
| A/quail/New York/11430/99 | H7N2 | AY240923 |
| A/turkey/Italy/13468/99 | H7N1 | AJ493216 |
| A/chicken/New York/30749-3/00 | H7N2 | AY240897 |
| A/turkey/Virginia/55/02 | H7N2 | AY240912 |
| A/chicken/Chile/4322/02 | H7N3 | AY303631 |
| A/Netherlands/219/03 | H7N7 | AY338459 |
| A/chicken/British Columbia/CN-7/04 | H7N3 | EF470587 |
HA genes of the UT/95, BC/04, and NC/93 viruses were sequenced, and others were obtained from GenBank to analyze the amino acid sequences. For sequencing, viral RNA of UT/95, BC/04, and NC/93 was extracted using the QIAmp viral RNA mini kit (QIAGEN, Valencia, CA). The full-length HA gene was amplified from viral RNA by reverse transcription-PCR using an Omniscript reverse transcription kit (QIAGEN, Germantown, MD) and an advantage high-fidelity PCR kit (Clontech, Mountain View, CA). PCR fragments were purified using a QIAquick PCR purification kit (QIAGEN). Sequencing was carried out using the big dye terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 3100 genetic analyzer (Applied Biosystems). The Sequencher 4.7 software package was used for the editing, assembly, and analysis of nucleotide sequence data. Amino acid sequences of the HA protein of the 10 selected H7 viruses were predicted from full-length nucleotide sequences. The percentage of amino acid identity among the selected viruses and the sequence of the cleavage site of the HA were determined using the MacVector 9.0 software package.
The 10 selected H7 viruses were propagated in the allantoic cavity of 10-day-old embryonated chicken eggs at 37°C and were stored at −80°C until use. Fifty percent tissue culture infectious doses (TCID50) were determined by serial titration of viruses in MDCK cells, and titers were determined by the method described by Reed and Muench (21).
All experiments, including animal studies with LP and HP H7 avian influenza viruses, were conducted in biosafety level 3+ containment laboratories approved for use by the U.S. Department of Agriculture and Centers for Disease Control and Prevention. Animal experiments were approved by the National Institutes of Health Animal Care and Use Committee.
Pathogenicity in mice.
To assess the lethality of the viruses in mice, groups of five 4- to 6-week-old female BALB/c mice (Taconic Farms, Hudson, NY) were anesthetized and infected intranasally (i.n.) with each of the selected H7 viruses at a dose of 105 TCID50 per mouse, administered in a 50-μl volume. Mice were monitored daily for mortality for 14 days. HP viruses BC/04, NL/03, and EG/63 and LP viruses VA/02 and UT/95 were further evaluated in mice. To determine the 50% lethal doses (LD50) of the HP viruses, groups of five female BALB/c mice were anesthetized and infected i.n. with serial 10-fold dilutions ranging from 100 to 106 TCID50 of each virus in a 50-μl volume. Mice were monitored daily for 14 days. Mice that lost 20% of their body weight were sacrificed according to the animal study protocol. The LD50 was determined by the Reed and Muench method (21). Similarly, groups of five female BALB/c mice were inoculated with 105 TCID50 of the LP viruses in 50 μl and were monitored and weighed daily for 14 days.
Kinetics of virus replication in mice.
To study the kinetics of replication of the viruses in different tissues, groups of 24 female BALB/c mice were inoculated i.n. with 105 TCID50 of each virus. Four mice from each group were euthanized 1, 2, 3, 4, 6, and 7 days postinfection (dpi). Organs were collected at up to 4 dpi for NL/03 and EG/63 viruses and at 6 dpi for BC/04 virus, because the mice did not survive beyond these time points. Nasal turbinates (the intact nasal cavity containing the nasal turbinates, nasal bones, and the upper jaw), lungs, brains, and spleens were harvested, weighed, and homogenized in L-15 medium (Invitrogen-GIBCO) containing a 2× concentration of antibiotic-antimycotic (penicillin, streptomycin, and amphotericin B) (Invitrogen-GIBCO) to make 5% (wt/vol) (nasal turbinates) or 10% (wt/vol) (lungs, brain, and spleen) tissue homogenates. Tissue homogenates were clarified by centrifugation and titrated in 24- and 96-well tissue culture plates containing MDCK cells. The virus titer for each organ was determined by the method described by Reed and Muench (21) and was expressed as the log10 TCID50/gram of tissue.
Generation of antisera. (i) Mice.
To generate specific postinfection sera, groups of five 4- to 6-week-old female BALB/c mice were anesthetized and infected i.n. with 50 μl of a dose of virus selected on the basis of the results of the lethality study. Mice received 105 TCID50 per mouse for RK/34, CH/02, QL/95, VA/02, UT/95, and OR/71, 103 TCID50 per mouse for NC/93, and 102 TCID50 per mouse for BC/04, NL/03, and EG/63. Mice were monitored for the development of disease and mortality. All of the mice were bled before virus administration, and postinfection sera were collected at one or more of the following time points: 28, 42, and 56 dpi.
(ii) Ferrets.
Two 4- to 5-week-old ferrets (Harlan Sprague Dawley, Inc., Hanover, NH) that were seronegative for antibodies to currently circulating H3N2 and H1N1 human influenza viruses were used to generate antiserum for each virus. Since pathogenicity data for H7 subtype viruses in ferrets are not available, the dose administered was based on data from mice and chickens. The inoculum for viruses that were not lethal for mice and/or chickens was 107 TCID50, and lower doses (105 and 102 TCID50) were used as inocula for viruses that were lethal in these species. If the ferret that received 105 TCID50 survived until day 21, serum from this ferret was used in serologic assays. If a dose of 105 TCID50 was lethal, serum from the ferret that received 102 TCID50 was used. Therefore, lightly anesthetized ferrets were inoculated i.n. with 107 TCID50 of VA/02, UT/95, and OR/71 or with 105 and 102 TCID50 of NL/03, CH/02, QL/95, EG/63, RK/34, NC/93, and BC/04 in a volume of 500 μl (250 μl per nostril). Ferrets were monitored daily for clinical signs of influenza infection, and body temperatures were recorded twice daily. Postinfection sera were collected at 14 and 21 dpi. For NL/03, CH/02, QL/95, EG/63, RK/34, and NC/93, postinfection sera from ferrets that received 105 TCID50 were used in serologic assays, and for BC/04 the serum from the ferret that received 102 TCID50 was used. For the remaining three viruses, sera from ferrets that received 107 TCID50 were used in serologic assays.
HI assay.
Antibody titers for postinfection mouse and ferret sera were determined by hemagglutination inhibition (HI) assays. Serum samples were treated with receptor-destroying enzyme (Denka Seiken Co., Ltd., Tokyo, Japan) for 22 h to remove nonspecific inhibitors. Serum samples were tested by standard methods using four HA units of the virus in V-bottom 96-well microtiter plates with 0.5% turkey erythrocytes and a 1% suspension of horse erythrocytes, as previously described (11, 23). HI titers were expressed as the reciprocals of the highest dilution of the serum that completely inhibited agglutination of erythrocytes by four HA units of the virus. HI titers were converted to log2 and were expressed as the arithmetic means ± standard errors.
Virus neutralization assay.
Neutralizing antibody titers for postinfection ferret and mouse sera were evaluated in a microneutralization (MN) assay. Postinfection ferret sera collected at 21 dpi from a single ferret and mouse sera collected at 28 dpi for VA/02 and RK/34 and at 56 dpi for the remaining eight viruses were used in the MN assay. Postinfection mouse sera from two to three mice were pooled to obtain a sufficient quantity for the MN assay. Serial twofold dilutions of heat-inactivated serum were prepared, and equal volumes of serum and virus were mixed and incubated for 60 min at room temperature. The residual infectivity of the virus-serum mixture was determined in MDCK cells in four replicates for each dilution. The neutralizing titer was defined as the reciprocal of the highest dilution of serum that completely neutralized the infectivity of 100 TCID50 of the virus as determined by the absence of cytopathic effect at day 4.
Nucleotide sequence accession numbers.
The nucleotide sequences presented in this article (accession numbers EF470585, EF470586, and EF470587) have been deposited in GenBank.
RESULTS
Selection of viruses.
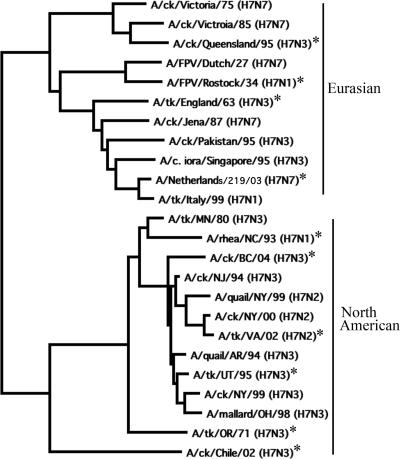
On phylogenetic analysis of the HA1 domain, the HA genes of 24 H7 subtype viruses clustered into Eurasian and North American lineages (Fig. 1). From these 24 H7 subtype viruses (Fig. 1 and Table 1), 10 viruses were selected on the basis of their phylogenetic relationships and geographic locations for evaluation in a mouse model and to identify viruses for vaccine development. Four viruses belonged to the Eurasian lineage, and six viruses belonged to the North American lineage (Fig. 1). Amino acid sequence identity of all pairwise comparisons of the predicted HA protein sequences ranged from 88.6 to 98%. The selected viruses included different subtypes of NA proteins, as shown in Table 2. The group of 10 viruses included six HP and four LP viruses on the basis of their previously established pathogenicity in chickens (Table 2) (1, 3, 8-10, 13, 18, 22, 24, 26, 28).
FIG. 1.
Phylogenetic relationships among selected avian influenza A H7 subtype viruses. The phylogram was generated by the neighbor-joining method, with the best tree option and midpoint rooting, using the MacVector 9.0 software package. The horizontal branch lengths are proportional to the nucleotide differences between the sequences. An asterisk indicates the viruses that were selected for further study.
TABLE 2.
Pathogenicity of selected H7 viruses for different speciesa
| Virus (subtype) | Pathogenicity forc:
|
HA cleavage site sequenceb | ||
|---|---|---|---|---|
| Chicken | Mouse | Human | ||
| A/ck/BC/CN-7/04 (H7N3) | + | + | + | ENPKQAV...RKRMTR/GLF |
| A/Netherlands/219/03 (H7N7) | + | + | + | EIPKRR........RR/GLF |
| A/tk/England/63 (H7N3) | + | + | − | ETPKRR........RR/GLF |
| A/Rostock/34 (H7N1) | + | + | − | EPSKKRE.......KR/GLF |
| A/ck/Queensland/95 (H7N3) | + | − | − | EIPRKR........KR/GLF |
| A/ck/Chile/02 (H7N3) | + | − | − | EKPKTCSPLSRCRETR/GLF |
| A/rhea/NC/93 (H7N1) | − | + | − | ENPK..........TR/GLF |
| A/tk/OR/71 (H7N3) | − | − | − | ENPK..........TR/GLF |
| A/tk/UT/95 (H7N3) | − | − | − | ENPK..........TR/GLF |
| A/tK/VA/02 (H7N2) | − | − | − | EKPK..........PR/GLF |
The pathogenicity for chickens and humans are historical data, and those for mice are part of the present study.
The slash indicates the site of cleavage of HA into HA1 and HA2.
A plus sign for chickens means that the virus is HP. A plus sign for mice indicates the virus is pathogenic as defined by mortality. A plus sign for humans indicates the ability of the virus to cause disease (illness and/or death). A minus sign indicates the absence of the ability to cause lethal infection in mice and chickens and the lack of a known ability to infect humans.
Lethality of H7 viruses for mice.
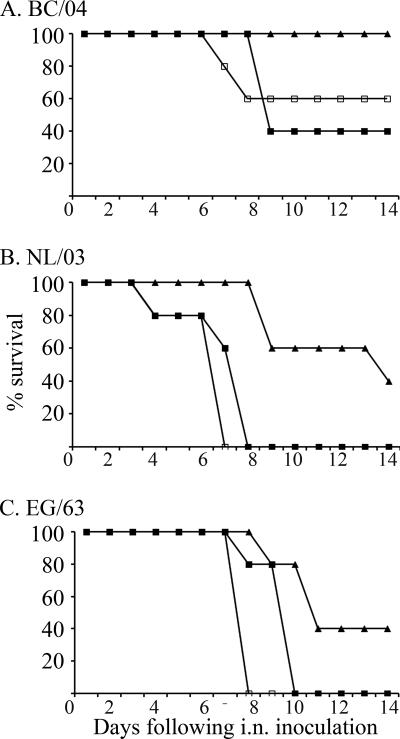
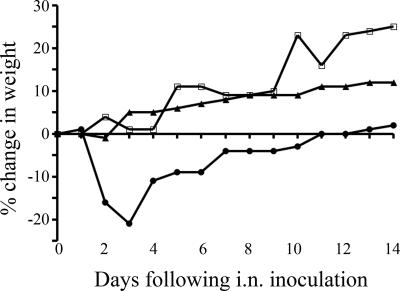
Differences in lethality among the 10 viruses were noted for BALB/c mice (Table 2). The mice inoculated with 105 TCID50 of the BC/04, NL/03, and EG/63 viruses succumbed by 5 to 6 dpi. However, the HP viruses CH/02 and QL/95 were not lethal for mice, even at the maximum dose of virus tested (105 TCID50). In contrast, the LP virus NC/93 was highly lethal for mice when it was used at 105 TCID50 per mouse. On the basis of their phylogenetic relationships and virulence for mice, BC/04 (an HP virus from the North American lineage), NL/03 and EG/63 (HP viruses from the Eurasian lineage), UT/95 (an LP virus from the North American H7N3 group), and VA/02 (an LP virus from the North American H7N2 group) viruses were selected for further study (Fig. 1 and Table 2). The three selected HP viruses, BC/04, NL/03, and EG/63, were lethal to mice at very low doses. Most of the mice that received 102 TCID50 of the virus were dead within 8 to 10 days (Fig. 2). On the basis of the lethality data for mice that received 102 TCID50 and 103 TCID50, the LD50 of BC/04 virus was determined to be 102.4 TCID50 (Fig. 2). Sixty percent of the mice that received 101 TCID50 of NL/03 died (Fig. 2), and all of the mice that received 100 TCID50 of the virus survived (not shown in Fig. 2), resulting in an LD50 of 100.8 for this virus. For the EG/63 virus, 60% of the mice that received 101 TCID50 died (Fig. 2), and only 50% of the mice that received 100 TCID50 of the virus survived (not shown in Fig. 2), resulting in an LD50 of 100.5. The LP virus UT/95 caused significant weight loss by 3 dpi, but the mice recovered; the VA/02 virus did not cause weight loss during the study period (Fig. 3).
FIG. 2.
Lethality of HP viruses BC/04 (A), NL/03 (B), and EG/63 (C) for mice following i.n. inoculation with 1,000 (□), 100 (▪), and 10 (▴) TCID50 of the indicated virus per mouse.
FIG. 3.
Percent change in weight following i.n. inoculation with LP viruses UT/95 (•) and VA/02 (▴). Mice that were mock infected (□) received L-15 medium.
Kinetics of replication of H7 viruses in mouse tissues.
The kinetics of replication of the five selected H7 viruses are summarized in Fig. 4. With the exception of UT/95 virus, all of the viruses replicated to high titers in the upper respiratory tract (nasal turbinates) of mice. UT/95 showed only a moderate level of replication in the upper respiratory tract, but it replicated to a higher titer in the lungs (Fig. 4A and B). All of the H7 viruses replicated to high titers in the lungs. The LP viruses showed peak replication in the lungs by 4 dpi and declined by 6 dpi, whereas the HP viruses maintained high titers in the lungs until the mice succumbed (Fig. 4B).
FIG. 4.
Replication kinetics of H7 viruses in mice following i.n. inoculation with 105 TCID50/mouse. Shown are the upper respiratory tract (A), lower respiratory tract (B), brain (C), and spleen (D) of mice at 1 (cross-hatched bars), 2 (open bars), 3 (diagonally hatched bars), 4 (black bars), 6 (heavily shaded bars), and 7 (lightly shaded bars) dpi. Virus titers are expressed as means ± standard errors in log10 TCID50/gram of tissue. The dashed horizontal line indicates the lower limit of detection. Organs were collected up to 4 dpi for NL/03 and EG/63 viruses and 6 dpi for BC/04 virus, because the mice did not survive beyond these time points.
HP H7 viruses also disseminated to the brain and spleen (Fig. 4C and D). In the brain, the titer of BC/04 peaked by 4 dpi. In the cases of NL/03 and EG/63, similar titers of virus were observed in the brain throughout the study period. In general, all of the HP viruses showed peak replication by 2 and 3 dpi in the spleen. The titers in the spleen were similar for NL/03 and EG/63 on days 2 and 3 postinfection. The BC/04 virus showed a moderate level of replication by 2 dpi in the spleen and showed limited replication by 4 and 6 dpi. The LP viruses UT/95 and VA/02 showed only limited and inconsistent replication in the brain, and neither was detected in the spleen (Fig. 4D).
Generation of postinfection sera in mice and ferrets.
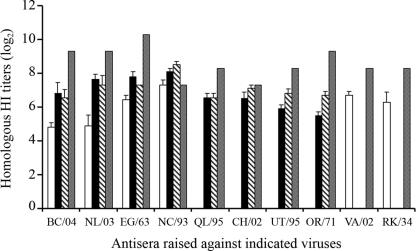
Postinfection sera were generated in mice and ferrets for the 10 selected H7 viruses. In an HI assay using postinfection mouse sera raised against NL/03, UT/95, NC/93, OR/71, EG/63, and RK/34, a two- to fourfold higher titer was observed when horse erythrocytes were used instead of turkey erythrocytes (Table 3). Therefore, horse erythrocytes were used in subsequent HI assays. Most of the H7 viruses induced serum HI antibody responses in mice that peaked by 42 or 56 dpi. The HI antibody titers observed in ferret sera by 21 dpi were similar to or higher than the HI antibody titers observed in mouse sera by 42 or 56 dpi for all viruses except the NC/93 virus (Fig. 5).
TABLE 3.
HI antibody titers induced by H7 subtype viruses in postinfection mouse sera using horse and turkey erythrocytesa
| Virus (subtype) | HI titers in homologous postinfection mouse sera using erythrocytes from:
|
Fold difference | |
|---|---|---|---|
| Horse | Turkey | ||
| A/Netherlands/219/03(H7N7) | 320 | 80 | 4 |
| A/tk/England/63 (H7N3) | 320 | 80 | 4 |
| A/rhea/NC/93 (H7N1) | 640 | 160 | 4 |
| A/tk/UT/95 (H7N3) | 160 | 40 | 4 |
| A/FPV/Rostock/34 (H7N1) | 80 | 40 | 2 |
| A/tk/OR/71 (H7N3) | 320 | 160 | 2 |
| A/ck/BC/CN-7/04 (H7N3) | 160 | 160 | 0 |
| A/tk/VA/02 (H7N2) | 160 | 160 | 0 |
| A/ck/Chile/02 (H7N3) | 80 | 80 | 0 |
| A/ck/Queensland/95 (H7N3) | 80 | 80 | 0 |
HI titers are expressed as the reciprocal of the highest dilution of serum that completely inhibited the agglutination of the indicated erythrocyte suspension by four HA units of the virus titrated. The HI titers in all of the prebleed sera were <10 with both horse and turkey erythrocytes. Antisera were collected at 28 dpi for VA/02 and RK/34 viruses and at 56 dpi for the remaining eight viruses. Postinfection sera from three to four mice were pooled and used in the HI assay.
FIG. 5.
HI antibody titers induced by H7 subtype viruses at 28 (open bars), 42 (black bars), and 56 (diagonally hatched bars) dpi in mice and at 21 (shaded bars) dpi in ferrets. HI titers were expressed as the reciprocal of the highest dilution of the serum that completely inhibited agglutination of horse erythrocytes by four HA units of the indicated virus. HI titers were converted to log2 and were expressed as the arithmetic means ± standard errors from three to five mice. Sera for CH/02, QL/95, UT/95, and OR/71 viruses at 28 dpi and sera for VA/02 and RK/34 viruses at 42 and 56 dpi were not obtained from mice. HI titers for the ferret sera are from a single ferret.
Homologous neutralization titers also were determined by an MN assay. Most of the viruses elicited homologous neutralization titers that were within a twofold difference from the level found for mice and ferrets. However, EG/63 and NC/93 showed a 4- and 10-fold difference, respectively, in homologous neutralization titers between mice and ferrets (Tables 4 and 5).
TABLE 4.
Titers of cross-neutralizing antibodies induced by H7 subtype viruses in postinfection mouse sera
| Virus (subtype) | Titers of mouse antisera raised against indicated virusa:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NL/03 | EG/63 | RK/34 | QL/95 | OR/71 | UT/95 | NC/93 | BC/04 | CH/02 | VA/02 | |
| A/Netherlands/219/03 (H7N7) | 453 | 10 | 50 | 40b | 16b | 20b | 254b | 20b | 63b | 7b |
| A/tk/England/63 (H7N3) | 254 | 202 | 16b | 63 | 40b | 28b | 127b | 25b | 101b | 10b |
| A/Rostock/34 (H7N1) | 254 | 127 | 160 | 57 | 25b | 28b | 453 | 16b | 99b | 7b |
| A/ck/Queensland/95 (H7N3) | 127 | 202 | 50 | 226 | 127b | 226 | 202b | 254 | 113b | 8b |
| A/tk/OR/71 (H7N3) | 640 | 905 | 160 | 403 | 1280 | 1016 | 2032 | 1016 | 453 | 160 |
| A/tk/UT/95 (H7N3) | 254 | 320 | 40b | 202 | 403 | 320 | 640 | 320 | 226b | 50b |
| A/rhea/NC/93 (H7N1) | 1016 | 320 | 202 | 160 | 254b | 254 | 1613 | 320 | 50b | 50b |
| A/ck/BC/CN-7/04 (H7N3) | 640 | 226 | 57 | 113 | 254b | 320 | 640 | 254 | 160b | 28b |
| A/ck/Chile/02 (H7N3) | 806 | 320 | 101 | 226 | 202b | 202 | 806 | 113 | 1016 | 25b |
| A/tk/VA/02 (H7N2) | 160 | 127 | 50 | 101 | 254b | 160 | 806 | 226 | 226b | 320 |
Mouse antisera were collected at 28 dpi for VA/02 and RK/34 viruses and at 56 dpi for the remaining eight viruses. Postinfection sera from three or four mice were pooled and used in the MN assay. Homologous neutralization titers are indicated in boldface.
Neutralization titers that are fourfold or lower than the homologous titers.
TABLE 5.
Titers of cross-neutralizing antibodies induced by H7 subtype viruses in postinfection ferret sera
| Virus (subtype) | Titers of ferret antisera raised against indicated virusa:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NL/03 | EG/63 | RK/34 | QL/95 | OR/71 | UT/95 | NC/93 | BC/04 | CH/02 | VA/02 | |
| A/Netherlands/03/219 (H7N7) | 254 | 57b | 101b | 63b | 28b | 28b | 63b | 101b | 160b | 8b |
| A/tk/England/63 (H7N3) | 226 | 806 | 101b | 127b | 101b | 63b | 63b | 50b | 226b | 8b |
| A/Rostock/34 (H7N1) | 453 | 127b | 453 | 226 | 57b | 63b | 113 | 80b | 508 | 20b |
| A/ck/Queensland/95 (H7N3) | 202 | 127b | 80b | 522 | 160b | 160 | 13b | 57b | 202b | 20b |
| A/tk/OR/71 (H7N3) | 508 | 403 | 127 | 403 | 806 | 806 | 160 | 320 | 640 | 254 |
| A/tk/UT/95 (H7N3) | 226 | 202 | 40b | 202 | 403 | 403 | 50b | 202 | 453 | 113b |
| A/rhea/NC/93 (H7N1) | 254 | 80b | 127 | 80b | 57b | 80b | 160 | 127 | 320 | 50b |
| A/ck/BC/CN-7/04 (H7N3) | 403 | 254 | 101b | 254 | 254 | 453 | 63b | 453 | 1016 | 226 |
| A/ck/Chile/02 (H7N3) | 113 | 50b | 101b | 403 | 160b | 226 | 28b | 113b | 905 | 32b |
| A/tk/VA/02 (H7N2) | 320 | 160b | 127 | 160 | 127b | 202 | 113 | 127 | 508 | 508 |
Antisera were collected at 21 dpi from a single ferret. Homologous neutralization titers are indicated in boldface.
Neutralization titers that are fourfold or lower than the homologous titers.
Cross-neutralization assay. (i) Mouse antisera.
Antigenic relationships among selected viruses were assessed using an MN assay, and the levels of cross-reactivity of antibodies elicited by different viruses in mice are summarized in Table 4. A neutralization titer that is fourfold or lower than the homologous neutralization titer was considered significantly different and indicated a lack of cross-reactivity. The Eurasian viruses elicited broadly cross-reactive antibodies that neutralized viruses from both Eurasian and North American lineages, but the converse was not true. NL/03 and EG/63 from the Eurasian lineage and BC/04, UT/95, and NC/93 from the North American lineage elicited postinfection mouse sera with a broader range of cross-reactivity than the other viruses from each group. Antibodies elicited by VA/02, CH/02, and OR/71 were poorly cross-reactive with other viruses, despite showing high homologous neutralization titers.
(ii) Ferret antisera.
All of the selected viruses, with the exception of NC/93, induced a more rapid neutralizing antibody response in ferrets than in mice. The levels of cross-reactivity of antibodies elicited by the different viruses in ferrets are summarized in Table 5. Seventy-four percent of the cross-MN titers were concordant between postinfection mouse and ferret sera. EG/63 and NC/93 viruses did not elicit the broad cross-reactivity in ferret sera that they did in the postinfection mouse sera. In contrast to postinfection mouse serum collected at 56 dpi, CH/02 virus elicited a broadly cross-reactive postinfection ferret serum collected at 21 dpi. A comparison of the cross-reactivity of postinfection mouse and ferret sera is shown in Table 6.
TABLE 6.
Concordance of cross-neutralization assays between postinfection mouse and ferret sera
| Virus (subtype) | Antisera raised against indicated virusa:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NL/03 | EG/63 | RK/34 | QL/95 | OR/71 | UT/95 | NC/93 | BC/04 | CH/02 | VA/02 | |
| A/Netherlands/219/03 (H7N7) | + | − | − | + | + | + | + | + | + | + |
| A/tk/England/63 (H7N3) | + | + | + | − | + | + | + | + | + | + |
| A/Rostock/34 (H7N1) | + | − | + | − | + | + | + | + | − | + |
| A/ck/Queensland/95 (H7N3) | + | − | − | + | + | + | + | − | + | + |
| A/tk/OR/71 (H7N3) | + | + | + | + | + | + | + | + | + | + |
| A/tk/UT/95 (H7N3) | + | + | + | + | + | + | − | + | − | + |
| A/rhea/NC/93 (H7N1) | + | − | + | − | + | − | + | + | − | + |
| A/ck/BC/CN-7/04 (H7N3) | + | + | − | + | − | + | − | + | − | − |
| A/ck/Chile/02 (H7N3) | + | − | − | + | + | + | − | − | + | + |
| A/tk/VA/02 (H7N2) | + | − | + | + | + | + | + | + | − | + |
A plus sign indicates concordant results; a minus sign indicates discordant results.
DISCUSSION
Avian influenza H7 subtype viruses have caused large outbreaks of disease in domestic poultry and several human infections in recent years (8, 13, 28). There is a need for the development of vaccines against H7 subtype viruses, because the ability of these viruses to transmit to humans may indicate pandemic potential. Animal models are needed for the study of pathogenesis to characterize the immune response and for the evaluation of vaccines against H7 subtype viruses.
In this study, we used 10 H7 subtype viruses that were selected on the basis of phylogenetic relationships and locations from which the viruses were isolated. On phylogenetic analysis of the HA genes, the viruses clustered into Eurasian and North American lineages, as has been reported previously (3). The percentage of amino acid sequence identity among the viruses ranged from 88.6 to 98%, and the VA/02 (H7N2) virus showed the lowest percentage of identity due to a deletion of eight amino acid residues in the HA1 domain of the HA protein (22).
In order to develop a mouse model of infection for H7 subtype viruses, NL/03 and EG/63 viruses from the Eurasian lineage and BC/04, VA/02, and UT/95 viruses from the North American lineage were evaluated in greater detail. LP and HP viruses showed a marked difference in their patterns of replication in the respiratory tract of mice. While the levels of replication of LP viruses increased and then declined in the respiratory tract of mice, the HP viruses that were lethal to mice maintained a high titer until the mice succumbed. The HP H7 viruses also were characterized by their spread to extrapulmonary tissues, as evidenced by their presence in the brain and the spleen, which is seen with some H5N1 avian influenza viruses (16). However, the H7 viruses were detected in the brain as early as 1 dpi, while H5N1 viruses were detected at 4 to 6 dpi. The lethality and kinetics of replication of NL/03 in mice were consistent with those of previously published studies (6). The differences in the levels and patterns of replication correlated with the differences in lethality of the viruses in mice. Very low doses of the HP viruses were lethal to mice, whereas LP viruses did not kill mice even at high doses. Interestingly, in contrast to the UT/95 virus, the VA/02 virus that replicated to a high titer in the upper and lower respiratory tracts of mice did not cause significant weight loss during the course of infection.
Most of the H7 subtype viruses were able to infect mice without adaptation and manifested different levels of lethality. The molecular basis of the difference between the pathogenicity of HP and LP avian influenza viruses for chickens has been attributed mainly to the cleavability of precursor HA0 into HA1 and HA2 subunits. The cleavability of the HA0 of LP viruses is restricted to tissues in which trypsin and trypsin-like proteases are present (respiratory and intestinal tracts), and this limits the tissue range of the virus. The HA0 of HP viruses that possess multiple basic amino acids at the cleavage site can be cleaved by ubiquitous intracellular proteases, like furins and non-trypsin-like proteases. This enables the virus to disseminate systemically and replicate in extrapulmonary organs, including the brain, causing fatal disease and death of the birds (12, 19). Our results indicate that the virulence motif of H7 viruses that makes viruses highly pathogenic to chickens may not be significant in mammals. Among the HP viruses, CH/02 and QL/95 were not lethal for mice, despite the presence of a multibasic amino acid cleavage site in the HA protein. In contrast, the LP virus NC/93, which does not have a multibasic cleavage site in the HA protein, was lethal for mice, and another LP virus, UT/95, caused significant weight loss in mice. A recent study also showed that an H7N2 avian influenza LP virus isolated from chickens in China was able to infect mice without adaptation, causing significant weight loss following i.n. inoculation (15). These observations suggest that some H7 viruses are capable of infecting mammals without adaptation, and a determinant(s) other than the presence of a multibasic cleavage site in the HA protein is involved in their pathogenicity for mammalian species. Further studies are required to understand the molecular basis for the lethality of H7 viruses in mice.
In order to understand the extent to which cross-protective antibody responses were induced by viruses from the two lineages, we generated postinfection mouse and ferret sera against 10 selected H7 viruses. Stephenson et al. (23) and Meijer et al. (17) have reported that the use of horse erythrocytes in the HI assay improved the sensitivity of detection of antibodies against H5 and H7 avian influenza subtype viruses, presumably due to the preference of avian HAs for α(2,3)-galactase-linked sialic acid receptors present on horse erythrocytes. Consistent with these studies, 6 of the 10 H7 viruses in this study showed two- to fourfold higher HI titers when horse erythrocytes were used in HI assays instead of turkey erythrocytes. However, the source of erythrocytes did not influence the HI titers of the remaining four viruses.
This study also provided an opportunity to compare the immune responses of mice to avian influenza H7 subtype viruses to those of ferrets. The levels of cross-reactivity of antibodies elicited by the 10 selected viruses in mice and ferrets in virus neutralization generally were consistent. Although ferret antiserum is the WHO standard for antigenic characterization of influenza viruses, our study suggests that postinfection mouse antiserum provides comparable data and could be used for analysis of H7 subtype viruses.
A delayed maturation of the functional antibodies in mice following infection with avian influenza A H5N1 viruses has been demonstrated previously (25). As was seen with H5N1 viruses, HI assays using horse erythrocytes showed that most of the H7 viruses in this study also induced serum HI antibody responses in mice that gradually increased and peaked at 42 or 56 dpi. This observation has implications for the preclinical evaluation of H7 virus vaccines given to mice. Ferrets developed a more rapid antibody response than mice, with similar or higher titers of HI antibodies by 21 dpi. Postinfection ferret and mouse sera were used in cross-HI and cross-MN assays to establish the extent of antigenic relatedness of the selected H7 viruses. The results from the two assays did not correlate well (data not shown). Since the HI assay using horse erythrocytes is not standardized and the MN assay may be a more relevant functional assay than the HI assay, the level of cross-reactivity of antibodies was assessed using MN titers.
Cross-neutralization studies showed that phylogenetic relationships or the percentage of amino acid identity may not accurately reflect antigenic relationships among H7 subtype viruses. In this study, viruses from the Eurasian lineage elicited broadly cross-reactive antibodies that neutralized viruses from both Eurasian and North American lineages, but the converse was not true. These data suggest that vaccines generated using viruses from the Eurasian lineage may provide a broader range of protection than vaccines generated from North American strains. The cross-neutralization titers suggest that NL/03 is a suitable candidate virus for vaccine development, because it elicited broadly cross-reactive antibodies that neutralized viruses from both lineages. Additionally, NL/03 was isolated from a fatal human infection during the H7N7 avian influenza outbreak in The Netherlands in 2003 (8, 13). However, it may be appropriate to select viruses from both lineages for the development of vaccines. Although the UT/95 and BC/04 viruses elicited a broader range of cross-reactive antibodies among the viruses from the North American lineage, the BC/04 virus was selected for vaccine development, because it is a recently identified isolate that has caused human infections (9, 28). Taking these results together, we selected NL/03 (H7N7) and BC/04 (H7N3) viruses for vaccine development.
In summary, we have developed a BALB/c mouse model for the evaluation of pathogenicity and antibody responses to avian influenza A H7 subtype viruses that can be used for the preclinical evaluation of vaccines. We have shown that several H7 viruses induced antibodies in postinfection mouse and ferret sera with comparable cross-reactivity. On the basis of phylogenetic data and the induction of broadly cross-neutralizing antibodies in postinfection mouse and ferret sera, we have selected NL/03 (H7N7) and BC/04 (H7N3) viruses for the development of vaccines against H7 pandemic influenza.
Acknowledgments
This research was supported by the Intramural Research Program of the NIAID, NIH. This research was performed as part of a Cooperative Research and Development Agreement (CRADA no. AI-0155) between the Laboratory of Infectious Diseases, NIAID, and MedImmune, Inc.
We thank Jadon Jackson and the staff of SoBran, Inc., and the Comparative Medicine Branch, NIAID, for excellent technical support for animal studies. We thank Nick Nguyen of MedImmune, Inc., for assistance with ferret studies. We are grateful to Robert G. Webster, David Swayne, Nancy Cox, and John Pasick for providing the viruses used in this study.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Akey, B. L. 2003. Low-pathogenicity H7N2 avian influenza outbreak in Virgnia during 2002. Avian Dis. 47:1099-1103. [DOI] [PubMed] [Google Scholar]
- 2.Banks, J., E. Speidel, and D. J. Alexander. 1998. Characterisation of an avian influenza A virus isolated from a human—is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch. Virol. 143:781-787. [DOI] [PubMed] [Google Scholar]
- 3.Banks, J., E. C. Speidel, J. W. McCauley, and D. J. Alexander. 2000. Phylogenetic analysis of H7 haemagglutinin subtype influenza A viruses. Arch. Virol. 145:1047-1058. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, C. H., R. G. Webster, and S. S. Breese, Jr. 1970. Fowl plague virus from man. J. Infect. Dis. 122:513-516. [DOI] [PubMed] [Google Scholar]
- 5.Capua, I., and D. J. Alexander. 2004. Avian influenza: recent developments. Avian Pathol. 33:393-404. [DOI] [PubMed] [Google Scholar]
- 6.de Wit, E., V. Munster, M. I. Spronken, T. M. Bestebroer, C. Baas, W. E. Beyer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2005. Protection of mice against lethal infection with highly pathogenic H7N7 influenza A virus by using a recombinant low-pathogenicity vaccine strain. J. Virol. 79:12401-12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., V. Munster, A. Wallensten, T. M. Bestebroer, S. Herfst, D. Smith, G. F. Rimmelzwaan, B. Olsen, and A. D. Osterhaus. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouchier, R. A., P. M. Schneeberger, F. W. Rozendaal, J. M. Broekman, S. A. Kemink, V. Munster, T. Kuiken, G. F. Rimmelzwaan, M. Schutten, G. J. Van Doornum, G. Koch, A. Bosman, M. Koopmans, and A. D. Osterhaus. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. USA 101:1356-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirst, M., C. R. Astell, M. Griffith, S. M. Coughlin, M. Moksa, T. Zeng, D. E. Smailus, R. A. Holt, S. Jones, M. A. Marra, M. Petric, M. Krajden, D. Lawrence, A. Mak, R. Chow, D. M. Skowronski, S. A. Tweed, S. Goh, R. C. Brunham, J. Robinson, V. Bowes, K. Sojonky, S. K. Byrne, Y. Li, D. Kobasa, T. Booth, and M. Paetzel. 2004. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg. Infect. Dis. 10:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, Y. L., and D. E. Swayne. 2004. Comparative pathobiology of low and high pathogenicity H7N3 Chilean avian influenza viruses in chickens. Avian Dis. 48:119-128. [DOI] [PubMed] [Google Scholar]
- 11.Kendal, A. P., M. S. Pereira, and J. J. Skehel. 1982. Concepts and procedures for laboratory-based influenza surveillance. Centers for Disease Control, U.S. Department of Health and Human Services, Atlanta, GA.
- 12.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 13.Koopmans, M., B. Wilbrink, M. Conyn, G. Natrop, H. van der Nat, H. Vennema, A. Meijer, J. van Steenbergen, R. Fouchier, A. Osterhaus, and A. Bosman. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in The Netherlands. Lancet 363:587-593. [DOI] [PubMed] [Google Scholar]
- 14.Kurtz, J., R. J. Manvell, and J. Banks. 1996. Avian influenza virus isolated from a woman with conjunctivitis. Lancet 348:901-902. [DOI] [PubMed] [Google Scholar]
- 15.Li, Y., C. Li, L. Liu, H. Wang, C. Wang, G. Tian, R. G. Webster, K. Yu, and H. Chen. 2006. Characterization of an avian influenza virus of subtype H7N2 isolated from chickens in northern China. Virus Genes 33:117-122. [DOI] [PubMed] [Google Scholar]
- 16.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijer, A., A. Bosman, E. E. van de Kamp, B. Wilbrink, R. van Beest Holle Mdu, and M. Koopmans. 2006. Measurement of antibodies to avian influenza virus A (H7N7) in humans by hemagglutination inhibition test. J. Virol. Methods 132:113-120. [DOI] [PubMed] [Google Scholar]
- 18.Pasick, J., K. Handel, J. Robinson, J. Copps, D. Ridd, K. Hills, H. Kehler, C. Cottam-Birt, J. Neufeld, Y. Berhane, and S. Czub. 2005. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J. Gen. Virol. 86:727-731. [DOI] [PubMed] [Google Scholar]
- 19.Perdue, M. L., M. Garcia, D. Senne, and M. Fraire. 1997. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 49:173-186. [DOI] [PubMed] [Google Scholar]
- 20.Puzelli, S., L. Di Trani, C. Fabiani, L. Campitelli, M. A. De Marco, I. Capua, J. F. Aguilera, M. Zambon, and I. Donatelli. 2005. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J. Infect. Dis. 192:1318-1322. [DOI] [PubMed] [Google Scholar]
- 21.Reed, L. J., and H. Muench. 1938. A simple method of estimation fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 22.Spackman, E., D. A. Senne, S. Davison, and D. L. Suarez. 2003. Sequence analysis of recent H7 avian influenza viruses associated with three different outbreaks in commercial poultry in the United States. J. Virol. 77:13399-13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephenson, I., J. M. Wood, K. G. Nicholson, and M. C. Zambon. 2003. Sialic acid receptor specificity on erythrocytes affects detection of antibody to avian influenza haemagglutinin. J. Med. Virol. 70:391-398. [DOI] [PubMed] [Google Scholar]
- 24.Suarez, D. L., D. A. Senne, J. Banks, I. H. Brown, S. C. Essen, C. W. Lee, R. J. Manvell, C. Mathieu-Benson, V. Moreno, J. C. Pedersen, B. Panigrahy, H. Rojas, E. Spackman, and D. J. Alexander. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg. Infect. Dis. 10:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suguitan, A. L., J. McAuliffe, K. L. Mills, H. Jin, G. Duke, B. Lu, C. J. Luke, B. Murphy, D. E. Swayne, G. Kemble, and K. Subbarao. 2006. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 3:e360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swayne, D. E., J. R. Beck, M. L. Perdue, M. Brugh, and R. D. Slemons. 1996. Assessment of the ability of ratite-origin influenza viruses to infect and produce disease in rheas and chickens. Avian Dis. 40:438-447. [PubMed] [Google Scholar]
- 27.Taylor, H. R., and A. J. Turner. 1977. A case report of fowl plague keratoconjunctivitis. Br. J. Ophthalmol. 61:86-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tweed, S. A., D. M. Skowronski, S. T. David, A. Larder, M. Petric, W. Lees, Y. Li, J. Katz, M. Krajden, R. Tellier, C. Halpert, M. Hirst, C. Astell, D. Lawrence, and A. Mak. 2004. Human illness from avian influenza H7N3, British Columbia. Emerg. Infect. Dis. 10:2196-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webster, R. G., J. Geraci, G. Petursson, and K. Skirnisson. 1981. Conjunctivitis in human beings caused by influenza A virus of seals. N. Engl. J. Med. 304:911. [DOI] [PubMed] [Google Scholar]
- 31.Wright, P. F., and R. G. Webster. 2001. Orthomyxoviruses, p. 1533-1579. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.