Abstract
VP90, the capsid polyprotein precursor of human astrovirus Yuc8, is assembled into viral particles, and its processing at the carboxy terminus by cellular caspases, to yield VP70, has been correlated with the cell release of the virus. Here, we characterized the effect of the VP90-VP70 processing on the properties of these proteins, as well as on their intracellular distribution. VP90 was found in membrane-enriched fractions (mVP90), as well as in fractions enriched in cytosolic proteins (cVP90), while VP70 was found exclusively in the latter fractions. Upon trypsin activation, infectivity was detected in all VP90-containing fractions, confirming that both mVP90 and cVP90 are able to assemble into particles; however, the two forms of VP90 showed differential sensitivities to trypsin, especially at their carboxy termini, which in the case of mVP90 was shown to remain membrane associated after protease digestion. Structural protein oligomers were detected in purified VP70-containing viruses, as well as in membrane-enriched fractions, but they were less evident in cytosolic fractions. Ultrastructural studies of infected cells revealed different types of viral particles, some of which appeared to be associated with membranes. By immunoelectron microscopy, structural proteins were shown to form virus particles in clusters and to associate with the edges of vesicles induced during infection, which also appear to contain subviral particles inside. Nonstructural proteins and viral RNA colocalized with mVP90, but not with cVP90, suggesting that mVP90 might represent the form of the protein that is initially assembled into particles, at the sites where the virus genome is being replicated.
Human astroviruses (HAstVs) are recognized as the second major cause of childhood viral gastroenteritis around the world (6). Eight astrovirus serotypes have been identified in humans (HAstV-1 to HAstV-8), which differ mainly in the amino acid sequence of the carboxy-terminal half of the capsid polyprotein (12, 13). The astrovirus genome, of around 6.8 kb, has three open reading frames (named ORF1a, -1b, and -2) (11, 18), each encoding a polyprotein which is processed during infection (7, 15, 16, 25). ORF1a and ORF1b encode the nonstructural protein precursors, which are processed by cellular and viral serine proteases (7, 16). Nonstructural proteins most likely participate in the synthesis of viral RNA species (11). ORF2 encodes the capsid polyprotein precursor, which contains around 785 amino acid residues, depending on the virus strain, and has a molecular mass of 87 to 90 kDa (90K protein) (11, 26). The processing of this protein, required to yield the infectious mature virus, occurs inside and outside the cell and has been best studied in the HAstV-8 strain Yuc8 (14). The VP90 polyprotein of this virus is cleaved intracellularly at its acidic carboxy-terminal region in at least four positions, to yield intermediate protein precursors of 82K, 78K, and 75K, as well as the mature intracellular cleavage product VP70 (15). These cleavages are carried out by caspases, proteases involved in regulation of cell death by apoptosis that are induced during HAstV infection of tissue-cultured Caco-2 cells (9, 15).
The processing of VP90 to VP70 has been correlated with release of the virus from the cell (15). The released astrovirus particles, formed by VP70, are either noninfectious or poorly infectious, and the activation of their infectivity requires that VP70 be processed by trypsin. The enhancement of infectivity induced by treatment with this protease can be more than 100-fold in the case of HAstV-8 strains (3, 14, 22). Upon trypsin treatment, VP70 is initially processed at Arg393 to yield polypeptides VP41 and VP28. These proteins are further cleaved in a sequential manner at their carboxy and amino termini, respectively, to yield the final products VP34 (derived from VP41) and VP27 and VP25 (both derived from VP28). Although the pathway for processing of VP70 by trypsin has been characterized in detail only for astrovirus Yuc8, it is thought that all other HAstV strains have a similar processing pathway, because the sizes of the final protein products observed in different strains are very similar (3, 14, 22).
The intracellular processing of the VP90 capsid protein is not required for virus assembly, since HAstV-8 particles formed by VP90 have been detected, and the assembly of virus progeny is not negatively affected when the processing of VP90 is drastically reduced, for instance, in the presence of the pancaspase inhibitor Z-VAD-FMK (15). In addition, the expression of the primary ORF2 polyprotein product of HAstV-1 and HAstV-2 using either baculovirus (4) or vaccinia virus (5) as an expression vector produces uncleaved products that assemble as virus-like particles. The amino-terminal region of the capsid protein has been reported as dispensable for astrovirus assembly, since the expression of capsid mutants lacking the first 70 amino acid residues of HAstV-1 results in the formation of virus-like particles in insect cells (4), and virus mutants lacking amino acid residues 11 to 30 of the precursor capsid protein assemble as efficiently as do wild-type viruses (8).
To characterize the relationship of the intracellular processing of the capsid polyprotein precursor with the morphogenetic pathway of astrovirus, we analyzed biochemically and ultrastructurally the intracellular distribution of VP90 as well as that of its cleavage product VP70. We found that, unlike VP70, a fraction of VP90 associates with membranes, and we present evidence that suggests that this cell compartment could be the site where virus particles initially assemble.
MATERIALS AND METHODS
Cells and virus.
The colon adenocarcinoma Caco-2 cell line from the American Type Culture Collection was used in this work. Cells were cultured in a 10% CO2 atmosphere at 37°C with advanced Dulbecco modified Eagle medium (Invitrogen), supplemented with 2 mM glutamine and 3% fetal bovine serum. Stocks of the HAstV serotype 8 strain Yuc8 (17) were prepared, and titers were determined as focus-forming units using anti-Yuc8 antibodies, as previously described (15). Titer was reported as focus-forming units/ml, and multiplicity of infection was calculated based on this titer.
Sera and reagents.
A synthetic peptide with the sequence TYVDAPLPEEPPIEEEETD (abbreviated TYVD), corresponding to the amino acid residues 654 to 672 of Yuc8 VP90, was used to generate hyperimmune rabbit polyclonal antibodies, as described previously (14, 16). Rabbit polyclonal antibodies to strain Yuc8 and to the recombinant protein E4 (residues 666 to 782 of VP90) have been previously described (15). Antibodies to the recombinant proteins 1a-1 and 1a-3 (amino acid residues 41 to 257 and 401 to 638 of nsp1a, respectively), and 1b-2 (residues 201 to 362 of nsp1b) (16) recognize the 20-kDa amino-terminal product of nsp1a, the protease, and the RNA polymerase motifs, respectively (16). Z-VAD-FMK (a pancaspase inhibitor) and TRAIL were purchased from BIOMOL Research Lab (Plymouth, PA). The peptide Z-VAD-FMK stock was prepared in dimethyl sulfoxide at 50 mg/ml, and it was used at 50 μM while TRAIL was used at 2 μg/ml during infection.
Cell infection.
Caco-2 cells were infected with astrovirus essentially as previously described (15). Prior to infection, all virus samples were activated with 200 μg/ml of trypsin, and this enzyme was maintained at 3 μg/ml after the adsorption period. Soybean trypsin inhibitor (Sigma) was added to the culture medium at 400 μg/ml during the adsorption period to avoid detachment of the cells. When used, Z-VAD-FMK and TRAIL were added immediately after the adsorption period and kept until the cells were harvested. For protein analysis, cells were harvested in TNS buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% sodium dodecyl sulfate [SDS], 20 μg/ml phenylmethylsulfonyl fluoride, and 100 μg/ml leupeptin). Virus purification was carried out from infected cells maintained with fetal bovine serum and purified in cesium chloride density gradients, using TNE buffer (10 mM Tris, pH 7.4, 150 mM sodium chloride, and 10 mM EDTA), as described previously (3, 14, 22).
Immunoassays.
Cell extracts were mixed with Laemmli sample buffer (50 mM Tris, pH 7.5, 2% SDS, 2% β-mercaptoethanol, 10 mM EDTA, and 01% bromophenol blue), and proteins were separated in SDS-polyacrylamide gels. Proteins were transferred to a nitrocellulose membrane (Millipore) and detected with the indicated primary antibodies, as described previously (15).
Cell fractionation.
Cells grown in 150-cm2 flasks were infected with Yuc8 at a multiplicity of infection of 5; 20 to 24 h postinfection (hpi), cell monolayers were washed three times with phosphate-buffered saline (PBS) and twice with cold HEPES-sucrose buffer (HSB; 50 mM HEPES, pH 7.4, 250 mM sucrose). Cells were kept for 10 min on ice with 5 ml of HSB, scraped, and centrifuged at 1,500 × g for 10 min; the cell pellet was resuspended in 5 ml of HSB and centrifuged again. The cell pellet was then resuspended in 0.8 ml of cold HSB with a protease inhibitor cocktail (Roche), and the cells were disrupted by 50 strokes in a Dounce homogenizer. Nuclei and unbroken cells were separated by centrifugation at 1,500 × g for 10 min, and the nucleus-free cell extract was mixed with iodixanol (OptiPrep; Axis-Shield PoC AS, Norway), to reach 40% iodixanol. Discontinuous gradients, used to fractionate the cell extract, were prepared by diluting the iodixanol stock solution with HSB. Gradients were set up in 5-ml Beckman SW50.1 centrifuge tubes from the bottom to the top as follows: 1.2 ml of 40% iodixanol (this fraction included the cell extract), 1.5 ml of 35% iodixanol, 0.8 ml of 30% iodixanol, 0.8 ml of 20% iodixanol, and 0.8 of 15% iodixanol. The gradients were centrifuged for 4 h at 150,000 × g (SW50.1 Beckman rotor) at 4°C, and fractions of approximately 0.35 ml were collected from the bottom of the tube, the pelleted insoluble material being discarded. In this type of gradient, the cellular cytosolic components are enriched in the bottom fractions of the tube upon centrifugation, while the membrane-enriched material floats and is found in the density-light, upper fractions of the gradient. Proteins from each fraction were analyzed by immunoblotting with the indicated antibodies, while titers of the infectious virus particles were determined in Caco-2 cells. No protease inhibitors were used in the gradient before titration of virus infectivity.
In vitro translation.
ORF1a and ORF2 of HAstV-8 (Yuc8 strain) were cloned in the plasmid pCDNA3.1 Hygro (−), and these constructs were used to translate nsp1a and VP90, using the TNT quick-coupled transcription-translation system (Promega) in the presence of Express-[35S]Met labeling (New England Nuclear). When used, canine pancreatic microsomal membranes (Promega) were included in the reaction mixture according to the manufacturer's instructions. The plasmid expressing ORF2 from HAstV-1 (Oxford strain) was a generous gift of S. Matsui and U. Geigenmuller (Stanford University). For density gradient centrifugation, the total translation reaction mixture was combined with iodixanol to reach a 40% concentration in a volume of 0.48 ml. The iodixanol gradients were set up as described in the section above using the indicated volumes for each iodixanol concentration (0.6 ml of 35%, 0.32 ml of 30%, 0.32 ml of 20%, and 0.32 of 15% iodixanol) in a tube for a TLA100.2 rotor (Beckman). Gradients were centrifuged for 2 h at 80,000 rpm, and fractions of 170 μl were collected.
Electron microscopy of astrovirus-infected cells. (i) Thin-section analysis.
Astrovirus-infected cells harvested 24 hpi were embedded in EMbed-812 epoxy resin (Electron Microscopy Sciences) using a conventional protocol. Cells were fixed with 4% p-formaldehyde and 2.5% glutaraldehyde in PBS buffer and treated with 1% osmium tetroxide in PBS for 1 h at 4°C. A short dehydration in an ethanol series followed by propylene oxide at room temperature was carried out. Cells were preembedded with a propylene oxide-EMbed-812 mixture at room temperature. Later, polymerization with EMbed-812 alone was done at 60°C for 2 days. Ultrathin sections, uniform in width (60 nm), were stained with uranyl acetate and lead citrate.
(ii) Ultrastructural labeling of specific components.
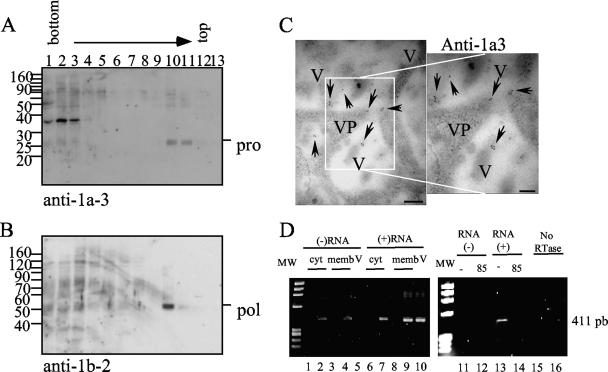
Astrovirus proteins were immunolabeled on thin sections of LR White-embedded virus-infected cells, previously fixed with 4% p-formaldehyde and 0.1% glutaraldehyde in PBS buffer, and followed by a short dehydration ethanol series. Immunolabeling was performed in thin sections collected on electron microscopy nickel grids covered with Formvar and carbon. Thin sections of 60 nm were prepared with an ultramicrotome (Leica Ultracut R; Vienna, Austria). Grids were incubated for 60 min at room temperature with the primary antibody (anti-Yuc8, anti-E4, or anti-1a-3). Grids were washed five times, and the secondary goat anti-rabbit immunoglobulin G (IgG) labeled with 10-nm colloidal gold diluted 1:10 in PBS was added. After being washed with PBS and distilled water, samples were stained with uranyl acetate.
(iii) IEM of membrane-enriched fractions.
Samples of fraction 9 of iodixanol gradients were loaded on grids covered with Formvar and carbon and processed for immunoelectron microscopy (IEM) using anti-E4 antibodies as primary antibodies. Finally, samples were negatively stained with uranyl acetate. Samples were observed in a Zeiss EM900 electron microscope operating at 80 kV.
Viral RNA analysis.
RNA was obtained from cytosolic and membrane fractions of iodixanol gradients and from the supernatant of Yuc8-infected cell cultures by extraction with TRIzol (Invitrogen). Reverse transcription (RT) was carried out with Superscript III (Invitrogen) and oligonucleotide Mon244 or Mon245 (19) at 42°C for 40 min. After this period of incubation, the enzyme activity was inactivated by heating the sample at 85°C for 10 min. PCR was carried out with the same pair of oligonucleotides and Vent polymerase (New England Biolabs) using equal volumes of the RT reaction mixtures. The reaction conditions were 2 min at 94°C (1 cycle), 20 s at 94°C, 20 s at 52°C, 30 s at 72°C (25 cycles), and 1 min at 72°C. As controls, RT reactions were carried out after heating the sample at 85°C for 10 min before adding the corresponding oligonucleotide. Additionally, a mock RT-PCR was carried out in the absence of Superscript. RT-PCR products were separated in 2% agarose gels and stained with ethidium bromide. Images were obtained by scanning the gels with a Thyphoon 4900 phosphorimager (Amersham Biosciences).
RESULTS
Proteins VP70 and VP90 are differentially localized in the cell.
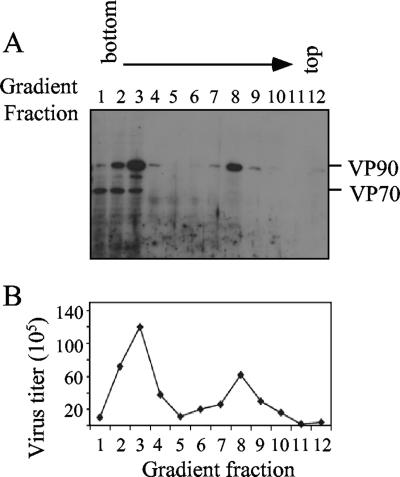
Since the intracellular VP90-VP70 processing has been related to release of the virus from the cells (2, 15), the association of these two polyproteins with different intracellular structures was investigated. This was carried out by density gradients of iodixanol obtained from nucleus-free infected-cell extracts. This method has been successfully used to separate cell organelles from cytosolic proteins, especially those containing membranes, since these float after ultracentrifugation in this type of gradient (27). After fractionation, protein VP90 was detected in fractions 1 to 4, which contain the cytosolic proteins, as well as in fractions 7 to 9, which contain proteins associated with density-light cell structures, mainly membranes (Fig. 1A). In contrast to VP90, most VP70 was found in the cytosolic fractions (Fig. 1A), suggesting that at least a fraction of the VP90 molecules were associated with cellular structures different from that of the processed protein VP70. After trypsin treatment, virus infectious particles were detected in the fractions where either VP70 or VP90 was present, i.e., fractions 2 to 4 and 7 to 9 (Fig. 1B). The fact that infectivity was obtained in fractions where VP70 was not detected confirmed that particle assembly was independent of the proteolytic cleavage of VP90, as previously observed for HAstV-8 strains as well as for astroviruses belonging to other serotypes (4, 5, 8, 15).
FIG. 1.
Structural astrovirus proteins VP90 and VP70 are differentially located in the cell. Cytoplasmic extracts were fractionated by ultracentrifugation in density gradients, as described in Materials and Methods. Twelve fractions were collected and analyzed for immunoblotting with anti-Yuc8 antibodies in a 7.5% polyacrylamide gel (A) and for the presence of infectious particles after trypsin treatment (B). Viral proteins are marked on the right.
VP90 associates with membranes through its carboxy terminus.
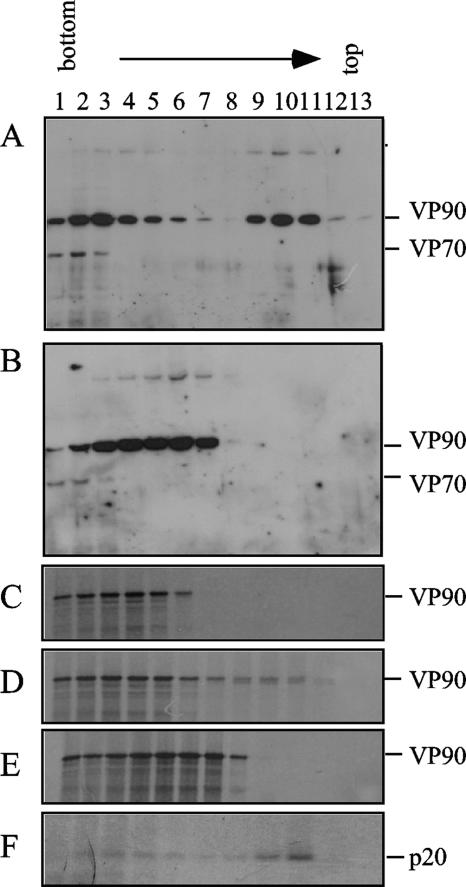
The ability of VP90 to float on density gradients suggests that it could be associated with cellular membranes. To test this possibility, the cytoplasmic extracts were treated with Triton X-100 (TX-100) before the density gradient was set up. Treatment with TX-100 shifted the position of VP90 from the lighter fractions of the gradient to the bottom fractions that contain cytosolic material (Fig. 2A and B), indicating that the detergent abolished its association with membranes. This association could occur either directly or through interactions with cellular or viral nonstructural proteins, since the latter have been found associated with the endoplasmic reticulum (10). To determine if viral nonstructural proteins were involved in this interaction, VP90 was translated in vitro in the presence of canine pancreatic microsomes and its association with membranes was analyzed. The amino-terminal protein product of ORF1a, p20, which is known to associate with membranes (7, 16), was used as a positive control for membrane association (Fig. 2F). In vitro-translated VP90 was found in the upper fractions of the gradients when it was translated in the presence of microsomes (fractions 8 to 10, Fig. 2D) but not in their absence (Fig. 2C). Association of VP90 with the floating fractions was again diminished by treatment with TX-100 (Fig. 2E). These observations indicate that VP90 is able to interact with membranes in the absence of any other viral protein.
FIG. 2.
Protein VP90 is found soluble and associated with membranes in the cells. Cytoplasmic extracts of untreated and infected cells (A) or cells treated for 30 min with TX-100 at room temperature (B) were fractionated by density gradients, and fractions were separated by 7.5% SDS-PAGE and immunoblotted with anti-TYVD antibodies. In vitro-translated VP90 labeled with 35S-Express label in the absence (C) or in the presence (D) of microsomes or in the presence of microsomes but with previous treatment with TX-100 (E) was loaded in the density gradients. ORF1a was in vitro translated in the presence of microsomes, and the p20 amino-terminal product of nsp1a was immunoprecipitated with anti-1a-1 antibodies (F) (16). Viral proteins in panels C to F were separated by SDS-PAGE and detected by autoradiography. The viral proteins VP90, VP70, and p20 are marked at the right.
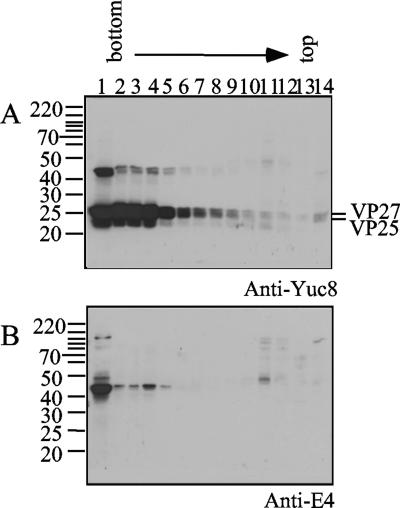
While at least a fraction of VP90 was found to associate with membranes, this was not the case for VP70. Thus, it is possible that the carboxy terminus of VP90 would be involved in this interaction. To investigate this possibility, cytoplasmic extracts were partially digested with trypsin and fractionated by density gradients, and the VP90 products were identified with antibodies to its carboxy terminus. Most of the viral proteins were found in the cytosolic fraction of the gradient, as detected with anti-Yuc8 and anti-E4 antibodies (fractions 1 to 6, Fig. 3A and B). However, a small proportion of the proteins detected with anti-E4 antibodies, which recognize polypeptides containing the carboxy terminus of VP90 (15), floated in the gradient (Fig. 3B). The product of approximately 45 kDa detected with those antibodies was also recognized by anti-Yuc8, although to a lesser extent (Fig. 3A). These data suggest that the carboxy terminus of VP90 interacts with membranes and that it could be involved in the association of the precursor protein with these structures. In spite of this association, most of the VP90 protein, either from infected cells or from in vitro translation, remained in the cytosolic fractions of the gradient, suggesting either a weak VP90 membrane association or the existence of two different subpopulations of VP90.
FIG. 3.
The carboxy terminus of VP90 is involved in membrane association. Cytoplasmic extracts treated with trypsin (200 μg/ml) for 30 min at 37°C were fractionated by density gradients and immunoblotted with the indicated antibodies. Viral proteins are marked at the right and molecular mass markers (in kilodaltons) at the left.
The membrane-associated form of VP90 is less susceptible to trypsin cleavage.
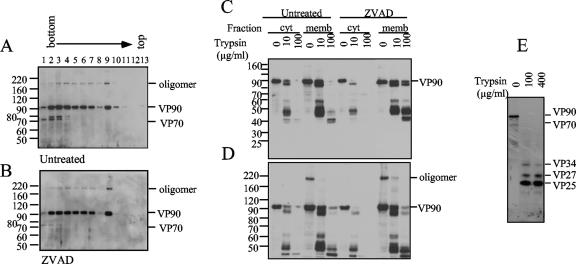
To distinguish putative differences between the membrane-associated and the cytosolic forms of VP90 (named mVP90 and cVP90, respectively), these proteins were analyzed by enzyme digestion accessibility. To detect VP90 and the digestion products derived from its carboxy terminus, anti-E4 antibodies and Z-VAD-FMK-treated cells were used. The effectiveness of the Z-VAD-FMK treatment was confirmed with anti-TYVD antibodies, which recognize both VP70 and VP90. As expected, the amount of VP70 drastically decreased in the soluble fractions upon Z-VAD-FMK treatment; however, the distribution of VP90 was essentially unaffected (Fig. 4A and B). A high-molecular-weight band was observed under these conditions, which was more evident in the membrane-enriched fractions (fraction 9, Fig. 4A and B). This band was evident with anti-TYVD or anti-E4 (Fig. 4A, B, and D) but not with anti-Yuc8 (Fig. 1A) and probably represents a dimer of VP90 (see below). Additional differences between cVP90 and mVP90 were found by partial digestion with trypsin. Treatments of cytosolic fractions with 100 μg/ml of trypsin resulted in the loss of reactivity with anti-E4 antibodies (Fig. 4C and 4D), suggesting complete processing of the VP90 carboxy terminus due to its accessibility to the enzyme. Under these conditions, specific cleavages occurred on cVP90, generating VP34, VP27, and VP25 (Fig. 4E). In contrast, mVP90 was more resistant to trypsin, since products of 40 to 50 kDa recognized by anti-E4 antibodies were still present after treatment with 100 μg/ml of the enzyme (Fig. 4C and 4D), indicating that the carboxy terminus of VP90 present in membrane fractions is protected from trypsin digestion. Products of similar size were also present in the floating fractions when cell fractionation was carried out with trypsin-treated extracts (shown in Fig. 3A and 3B). mVP90 from Z-VAD-FMK-treated cell extracts was even more resistant than the protein from untreated cells, since uncleaved mVP90 was still observed at the highest trypsin concentration used (Fig. 4C and 4D). Thus, the carboxy end of VP90 seems to be exposed on cVP90 but protected on mVP90, likely due to its association with membranes.
FIG. 4.
Membrane-associated VP90 is less susceptible to trypsin digestion. Density gradients of untreated (A) or Z-VAD-FMK-treated (B) infected cells were obtained as described for Fig. 2, and fractions were immunoblotted with anti-TYVD antibodies. Fractions 2 and 9, corresponding to the cytosolic (cyt) and membrane-associated (memb) fractions from these gradients, were treated with the trypsin concentrations and immunoblotted with anti-E4 antibodies, as indicated (C and D). Digestion mixtures were separated in 12.5% (C) and 7.5% (D) polyacrylamide gels to observe products in a wide molecular mass range. Trypsin products of cVP90 from Z-VAD-FMK-treated cells were immunoblotted also with anti-Yuc8 antibodies to ensure that the protein was not totally degraded by trypsin digestion (E). In every panel, viral proteins are marked at the right. A putative oligomer of VP90 is also marked. Numbers at left in panels A to D are molecular masses in kilodaltons.
The astrovirus capsid protein forms oligomers.
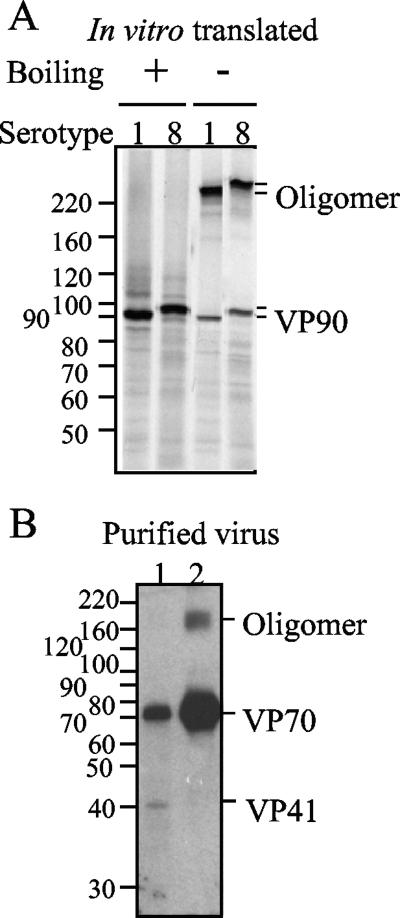
As mentioned in the previous section, the additional band migrating above the 160-kDa molecular mass marker could represent a VP90 homo-oligomer, probably a dimer; alternatively, this band could represent a complex of VP90 with other viral and/or cellular proteins. To discriminate between these possibilities, in vitro-translated VP90s of two different strains belonging to serotypes 1 and 8 were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). A high-molecular-mass band, larger than 220 kDa, was observed in the samples that were not boiled but not in the boiled samples (Fig. 5A), suggesting the formation of a homotrimer (based on its migration rate). The ability of structural proteins to form oligomers was confirmed with antibodies to the recombinant protein E2 (residues 209 to 341 of VP90) (14). These antibodies recognized a putative trimer in virus preparations grown in the absence of trypsin and purified by cesium chloride density gradients (Fig. 5B). The absence of oligomers in the purified viruses with lower density could be explained if these virions were formed by less-stable oligomeric forms of VP70. Thus, these data suggest that in the absence of any other viral protein, the astrovirus VP90 and VP70 capsid precursors form oligomers. The presence of VP90 bands migrating as putative dimers in cell lysates, which were not detected with anti-Yuc8 antibodies, suggests that this protein could be assembled in different high-order structures (either dimers or trimers), which could be present as part of different intermediates in virus assembly.
FIG. 5.
Astrovirus capsid proteins form oligomers. VP90s of HAstV serotypes 1 and 8 were translated in vitro in the presence of [35S]Met and electrophoresed in 7.5% polyacrylamide gels (A). Samples were boiled or not boiled, as indicated, in the presence of reducing agents and analyzed by autoradiography. Two fractions of cesium chloride gradients from HAstV-8-infected-cell lysates, obtained in the absence of trypsin and the presence of fetal bovine serum, were separated by PAGE and analyzed by immunoblotting with anti-E2 antibodies (B). Purified particles were from fractions of densities of around 1.30 (lane 1) or 1.36 (lane 2) g/cm3. The molecular weight markers (weights in thousands) and the viral proteins are marked.
Viral particles with different morphological features are present in infected cells.
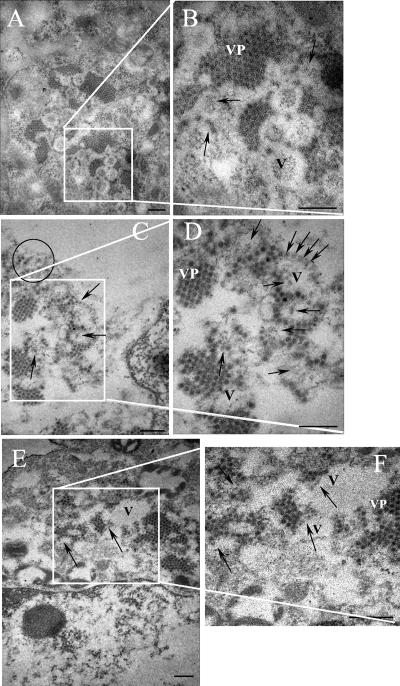
To characterize virus particles that could represent intermediates in morphogenesis, infected cells were analyzed by electron microscopy. Large groups of viral particles (VP in Fig. 6) surrounding “O-ring” structures (V [for vesicles] in Fig. 6) were clearly observed. These O-ring structures, which were not observed in mock-infected cells (not shown), probably correspond to the double-membrane vesicles previously reported by Guix et al. (9, 10), which were also reported to be in close association with virus agglomerates. More dispersed HAstV-8 particles were also detected in the cells' cytoplasm (circled, Fig. 6C). Additional virus-like particles, which could represent subviral particles at different stages of assembly, were observed inside and at the edges of O-ring structures (Fig. 6; more clearly observable in panels C, D, and F and denoted with arrows). Micrographs shown in Fig. 6 show viral particles with distinct features distributed in different locations inside the cell.
FIG. 6.
Electron micrographs of HAstV-8-infected Caco-2 cells. Cells were harvested at 24 hpi and processed for electron microscopy, as described in Materials and Methods. Panels A, C, and E represent three different cells, and panels B, D, and F represent enlargements of the corresponding areas. Astrovirus particles were observed in clusters (VP) and in isolated (circled) forms. Particles that look partially assembled inside or at the edges of vesicles (V) induced during infection are marked with arrows. Bars, 200 nm.
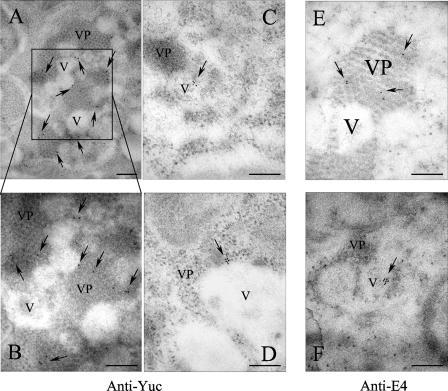
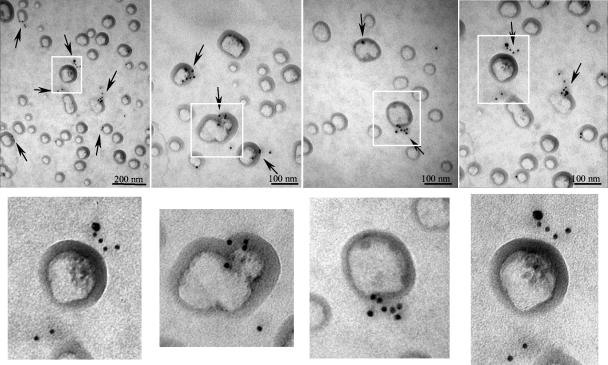
To localize the structural proteins in infected cells and on membrane-enriched fractions of iodixanol gradients, gold-labeled antibodies to VP90 (anti-E4) and to VP70/VP90 (anti-Yuc8) were used for IEM analysis. As expected, anti-Yuc8 antibodies detected the virus clusters, confirming that the structural proteins (VP70 and/or VP90) were present in these particles (Fig. 7A and B). To find out whether VP90 was present in such viral clusters, anti-E4 antibodies were used for IEM in Z-VAD-FMK-treated infected cells (Fig. 7E and F). VP90 was indeed detected in the particle aggregates (Fig. 7E), and of particular interest, the anti-E4 antibodies also interacted with material located in the interior of the vesicles (Fig. 7F), where virus-like particles are observed (Fig. 6B to D and F). Anti-Yuc8 antibodies also detected antigen at the edges of these vesicles (Fig. 7C and D). When the membrane-enriched fractions from the iodixanol gradients were analyzed by IEM with anti-E4 antibodies, specific to VP90, antigen was mainly localized at the periphery of vesicles (Fig. 8). Some of these vesicles also had associated small subviral particles, similar to those observed in the vesicles present in infected cells (Fig. 6, compare with enlarged images in Fig. 8). The protein recognized in these fractions most likely represents mVP90, while cVP90 likely forms part of the viral clusters observed in the cell. Thus, structural proteins of astrovirus were localized in virus particles but also in close association with vesicles, where apparent immature subviral particles were observed. The association of mVP90 with vesicle membranes, together with the presence of putative intermediate particles in these structures, suggests that this protein, but not cVP90, represents the form of VP90 that first assembles into particles.
FIG. 7.
IEM of Caco-2 cells infected with HAstV-8. Cells were infected with the Yuc8 astrovirus strain at a multiplicity of infection of 3 for 24 h, fixed, and processed for IEM with the indicated primary antibodies. Goat anti-rabbit IgG labeled with 10-nm gold particles was used as detection antibody. Cells in panels A to D were processed with anti-Yuc8, and cells in panels E and F were processed with anti-E4 antibodies. The photograph shown in panel E is from Z-VAD-FMK-treated cells. V and VP are used for vesicles and viral particles in clusters, respectively. Arrows indicate positive gold signals. Bars, 200 nm.
FIG. 8.
IEM of membrane-enriched fraction. Fraction 9 of iodixanol gradients was processed for IEM using anti-E4 antibodies and goat anti-rabbit IgG labeled with 10-nm gold particles, as described in Materials and Methods. Images of different preparations of vesicles with particles associated are shown. Arrows indicate positive immunogold signals. Images of vesicles containing associated particles in the upper panels (white rectangles) are enlarged in the lower corresponding panels.
Nonstructural proteins and viral RNA localize in membrane-enriched cell fractions, where mVP90 is present.
RNA replication of most, if not all, viruses with a single-stranded RNA positive-sense genome has been found to occur in complexes that are associated with membranes (1). Particularly, astrovirus nsp1a-derived proteins, which are likely involved in genome replication, have been reported to be associated with the endoplasmic reticulum (10). Based on the hypothesis that mVP90 initiates virus capsid assembly, it is possible that it colocalizes in the same cell compartment as the nonstructural proteins responsible for virus RNA replication. As shown in Fig. 9A and B, the nonstructural proteins that contain the serine protease and the RNA polymerase domains were present in the same membrane-associated fraction where mVP90 and the trypsin-generated VP90 carboxy-terminal product were detected. The additional cellular protein migrating above 30 kDa that was detected with anti-1a3 antibodies (Fig. 9A) is of cellular origin, since it has been detected in mock-infected cell lysates by Western blotting (not shown). Antibodies to the viral protease (serum anti-1a3) recognize antigen inside and at the edges of vesicles in infected cells (Fig. 9C). Although antibodies to the viral RNA polymerase did not work for IEM, the signal obtained by this method with antibodies to the protease and the presence of both nonstructural proteins in the same fraction of density gradients suggest that the viral polymerase could also be associated with the above-mentioned vesicles.
FIG. 9.
Nonstructural proteins and viral RNA colocalize with mVP90. (A and B) Fractions of the iodixanol gradients were immunoblotted with antibodies to the recombinant proteins 1a-3 (A) and 1b-2 (B), as indicated. Viral proteins and molecular weight markers (weights are in thousands) are marked. (C) IEM of Caco-2 cells using anti-1a3 as the primary antibody, as mentioned in Materials and Methods. V and VP are used for vesicles and viral particles, respectively. Arrows indicate positive signals. Bars, 200 nm. (D) Viral RNA was obtained from iodixanol gradient fractions 2 (cyt; lanes 1, 2, 6, and 7) and 9 (memb; lanes 3, 4, 8, and 9) of mock-infected (lanes 1, 3, 6, and 8) and HAstV-infected (lanes 2, 4, 7, and 9) cells or from supernatant of infected cells harvested 24 hpi (lanes 5 and 10). RT was carried out with oligonucleotide Mon244 to detect the negative-sense RNA (lanes 1 to 5, 11, 12, and 15) or Mon245 to detect the positive-sense RNA (lanes 6 to 10, 13, 14, and 16). As controls, Superscript was heated at 85°C for 10 min before the RT reaction (lanes 12 and 14) and no reverse transcriptase (RTase) was added to the RT-PCR (lanes 15 and 16). The molecular weight marker is φX174 digested with HaeIII.
To test our hypothesis that the floating fractions represent structures where RNA replication takes place, experiments directed to detect either the negative or the positive viral RNA strands were carried out. RT was performed with either oligonucleotide Mon244 or Mon245 (19), to produce cDNA from negative or positive RNA, respectively. After inactivation of the reverse transcriptase, the synthesized cDNA was amplified by PCR. Both positive and negative RNA species were found to be present in the cytosolic and in the membrane-enriched fractions (Fig. 9D, lanes 2, 4, 7, and 9). The presence of both viral RNA species in the membrane-enriched fraction supports the idea that replication occurs in this fraction. RNA of both polarities was also found in the more dense fractions; however, this is not surprising given that it is highly likely that negative RNA was released from vesicles during cell disruption. To validate these results, RNAs extracted from the same iodixanol gradient fractions of mock-infected cell lysates and from the supernatant of infected cells that should contain only mature particles with genomic positive RNA were analyzed. As expected, PCR products were amplified neither from mock-infected cell lysates (Fig. 9D, lanes 1, 3, 6, and 8) nor from virus supernatant when the presence of negative RNA was evaluated (Fig. 9D, lane 5). On the other hand, as expected, positive RNA was found in the supernatant of virus-infected cells (Fig. 9D, lane 10). The amplification reactions were specific, since no PCR DNA fragments were obtained when the reverse transcriptase was inactivated with heat before RT (Fig. 9D, lanes 12 and 14) or when no reverse transcriptase was added to the RT-PCR (Fig. 9D, lanes 15 and 16). Based on the intensity of the bands amplified, the positive RNA strand seems to be more abundant than the negative strand.
DISCUSSION
Fractionation by density centrifugation of cytoplasmic extracts from astrovirus-infected cells showed that VP70 was present exclusively in the cytosolic fractions of the cell, while VP90 was found in cytosolic fractions (cVP90), as well as in fractions of lower density (mVP90), presumably due to its association with cellular membranes. After trypsin activation, infectious particles were detected in all fractions of the gradients where either VP70/VP90 or VP90 alone was present. Of particular interest, in the case of the gradients prepared from Z-VAD-FMK-treated cells, infectious particles were present in fractions containing either cVP90 or mVP90, indicating that these two forms of the protein are able to assemble into particles.
The form of VP90 associated with membrane was more resistant to trypsin cleavage than was cVP90, since at 100 μg/ml of trypsin mVP90 yielded protein products of around 40 to 50 kDa that represent partial cleavage products derived from the carboxy-terminal end of the protein, instead of the fully processed VP27 and VP25 polypeptides corresponding to the same region of cVP90 (Fig. 4B to E). These data indicate that the carboxy-terminal end of cVP90 is more accessible to digestion by trypsin than the corresponding region of mVP90 (amino acid residues 672 to 782) and also probably to intracellular cleavage by caspases. The decreased susceptibility of the mVP90 carboxy terminus to trypsin is consistent with the observation that this region of the protein associates with membrane-enriched fractions.
Besides their differential susceptibilities to trypsin, mVP90 and cVP90 appear to differ in their ability to form oligomers. Putative dimers were detected in gradient fractions where mVP90 was present, but they were barely observed in fractions containing cVP90. On the other hand, capsid proteins translated in vitro in the absence of microsomal membranes are able to form trimers, which are also formed by the VP70 protein incorporated into purified particles. These observations suggest that the structural proteins VP90 and VP70 are able to form oligomers (trimers) in the absence of any other viral nonstructural proteins or membranous structures. However, it is possible that some cellular or viral protein can modulate oligomer formation in infected cells, yielding dimers. It is possible that VP90 adopts different conformations depending on its membrane/cytosolic status. In agreement with this observation, it has been reported that the HAstV-1 capsid protein expressed in insect cells is able to form distinct structures that show different morphology and sedimentation rates, although they are very similar by other biochemical criteria (4). In that case, it was suggested that the conformations of the carboxy-terminal domains of the proteins present in such structures were different, based on their reactivities with a monoclonal antibody to VP26.
The apparent flexibility of the astrovirus capsid protein in forming different structures is shared with the capsid protein of other icosahedral viruses, such as calicivirus, whose structural protein is able to form dimers with different structures but with similar biochemical properties (24). Changes in the association of VP90 with membranes and in its oligomerization could represent different steps in the maturation process of astrovirus particles. These putative structural changes in VP90 could facilitate its accessibility to caspase processing to yield VP70, with the consequent release of the virions from the cell. Changes in the oligomerization status and proteolytic processing of capsid proteins have been reported to be important events in the maturation of a variety of viruses, such as phages, herpesviruses, alphavirus, Ebola virus, and hepatitis B virus, among others (23, 28).
Ultrastructural analysis by electron microscopy and IEM revealed different types of viral particles in Yuc8-infected cells, some of which appear to be associated with membranes. Clustered virus particles, such as those previously observed in HAstV-2-infected cells (21), were observed and recognized with antibodies to structural proteins (anti-Yuc8 and anti-E4). In addition, particles that look partially assembled were also observed at sites where mVP90 was localized by IEM, at the edges of cytoplasmic vesicles, and in membrane-enriched cytoplasmic fractions. Subviral particles similar to those found in the present work have also been observed inside vesicles during morphogenesis of alphaviruses (20). These observations, together with the fact that astrovirus nonstructural proteins as well as both positive and negative viral RNA species are present in gradient fractions where mVP90 is detected, support the idea that these particles could represent intermediates of the astrovirus particle morphogenetic process.
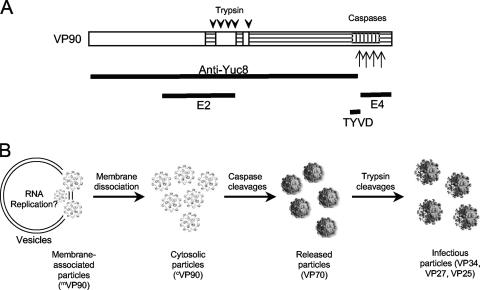
Our results suggest that the form of VP90 that associates with membranes through its carboxy-terminal region could participate in the initial steps of virus morphogenesis. In agreement with this idea, previous observations have demonstrated that infectious progeny of a recombinant astrovirus lacking the carboxy-terminal five amino acid residues of VP90 is drastically reduced (8). Our hypothesis is that mVP90 starts astrovirus assembly in membranes, at the same site where the nonstructural proteins replicate the genome. During this stage, the carboxy terminus of VP90 would not be accessible to caspase cleavage; these particles would then dissociate from membranes generating cVP90-containing particles, whose carboxy terminus would then be accessible to cleavage by caspases. This scenario is supported by the observation that trypsin digestion of total cytoplasmic extracts previously treated with detergent yields protein products similar to those obtained by digestion of cVP90, indicating that the carboxy end of mVP90 becomes accessible to proteases once the protein dissociates from membranes. Thus, particles containing cVP90 are the most likely substrate for caspases to produce the VP70-containing particles, which will be subsequently released from cells. The lower abundance of mVP90 than of cVP90 in the cell could indicate that the transit of virus particles through the membrane-associated stage is a rapid event. Based on the present results and additional published data (14, 15), a hypothetical model for astrovirus morphogenesis is presented in Fig. 10.
FIG. 10.
Model of the astrovirus morphogenesis pathway. (A) Scheme of VP90, in which the conserved (white box), the hypervariable (horizontally hatched), and the acidic-rich (vertically hatched) regions are indicated. Downward arrowheads and upward arrows indicate trypsin and caspase cleavages, respectively. The regions of VP90 comprised in the constructs employed to generate the antibodies used in this work are shown below the scheme of VP90. (B) Model for the astrovirus morphogenesis process (see text).
Our findings suggest that mVP90 has an intracellular distribution different from that of cVP90 and VP70. Since these proteins could form part of particles found at different steps of virus maturation, it is likely that they interact with different viral and/or cellular molecules. Thus, mVP90 would be present in membranes, interacting with molecules involved in assembly; cVP90 would interact with molecules involved in its processing; and VP70 would interact with molecules involved in the release of the virus. It is our intent to test this hypothesis and to identify the potential cellular molecules involved in the steps of virus morphogenesis and cell release.
Acknowledgments
We thank members of our laboratory for enriching discussions of this work.
This work was partially supported by grants 44884-Q from the National Council for Science and Technology-Mexico, IN227602 and IN226106 from DGAPA-UNAM, CRP.LA/MEX03-01 from ICGB-OPS-RELAB, and 55003662 and 55000613 from the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass, D. M., and S. Qiu. 2000. Proteolytic processing of the astrovirus capsid. J. Virol. 74:1810-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belliot, G., H. Laveran, and S. S. Monroe. 1997. Capsid protein composition of reference strains and wild isolates of human astroviruses. Virus Res. 49:49-57. [DOI] [PubMed] [Google Scholar]
- 4.Caballero, S., S. Guix, E. Ribes, A. Bosch, and R. M. Pinto. 2004. Structural requirements of astrovirus virus-like particles assembled in insect cells. J. Virol. 78:13285-13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalton, R. M., E. P. Pastrana, and A. Sanchez-Fauquier. 2003. Vaccinia virus recombinant expressing an 87-kilodalton polyprotein that is sufficient to form astrovirus-like particles. J. Virol. 77:9094-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennehy, P. H., S. M. Nelson, S. Spangenberger, J. S. Noel, S. S. Monroe, and R. I. Glass. 2001. A prospective case-control study of the role of astrovirus in acute diarrhea among hospitalized young children. J. Infect. Dis. 184:10-15. [DOI] [PubMed] [Google Scholar]
- 7.Geigenmuller, U., T. Chew, N. Ginzton, and S. M. Matsui. 2002. Processing of nonstructural protein 1a of human astrovirus. J. Virol. 76:2003-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geigenmuller, U., N. H. Ginzton, and S. M. Matsui. 2002. Studies on intracellular processing of the capsid protein of human astrovirus serotype 1 in infected cells. J. Gen. Virol. 83:1691-1695. [DOI] [PubMed] [Google Scholar]
- 9.Guix, S., A. Bosch, E. Ribes, L. D. Martinez, and R. M. Pinto. 2004. Apoptosis in astrovirus-infected CaCo-2 cells. Virology 319:249-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guix, S., S. Caballero, A. Bosch, and R. M. Pinto. 2004. C-terminal nsP1a protein of human astrovirus colocalizes with the endoplasmic reticulum and viral RNA. J. Virol. 78:13627-13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang, B., S. S. Monroe, E. V. Koonin, S. E. Stine, and R. I. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 90:10539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonassen, C. M., T. O. Jonassen, Y. M. Saif, D. R. Snodgrass, H. Ushijima, M. Shimizu, and B. Grinde. 2001. Comparison of capsid sequences from human and animal astroviruses. J. Gen. Virol. 82:1061-1067. [DOI] [PubMed] [Google Scholar]
- 13.Liu, C. Y., K. L. Shen, S. X. Wang, Y. Y. Liu, and G. T. Zhaori. 2004. Astrovirus infection in young children with diarrhea hospitalized at Beijing Children's Hospital. Chin. Med. J. (Engl. Ed.) 117:353-356. [PubMed] [Google Scholar]
- 14.Mendez, E., T. Fernandez-Luna, S. Lopez, M. Mendez-Toss, and C. F. Arias. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J. Virol. 76:7996-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendez, E., E. Salas-Ocampo, and C. F. Arias. 2004. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J. Virol. 78:8601-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendez, E., M. P. Salas-Ocampo, M. E. Munguia, and C. F. Arias. 2003. Protein products of the open reading frames encoding nonstructural proteins of human astrovirus serotype 8. J. Virol. 77:11378-11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendez-Toss, M., P. Romero-Guido, M. E. Munguia, E. Mendez, and C. F. Arias. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891-2897. [DOI] [PubMed] [Google Scholar]
- 18.Monroe, S. S., B. Jiang, S. E. Stine, M. Koopmans, and R. I. Glass. 1993. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J. Virol. 67:3611-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noel, J. S., T. W. Lee, J. B. Kurtz, R. I. Glass, and S. S. Monroe. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risco, C., J. L. Carrascosa, A. M. Pedregosa, C. D. Humphrey, and A. Sanchez-Fauquier. 1995. Ultrastructure of human astrovirus serotype 2. J. Gen. Virol. 76:2075-2080. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Fauquier, A., A. L. Carrascosa, J. L. Carrascosa, A. Otero, R. I. Glass, J. A. Lopez, C. San Martin, and J. A. Melero. 1994. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology 201:312-320. [DOI] [PubMed] [Google Scholar]
- 23.Steven, A. C., J. B. Heymann, N. Cheng, B. L. Trus, and J. F. Conway. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White, L. J., M. E. Hardy, and M. K. Estes. 1997. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J. Virol. 71:8066-8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willcocks, M. M., A. S. Boxall, and M. J. Carter. 1999. Processing and intracellular location of human astrovirus non-structural proteins. J. Gen. Virol. 80:2607-2611. [DOI] [PubMed] [Google Scholar]
- 26.Willcocks, M. M., T. D. Brown, C. R. Madeley, and M. J. Carter. 1994. The complete sequence of a human astrovirus. J. Gen. Virol. 75:1785-1788. [DOI] [PubMed] [Google Scholar]
- 27.Xia, W., J. Zhang, B. L. Ostaszewski, W. T. Kimberly, P. Seubert, E. H. Koo, J. Shen, and D. J. Selkoe. 1998. Presenilin 1 regulates the processing of beta-amyloid precursor protein C-terminal fragments and the generation of amyloid beta-protein in endoplasmic reticulum and Golgi. Biochemistry 37:16465-16471. [DOI] [PubMed] [Google Scholar]
- 28.Zlotnick, A. 2003. Are weak protein-protein interactions the general rule in capsid assembly? Virology 315:269-274. [DOI] [PubMed] [Google Scholar]