Abstract
Many RNA viruses, which replicate predominantly in the cytoplasm, have nuclear components that contribute to their life cycle or pathogenesis. We investigated the intracellular localization of the multifunctional nonstructural protein 2 (nsP2) in mammalian cells infected with Venezuelan equine encephalitis virus (VEE), an important, naturally emerging zoonotic alphavirus. VEE nsP2 localizes to both the cytoplasm and the nucleus of mammalian cells in the context of infection and also when expressed alone. Through the analysis of a series of enhanced green fluorescent protein fusions, a segment of nsP2 that completely localizes to the nucleus of mammalian cells was identified. Within this region, mutation of the putative nuclear localization signal (NLS) PGKMV diminished, but did not obliterate, the ability of the protein to localize to the nucleus, suggesting that this sequence contributes to the nuclear localization of VEE nsP2. Furthermore, VEE nsP2 specifically interacted with the nuclear import protein karyopherin-α1 but not with karyopherin-α2, -3, or -4, suggesting that karyopherin-α1 transports nsP2 to the nucleus during infection. Additionally, a novel nuclear export signal (NES) was identified, which included residues L526 and L528 of VEE nsP2. Leptomycin B treatment resulted in nuclear accumulation of nsP2, demonstrating that nuclear export of nsP2 is mediated via the CRM1 nuclear export pathway. Disruption of either the NLS or the NES in nsP2 compromised essential viral functions. Taken together, these results establish the bidirectional transport of nsP2 across the nuclear membrane, suggesting that a critical function of nsP2 during infection involves its shuttling between the cytoplasm and the nucleus.
Venezuelan equine encephalitis virus (VEE), a New World member of the genus Alphavirus in the Togaviridae family, is a pathogen that is responsible for significant disease in equines and can cause a wide range of human symptoms, from inapparent infection to acute encephalitis. In nature, VEE is transmitted between susceptible hosts through a mosquito vector and is responsible for periodic outbreaks of widespread human and equine disease throughout the Americas.
The alphaviruses are enveloped viruses with an 11.5-kb genome of single-stranded message-sense RNA. The viral genome, which resembles cellular messages because it contains 5′- and 3′-untranslated regions, a 5′-terminal methylguanylate cap, and a 3′-terminal polyadenylate tail, encodes four nonstructural proteins (nsP1 through 4) and three mature structural proteins (capsid, E2, and E1). Upon infection, this RNA genome is directly translated in the cell to produce the nonstructural polyproteins P1234 and P123, depending on whether there is readthrough at the opal stop codon present between nsP3 and nsP4 in most alphaviruses (10, 32). The polyprotein is processed by nsP2, which contains a protease domain in its carboxy terminus, to form the four individual nonstructural proteins, nsP1 to -4 (22). Early during infection, negative-sense viral RNA is synthesized by two complexes, the P123 intermediate in complex with nsP4 and the P23 intermediate in complex with nsP1 and nsP4 (31, 59, 66). In the late stage of infection, the four mature individual nonstructural proteins constitute a replication complex that synthesizes full-length positive-sense genomic RNA from the negative-sense template and also a subgenomic RNA that is expressed at high levels from the internal 26S promoter (33, 47). The 26S subgenomic RNA encodes the structural proteins and is translated as a polyprotein that is processed into individual structural proteins by both nascent viral proteins and host proteins in the endoplasmic reticulum. Alphavirus replication has been reviewed previously (60).
The alphavirus nsP2 protein is a quintessential multifunctional viral protein, with multiple critical activities in the replication cycle ascribed to it. nsP2 possesses multiple enzymatic activities and is required for a functional alphavirus replication complex. The N terminus of nsP2 contains a nucleoside triphosphate-dependent helicase believed to function as an RNA helicase that unwinds double-stranded RNA that forms during replication (19, 49). This region of nsP2 also has RNA 5′-triphosphatase activity responsible for cleaving the 5′ end of the viral RNA prior to the addition of the methylguanylate cap (63). The C terminus of nsP2 possesses a papain-like proteinase that is responsible for the cleavage of the nonstructural polyprotein precursor (22). This portion of nsP2 is also involved in the regulation of synthesis of the subgenomic 26S RNA and of negative-strand RNA (9, 55, 56, 61). The crystal structure of the multifunctional C terminus of VEE nsP2 was recently elucidated, and it is comprised of two distinct domains (53).
Aside from providing essential functions in viral replication, nsP2 may also have auxiliary functions during infection that affect its outcome. Throughout the course of infection, nsP2 is found in association with ribosomes, and the nsP2 proteins of both New World and Old World alphaviruses specifically interact with ribosomal protein S6, although the function of this interaction with an essential host protein has yet to be determined (36, 45) Additionally, although alphaviruses characteristically cause a lytic infection of mammalian cells, several independent studies have identified mutations in nsP2 that allow the establishment of a persistent alphavirus infection (11, 16, 43, 67). Recent studies have suggested a difference in the nsP2s of Old World and New World alphaviruses, as mature nsP2 proteins of the Old World alphaviruses Sindbis virus and Semliki Forest virus (SFV), but not that of VEE, cause transcription shutoff of the host, which may have a direct function in antagonizing the host's antiviral response (17, 18). Collectively, such observations suggest that there are undefined functions of nsP2 mediated through interactions with the host cell that affect the outcome of viral infection.
Similar to that of most positive-strand RNA viruses, the alphavirus replication cycle occurs in the cytoplasm, and it has been demonstrated that the mammalian cell nucleus is not required for alphavirus replication (12). In spite of this, approximately one-half of nsP2 is found in the nucleus of infected mammalian cells, and nsP2 is the only alphavirus nonstructural protein present in the nucleus (1, 20, 35, 42, 50, 51). Studies performed with SFV, an Old World alphavirus, identified PRRRV as the nuclear localization signal (NLS) in nsP2 and defined the central arginine residue in the sequence as essential for nsP2 nuclear localization. Moreover, SFV nsP2 localizes to the nucleolus, the cellular site of ribosome assembly, and a signal for nucleolar localization is present in the C terminus of SFV nsP2 (50). The NLS is not essential for the viral life cycle, but disruption of the ability of nsP2 to localize to the nucleus compromises the abilities of SFV to inhibit host DNA synthesis and to spread in the brain of an infected mouse (14, 48). To date, the molecular function of nuclear nsP2 remains elusive.
Some components and functions of eukaryotic cells are segregated to the nucleus, which is enveloped by a double-layered membrane. Translocation across this membrane barrier occurs at nuclear pores, through which bidirectional transport of macromolecules occurs. Theoretically, proteins of less than 30 to 40 kDa are able to diffuse passively through the nuclear pore complex (NPC), but larger proteins require active transport (4, 21, 40). The movement of larger proteins through the NPC is governed by importins and exportins, collectively termed karyopherins. The transport of these proteins into and out of the nucleus is an energy-dependent, signal-mediated process. Karyopherins specifically recognize NLSs or nuclear export signals (NESs), which dictate transport into or out of the nucleus, respectively, and are able to traverse the NPC with a cargo protein. The classic NLSs are termed monopartite or bipartite depending on how many stretches of basic amino acids are present; however, there are many examples of NLSs that do not fall into either category (3, 28). The SFV NLS is a monopartite, or simian virus 40 large T antigen-like, NLS. Just as NLSs confer the ability of a protein to be recognized by an importin and actively transported from the cytoplasm into the nucleus, NESs are recognized by exportins and allow proteins to be transported actively from the nucleus to the cytoplasm through the NPC (52). The most commonly identified NESs are short leucine-rich signals, although other hydrophobic residues, such as isoleucine, valine, methionine, and phenylalanine, can contribute to the signal as well (26, 38, 41).
While it once seemed counterintuitive for RNA viruses, in particular positive-strand RNA viruses, to enter the nucleus, there are now many examples of such viruses that possess a viral protein that interacts with the nucleus and/or nuclear trafficking. These include but are not limited to coronaviruses, dengue virus, hepatitis C virus, Newcastle disease virus, influenza virus, tobacco etch virus, poliovirus, Ebola virus, and human immunodeficiency virus (2, 15, 23, 29). By targeting the nucleus or nuclear trafficking, a virus can recruit host factors for viral replication or evade the host antiviral response by inhibiting host gene expression, inhibiting the export of host proteins or RNAs from the nucleus, and inducing cytoplasmic retention of nuclear proteins.
Although the nuclear localization of nsP2 has been examined with Old World alphaviruses, it has not been investigated with VEE, a New World alphavirus and an important pathogen. In this study, we show that VEE nsP2 localizes to both the nucleus and the cytoplasm of mammalian cells during infection. The subcellular distribution of nsP2 can be reproduced by expression of nsP2 in the absence of other viral proteins, which serves as a useful tool to more closely examine signals that direct the localization of nsP2 within the host cell. We show here that nsP2 possesses signals for both its import into and export from the nucleus. Furthermore, nsP2 interacts exclusively with karyopherin-α1, suggesting that the nuclear import of nsP2 is mediated by this cargo protein. Finally, inhibition of the CRM1-dependent nuclear export pathway led to nuclear accumulation of nsP2, indicating that nuclear export of nsP2 is mediated by CRM1. Evidence of nuclear import and export of VEE nsP2 suggests that the localization of nsP2 during alphavirus infection may represent a dynamic cycling of this multifunctional replicase protein between the cytoplasm and the nucleus.
MATERIALS AND METHODS
Cells.
BHK-21 (ATCC; passages 55 to 65), HEK-293 (ATCC; passages 30 to 45), and Vero (ATCC; passages 95 to 110) cells were grown at 37°C under 5% CO2. BHK-21 cells were maintained in alpha minimal essential medium (Gibco) containing 10% donor calf serum, 10% tryptose phosphate broth, 0.29 mg of l-glutamine per ml, 100 U of penicillin per ml, and 0.5 mg of streptomycin per ml. HEK-293 cells were maintained in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal bovine serum, 100 U of penicillin per ml, and 0.5 mg of streptomycin per ml. Vero cells were maintained in minimum essential medium (Eagle) with 2 mM l-glutamine and Earle's buffered salt solution adjusted to contain 1.5 g/liter sodium bicarbonate, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 10% fetal bovine serum.
Infection with VEE and VEE replicon particles (VRP).
The construction of a full-length VEE cDNA clone, pV3000, derived from the Trinidad donkey VEE isolate, has been described previously (7). The preparation of purified VEE stocks has also been described previously (6). Briefly, VEE was produced by electroporating in vitro-transcribed RNA of pV3000 into BHK-21 cells and harvesting virus particles from the supernatant 24 h later. Virus stocks were purified by centrifugation (10,000 × g, 30 min, 4°C) of the supernatant for clarification and then further concentrated by pelleting the clarified virus preparations through 20% (wt/vol) sucrose in low-endotoxin phosphate-buffered saline (PBS) at 72,000 × g for 5 h at 4°C. Plaque assays were performed on BHK-21 cells to determine viral titers, expressed in PFU per ml.
VRP expressing green fluorescent protein (GFP) were packaged using a split helper system as previously described (44). To synthesize VRP, three in vitro-synthesized RNA messages were coelectroporated into BHK-21 cells, including the replicon genome, which contains the four VEE nonstructural genes and the heterologous gene expressed from the viral 26S promoter, and two defective helper RNAs which provide either the wild-type capsid or the wild-type glycoprotein gene but lack the virus-specific packaging signal. The supernatant was harvested at 24 h postelectroporation and subsequently concentrated through a 20% (wt/vol) sucrose cushion. After the particles were resuspended, VRP titers were determined on BHK-21 cells by immunofluorescence and expressed in infectious units per ml.
For VEE and VRP infections, BHK-21 or HEK-293 cells were seeded 18 h prior to infection at 1 × 104 cells per well of an eight-well glass titer slide. BHK-21 cells were seeded on Lab-Tek glass slides (Nunc), but HEK-293 cells are fastidious and required Lab-Tek II CC2 chamber slides (Nunc). For infection, the growth medium was removed, and cell monolayers were infected at a multiplicity of infection of 1 in 80 μl of low-endotoxin PBS supplemented with 1% donor calf serum and Ca2+/Mg2+. After incubation at 37°C for 1 h to allow for adsorption, 200 μl of the appropriate complete growth medium was added back to each well.
Plasmid constructs.
The plasmid used to synthesize the replicon genome for packaging VRP was the wild-type replicon plasmid pV5005, which contains the gene for GFP mutant 2 (5) located directly downstream of the 26S promoter in place of the VEE structural protein genes (34). The K646D mutation, which replaces a lysine residue at VEE nsP2 position 646 with an aspartic acid, was made in the pV5005 plasmid by replacing the AAA codon with GAT.
All enhanced GFP (EGFP) fusions were constructed using the pEGFP-C3 vector (Clontech). To insert heterologous genes into the pEGFP-C3 vector, the XhoI and ApaI sites of the multicloning site were used and a translational stop codon was added twice in tandem immediately following the inserted heterologous sequence. By always using the same restriction sites to insert the heterologous sequence, all fusion proteins had the same 5-amino-acid (aa) linker, YSDLE, between EGFP and the inserted sequence. For all of the constructs containing a sequence from the VEE viral nonstructural protein nsP2 gene, the sequence was PCR amplified from pV5005 (7). The following list of fusion plasmids used in this study describes the heterologous sequence inserted into pEGF-C3 by naming the plasmid and the inserted amino acids: EGFP-1-265nsP2, G1 to L265; EGFP-266-530nsP2, I266 to S530; EGFP-531-794nsP2, G531 to C794; EGFP-506-769nsP2, T506 to R769; EGFP-531-769nsP2, G531 to R769; EGFP-506-794nsP2, T506 to C794; EGFP-511-794nsP2, S511 to C794; EGFP-516-794nsP2, L516 to C794; EGFP-521-794nsP2, V521 to C794; EGFP-526-794nsP2, L526 to C794; EGFP-VEECterm-nsP2, N399 to C794; and EGFP-fullnsP2, G1 to C794. For EGFP-SFVCterm-nsP2, the segment of the SFV nsP2 gene encoding N400 to C798 was PCR amplified from SFV4 (gift from P. Berglund) (36).
The construction of pCI-neo-nsP2 was described previously (36). Briefly, a consensus Kozak sequence and start codon were added to the 5′ end of the full-length nsP2 derived from pV5005 and inserted into the multicloning site of pCI-neo (Promega).
Full-length VEE infectious clones containing either the K646D mutation (pVK646D) or the L526/528A double mutation (pVL526/8A) were constructed with the described substitutions by site-directed mutagenesis of the previously described parental VEE cDNA clone pV3000 (7).
The plasmids expressing karyopherin-α1, -2, -3, and -4 with an N-terminal FLAG epitope tag were the generous gift of P. Palese, and their assembly has been described previously (58, 65). The yellow fluorescent protein (YFP) fragment complement assay was the generous gift of S. Michnick (39). PCR-amplified products encoding full-length nsP2 or aa 531 to 794 of nsP2 were cloned into the BspEI and XbaI sites at the 3′ end of Venus 1, and the karyopherin-α1 coding sequence, PCR amplified from the plasmid described above, was inserted at the 5′ end of Venus 2 at the NotI and ClaI sites.
Site-directed mutagenesis.
To construct all mutations in the EGFP-nsP2 fusion proteins and the full-length virus, complementary sense and antisense primers were designed to contain the desired mutation(s) flanked by 12 to 14 nucleotides (nt) of unmodified sequence. To construct the VEE C-terminal coding sequence containing the SFV substitution encoding RRR, GGCAAAATG was mutated to CGCAGAAGG, and to construct the SFV C-terminal coding sequence containing the VEE substitution encoding GKM, CGACGCAGG was mutated to GGAAAGATG (mutations are shown in italics). To construct the V521A mutant, the GTG codon was mutated to GCG; for the L526A mutant, CTC was mutated to GCC; and for the L528A mutant, CTG was mutated to GCG. Primers containing these mutations were used with a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All mutations were confirmed by sequencing.
To assemble the K646D mutation in pVR21, an overlap PCR mutagenesis method was employed. To mutate the lysine residue to an aspartic acid, overlapping primers containing a GAT substitution for the parental AAA codon were used to generate a PCR product that was inserted between the unique BstEII and BsaBI sites present in pVR21. The identity of this insertion was confirmed by sequencing.
Plasmid DNA transfections.
One day prior to transfection, 1 × 104 HEK-293 cells were seeded at 200 μl per well of an eight-well slide such that the monolayers would be approximately 50% confluent when transfected. For transfections performed in six-well plates, 5 × 105 HEK-293 cells were plated at 2 ml per well 24 h prior to transfection. Transfections were carried out using Fugene 6 (Roche) according to the manufacturer's directions. Briefly, 3 to 6 μl of Fugene 6 was added to 100 μl of Opti-MEM I reduced-serum medium (Gibco) and incubated at room temperature for 5 min. DNA was then added to the mixture at a ratio of approximately 3 μl of transfection reagent to 1 μg of DNA. Following incubation for 15 min at room temperature, 7 μl of the mixture was added to each well of an eight-well slide or 100 μl of the mixture was added to each well of a six-well dish. Cells were fixed in 2% paraformaldehyde or lysed at 36 to 48 h posttransfection.
Indirect immunofluorescence staining.
BHK-21 cells were fixed in 2% paraformaldehyde for 35 min at room temperature, whereas HEK-293 cells were fixed overnight at 4°C, washed three times with PBS, and permeabilized. Cells were permeabilized in acetone at −20°C for 20 min, rehydrated with three PBS washes, and blocked for 1 h in 7.5% bovine serum albumin (BSA) in PBS. After excess blocking solution was removed by three PBS washes, monolayers were stained with anti-histone H1 antibody (Leinco Technologies) at a 1:50 dilution in 0.1% BSA in PBS for 1 h. After PBS washes, cells were stained in 0.1% BSA in PBS for 1 h with the appropriate anti-immunoglobulin G (anti-IgG) secondary antibody directly conjugated to Alexa Fluor 594 or Alexa Fluor 488 (Invitrogen) diluted 1:400. After a final series of PBS washes, cells were mounted under a glass coverslip by using a polyvinyl alcohol-glycerol mounting medium with 2% N-propyl gallate and 0.25% 1,4-diazabicyclo[2,2,2]octane (a gift from the University of North Carolina Microscopy Services Laboratory) to resist photobleaching. For DAPI (4′,6′-diamidino-2-phenylindole) staining, coverslips were mounted using Vectashield containing DAPI (Vector Labs). For nsP2 staining, an antibody recognizing VEE nsP2 (a gift from AlphaVax, Inc.) was used according to a previously described protocol (24).
Nuclear purification.
Nuclei were purified from BHK-21 cells as previously described (36). Briefly, mock- or VRP-infected cells were harvested, resuspended in hypotonic buffer (20 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride), incubated on ice, and gently lysed using Nonidet P-40 at a final concentration of 1%. Nuclei were pelleted at 1,000 × g at 4°C, and the supernatant was collected as the cytoplasmic fraction. The nuclear pellet was resuspended in a 25% solution of Optiprep (Sigma) and purified through layers of 30% and 35% Optiprep. Nuclei were recovered from the Optiprep gradient, washed, pelleted, and lysed with nuclear extraction buffer (20 mM Tris [pH 8.0], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol). After lysis, the insoluble nuclear fraction was removed by centrifugation and the supernatant was collected as the nuclear fraction. Whole-cell lysates were made by using radioimmunoprecipitation assay lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 1% sodium deoxycholate, Complete protease inhibitor cocktail tablets [Roche]) and clarifying the lysates by centrifugation after lysis.
Immunoblotting.
Protein samples were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and subsequently transferred to Immun-blot polyvinylidene difluoride (Bio-Rad) membranes in transfer buffer (48 mM Tris, 39 mM glycine, 10% methanol) at 12 V for 60 min. Membranes were then blocked for 1 h in 5% dry milk in TBST (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.1% Tween 20) and incubated overnight at 4°C with primary antibodies diluted in blocking buffer (anti-GRP-78/BiP [Santa Cruz] at 1:750, anti-nsP2 [a gift from AlphaVax, Inc.] at 1:40,000, anti-EGFP [BD Biosciences] at 1:1,000, and anti-FLAG [Sigma] at 1:1,000). After being probed with primary antibodies, the membranes were washed three times in TBST and probed with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies diluted in blocking buffer (anti-goat-HRP [Novus] at 1:14,000 and anti-rabbit-HRP and anti-mouse-HRP [GE Healthcare] at 1:3,000) for 1 h at room temperature. Membranes were rinsed in a final series of TBST washes before detection of HRP-conjugated secondary antibodies by chemiluminescence using ECL detection reagents (GE Healthcare). Blots were exposed to film and developed.
Immunoprecipitation.
For each immunoprecipitation sample, lysates were combined from two confluent wells of a six-well tissue culture plate at 36 to 48 h posttransfection. Monolayers were rinsed once with PBS, and each well was lysed with 200 μl of 1% NP-40 lysis buffer (170 mM NaCl, 50 mM Tris, 15 mM EDTA, 1% NP-40, Complete protease inhibitor cocktail tablets [Roche]). After a 5-min incubation on ice, lysates were collected in microcentrifuge tubes and cleared by centrifugation for 5 min at 4°C at 12,000 × g. Immunoprecipitation against FLAG was performed using an EZView red protein A anti-FLAG affinity gel (Sigma) according to the manufacturer's directions. Briefly, 400 μl of clarified lysate was incubated with 40 μl of equilibrated beads overnight at 4°C with agitation. Beads were then washed three times in TBS (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) and eluted under acidic conditions (0.1 M glycine-HCl at pH 3.5). The dissociated protein eluates were neutralized with buffer (0.5 M Tris-HCl, pH 7.4, 1.5 M NaCl) before the addition of protein gel loading buffer for immunoblotting.
Microscopy.
Confocal fluorescence microscopic images were acquired with a Carl Zeiss LSM5 Pa confocal laser scanning microscope equipped with a 40× objective and a 63× oil immersion objective and were processed with imaging software. Pinholes were set to acquire optical sections of <0.6 μm, and images were averaged four times. After collecting signals from EGFP- and Alexa Fluor 594-labeled samples, the fluorescent signals were mapped to green and red, respectively. Fluorescence microscope images (showing DAPI staining) were acquired on a Leica Microsystems fluorescence microscope equipped with a 40× objective using Spot software. Unless otherwise indicated, images shown were acquired using a 63× objective and a magnification of ×2.
LMB treatment.
At 24 h posttransfection, cells were treated with leptomycin B (LMB; Sigma) at a final concentration of 20 ng/ml or left untreated in the appropriate growth medium and incubated for 6 h. Cells were then fixed, and the subcellular distributions of EGFP fusion proteins were examined by confocal fluorescence microscopy.
Specific infectivity of RNA.
An assay for specific infectivity was conducted based on previously described methods (8). Briefly, plasmid DNA was linearized by digestion with NotI, and equal amounts of DNA were used as a template to synthesize RNA transcripts, using a T7 mMessage mMachine in vitro transcription kit (Ambion) according to the manufacturer's directions. A portion of each transcription reaction was radiolabeled using 35S-UTP (GE Healthcare). To measure 35S-UTP incorporation, labeled transcripts were bound to glass filters, washed, and quantified using a scintillation counter. After examination and quantification of RNA transcripts, dilutions of equal amounts of RNA were transfected into BHK-21 cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. Under standard plaque assay conditions, plaques were stained and counted at 36 h posttransfection. The cpm/PFU ratio was determined for each clone to determine the specific infectivity of the RNA.
RESULTS
VEE nsP2 localizes to the nucleus and cytoplasm in infected mammalian cells.
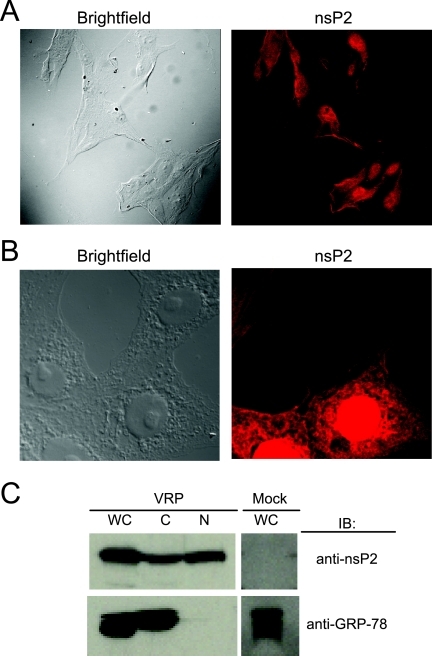
VEE nsP2 is a multifunctional protein that is likely to have as yet undefined activities. As one step in its characterization, its intracellular localization during infection was determined. BHK-21 cells were infected with VEE (V3000), stained for nsP2, and examined by indirect immunofluorescence and confocal fluorescence microscopy. At 8 h postinfection (hpi), VEE nsP2 accumulation was visible in both the cytoplasm and the nuclei of infected cells (Fig. 1A). VRP are nonpropagating viral vectors that contain a genome encoding the four nonstructural proteins but express a heterologous gene from the subgenomic promoter in place of the structural proteins. We confirmed the nuclear and cytoplasmic localization of nsP2 during VRP infection of multiple cell lines, including HEK-293 cells (Fig. 1B).
FIG. 1.
Cytoplasmic and nuclear distribution of nsP2 during infection. (A) During VEE infection, nsP2 is present in both the cytoplasm and the nucleus. BHK-21 cells were infected with VEE and examined by immunofluorescence staining using anti-nsP2 antibody (red) at 8 hpi. Confocal images were acquired using a 40× objective lens. (B) Similar to the case for VEE infection, nsP2 localizes to both the nucleus and the cytoplasm during VRP infection. HEK-293 cells were infected with VRP, and the localization of nsP2 was examined at 8 hpi by immunofluorescence staining using anti-nsP2 antibody (red). Confocal images were acquired using a 63× objective lens. (C) Confirmation of the intracellular distribution of nsP2 by using subcellular fractions. At 8 hpi, VRP-infected BHK-21 cells were fractionated into cytoplasmic (C) and nuclear (N) lysates, or whole-cell lysates were prepared (WC). Samples were examined by immunoblotting (IB) for nsP2 (top). The cytoplasmic marker GRP-78 (bottom) was used to confirm the purity of the nuclear fraction.
To confirm the subcellular localization pattern of nsP2 during infection, nuclear and cytoplasmic fractions of infected cells were examined for nsP2. VRP-infected BHK-21 cells were harvested at 8 hpi and subsequently fractionated to obtain cytoplasmic and nuclear fractions for analysis by immunoblotting for nsP2. VEE nsP2 was detected in both the cytoplasmic and nuclear extracts (Fig. 1C). The purity of the nuclear fraction was confirmed by the absence of GRP-78, an abundant cytoplasmic protein that is indicative of cytoplasmic contamination.
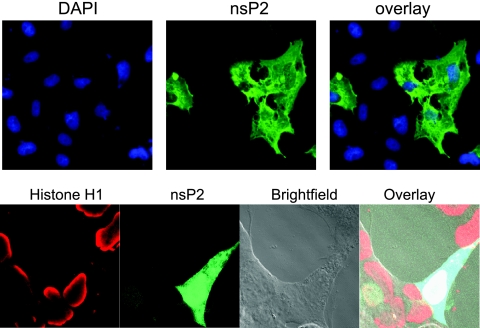
The localization of nsP2 expressed alone was also investigated, as the presence of other viral proteins could affect the localization of nsP2. Full-length nsP2 was expressed from the pCI-neo mammalian expression vector, and 48 h after it was transfected into cells, the distribution of nsP2 was assessed by indirect immunofluorescence staining. When expressed alone, nsP2 was distributed throughout the cytoplasm and the nucleus in a manner indistinguishable from that in VEE- or VRP-infected cells (Fig. 2). Therefore, the localization pattern of nsP2 was not dependent upon the presence of other viral proteins.
FIG. 2.
Cytoplasmic and nuclear distribution of nsP2 expressed alone. Vero (top) or HEK-293 (bottom) cells were transfected with pCI-neo-nsP2, a mammalian expression vector expressing VEE nsP2. Cells were stained for nsP2 (green) at 24 h posttransfection. DAPI (blue) or histone H1 (red) staining was used to visualize the nuclei of cells. Fluorescent images (top) were obtained using a 40× objective, and confocal images (bottom) were obtained using a 63× objective.
Analysis of nsP2 localization using EGFP fusion proteins.
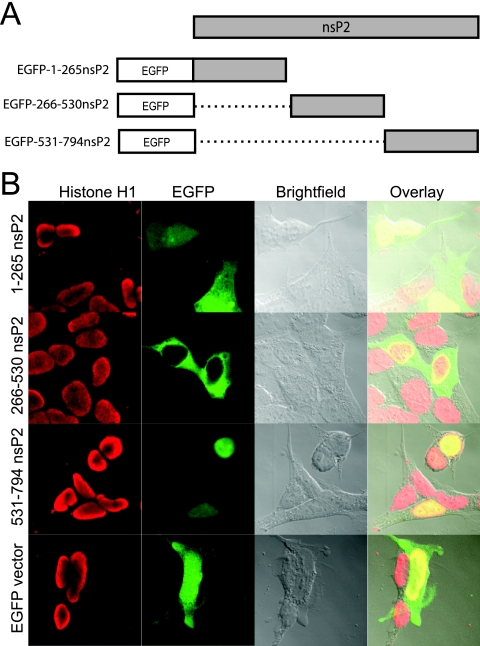
To identify intracellular transport signals contained within nsP2, a series of EGFP fusion proteins was employed. Sequential thirds of nsP2 were constructed so that EGFP was fused in frame to their C termini to form the fusion proteins EGFP-1-265-nsP2, EGFP-266-530nsP2, and EGFP-531-794nsP2 (Fig. 3A). The fusion constructs were transfected individually into cells, and their subcellular localization was analyzed by fluorescence microscopy using a confocal fluorescence microscope. Interestingly, each third of nsP2 displayed a dramatically different subcellular localization. The N-terminal third, EGFP-1-265-nsP2, localized to the nucleus and the cytoplasm; the middle third, EGFP-266-530nsP2, localized entirely to the cytoplasm; and the C-terminal third, EGFP-531-794nsP2, localized entirely to the nucleus (Fig. 3B). The distinct localization pattern of each fusion protein suggested that nsP2 contains multiple signals directing its transport within the cell.
FIG. 3.
Construction and subcellular distribution of EGFP-nsP2 fusion proteins spanning nsP2. (A) Illustration of the EGFP-nsP2 fusion constructs EGFP-1-264nsP2, EGFP-265-529nsP2, and EGFP-530-794nsP2. Dotted lines represent deleted regions of nsP2. There is a 5-aa linker between EGFP and any fused portion of nsP2. (B) Distribution of EGFP-nsP2 fusion proteins, as well as EGFP expressed alone, in HEK-293 cells by fluorescence confocal microscopy.
Identification of a C-terminal NLS in VEE nsP2.
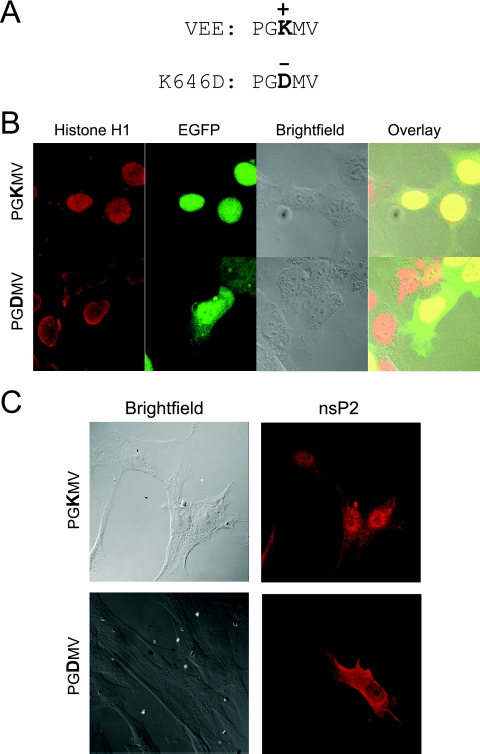
EGFP-531-794nsP2, which was entirely confined to the nucleus, spanned a region of nsP2 that contained a sequence homologous to the SFV NLS. Since the work of Rikkonen and colleagues demonstrated that the central positively charged residue of the signal (PRRRV) was crucial for its function, the equivalent VEE signal (PGKMV) was disrupted with the mutation K646D, where the positively charged lysine was replaced with a negatively charged aspartic acid (Fig. 4A) (50). When the K646D substitution was present in EGFP-531-794nsP2, the completely nuclear localization pattern was disrupted and the product was present in both the cytoplasm and the nucleus, suggesting that the mutation disrupted the function of an NLS (Fig. 4B).
FIG. 4.
Examination of an NLS within the C terminus of VEE nsP2. (A) The K646D mutation disrupts the putative NLS sequence PGKMV. (B) When the K646D mutation is present in EGFP-530-794nsP2, the protein is no longer contained entirely within the nucleus, as shown by confocal microscopy. (C) Confocal microscopy images of cells infected with VRP containing the K646D mutation in nsP2 and examined by indirect immunofluorescence staining for nsP2 also show a diminished amount of nuclear nsP2.
The K646D mutation was also placed in the context of the full-length nsP2 protein in the replicon genome. BHK-21 cells were infected with the K646D mutant VRP, and the localization of nsP2 was examined using indirect immunofluorescence staining. Although the cytoplasm and nucleus stained positive for K646D mutant nsP2, there appeared to be a diminished amount of nsP2 present in the nucleus in comparison to that in cells infected with VRP expressing wild-type nsP2 (Fig. 4C).
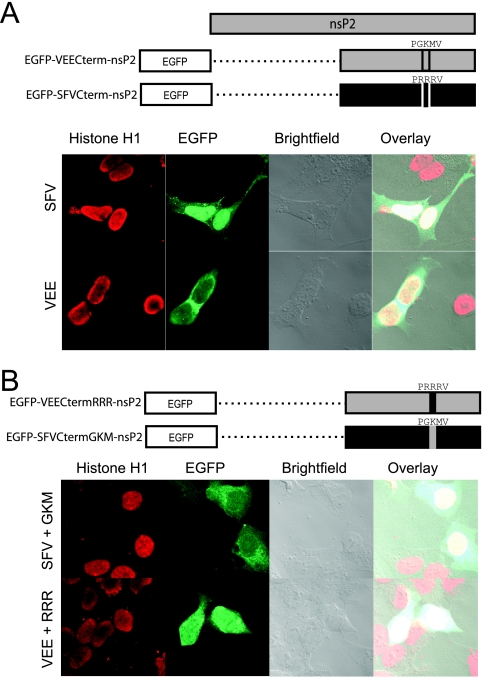
Comparison of the NLSs of SFV and VEE.
We sought to directly compare the PRRRV motif in SFV with the PGKMV motif in VEE. For this comparison, we constructed two EGFP fusion proteins. In the first, EGFP-VEECterm-nsP2, a region spanning the C-terminal half of VEE nsP2 (from N399 to C794) was fused to the C terminus of EGFP. In the second fusion protein, EGFP-SFVCterm-nsP2, a similar region of SFV nsP2 was fused to the C terminus of EGFP (SFV4 sequence from N400 to C798). When these constructs were transfected individually into cells, we observed that the C-terminal half of VEE nsP2 was distributed in the cytoplasm and diffusely throughout the nucleus, whereas the C-terminal half of SFV nsP2 was nearly entirely contained within the nucleus and only diffusely present in the cytoplasm, similar to what has previously been reported for SFV nsP2 (Fig. 5A) (50). To determine how the PGKMV motif in VEE and the PRRRV motif in SFV contributed to the subcellular distributions of the proteins, we constructed two substitution constructs. First, to obtain a VEE construct with the PGKMV motif modified to PRRRV, a substitution in which RRR replaced GKM was made in the VEE C terminus (EGFP-VEECtermRRR-nsP2). Second, the reciprocal SFV construct, with the PRRRV motif modified to PGKMV, was made by substituting GKM for RRR in the SFV C terminus (EGFP-SFVCtermGKM-nsP2). Interestingly, the substitution of the SFV NLS for the endogenous VEE sequence led to a more homogeneous distribution of the fusion protein throughout the cytoplasm and the nucleus (Fig. 5B) than that for the homologous VEE construct (Fig. 5A), but it did not cause a predominantly nuclear localization as observed with the natural SFV nsP2 C terminus (Fig. 5A). Substituting the VEE motif PGKMV for the SFV motif PRRRV (EGFP-SFVCtermGKM-nsP2) produced a distribution of the fusion protein that was identical to that of the VEE C terminus (EGFP-VEECterm-nsP2) (Fig. 5A): the fusion protein was found throughout the cytoplasm and diffusely throughout the nucleus (Fig. 5B). Thus, substituting PGKMV in SFV nsP2 led to a distribution that was similar to that of VEE nsP2, whereas substituting PRRRV in VEE nsP2 led to a distribution that, although appearing to increase the amount of nuclear nsP2, was not predominantly nuclear, like that observed in SFV.
FIG. 5.
Direct comparison of the nuclear localization of VEE and SFV nsP2 proteins. (A) As illustrated, the C-terminal half of either VEE nsP2 or SFV nsP2 was fused to the C terminus of EGFP and used to compare the nuclear localization of VEE and SFV nsP2 proteins. The C terminus of SFV nsP2 resulted in more intense green fluorescence in the nucleus than did that of VEE nsP2. (B) Substitutions were made in the NLS region of each of the SFV and VEE fusion proteins such that each protein contained the NLS of the other. Substitution of the VEE NLS in the C-terminal half of SFV nsP2 reduced the protein's ability to localize to the nucleus, while substitution of the SFV NLS in the C-terminal half of VEE nsP2 did not alter the localization pattern, as shown by confocal microscopy.
The nuclear import protein karyopherin-α1 binds nsP2.
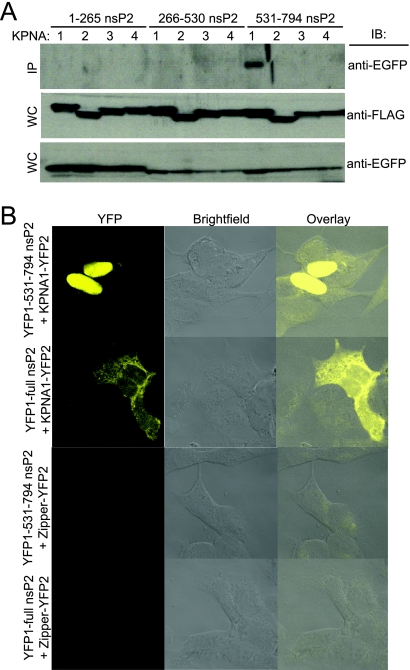
Given the ability of nsP2 to accumulate in the nucleus, we asked whether nsP2 was able to interact with karyopherin-α (also known as importin-α), a part of the nuclear import machinery that is responsible for recognizing cargo bearing an NLS. Plasmids expressing N-terminal FLAG-tagged versions of each of four individual karyopherin homologues (karyopherin-α1, -α2, -α3, and -α4) were cotransfected with plasmids expressing each of the nsP2 thirds. At 24 h posttransfection, cell lysates were prepared and immunoprecipitation was performed using an anti-FLAG monoclonal antibody and subsequently analyzed by Western blotting for EGFP. EGFP-531-794nsP2, which displays an entirely nuclear distribution, coimmunoprecipitated exclusively with karyopherin-α1 (Fig. 6A).
FIG. 6.
VEE nsP2 interacts with karyopherin-α1. (A) As indicated, 293 cells were cotransfected with two plasmids, including one expressing a portion of nsP2 fused to EGFP and one expressing one of the FLAG-tagged karyopherin-α (KPNA) proteins (1, 2, 3, or 4). After 24 h, lysates were made and protein complexes were immunoprecipitated with anti-FLAG antibody. Immunoprecipitated samples (IP) were subsequently examined by immunoblotting for EGFP (top). The expression of all constructs in whole-cell lysates (WC) was confirmed by immunoblotting for FLAG (middle) and EGFP (bottom). (B) The interaction between nsP2 and karyopherin-α1 (KPNA1) was investigated directly in living cells by using a YFP complementation reporter approach. 531-794nsP2 or full-length nsP2 was fused to the N terminus of YFP, and karyopherin-α1 was fused to the C terminus of YFP. HEK-293 cells were cotransfected with either of the nsP2-YFP1 fusion constructs and KPNA1-YFP2, and fluorescence was examined 36 h later by confocal microscopy. While the C-terminal one-third interacted with karyopherin-α1 exclusively in the nucleus, full-length nsP2 interacted with karyopherin-α1 throughout the cytoplasm and the nucleus. No fluorescence signal was detected when either nsP2-YFP1 fusion construct was cotransfected with YFP1-zipper.
In order to examine the interaction between nsP2 and karyopherin-α1 in living cells, we employed a YFP fragment complement assay (also called “split YFP” or “bimolecular fluorescence complementation assay”) (39). This approach is based on the reconstitution of a functional fluorescent complex when the two halves of YFP, each fused to a different protein, are brought into close proximity by the interaction of those proteins. Thus, the technique allows for the visualization of protein-protein interactions within a living cell. We constructed fusion proteins such that the C-terminal portion of YFP was fused to the N-terminal end of karyopherin-α1 (KPNA1-YFP2) and the N-terminal portion of YFP was fused to the C terminus of either 531-794nsP2 or full-length nsP2 (YFP1-531-794nsP2 and YFP1-nsP2, respectively). Complementary fusion plasmids were cotransfected into cells, and fluorescence was examined 36 h later by confocal microscopy. Yellow fluorescence was observed in the nucleus for 531-794nsP2 and throughout both the cytoplasm and the nucleus for full-length nsP2 (Fig. 6B). No fluorescent signal was observed when KPNA1-YFP2 was coexpressed with the leucine zipper-containing control YFP1-zipper or when either YFP1-531-794nsP2 or YFP1-nsP2 was coexpressed with zipper-YFP2.
This direct visualization demonstrates that karyopherin-α1 interacts with nsP2 in living cells. Moreover, it suggests that karyopherin-α1 is involved in the active transport of nsP2 into the nucleus, as both YFP1-531-794nsP2 and YFP1-nsP2 are too large to diffuse passively into the nucleus. The pattern of fluorescent signals produced by the interacting proteins duplicates the pattern of nsP2 distribution for the respective constructs. That is, the 531-794 fragment of nsP2 interacted with karyopherin-α1 exclusively in the nucleus, whereas the interaction of full-length nsP2 with karyopherin-α1 was detected throughout the nucleus and cytoplasm. The diffuse nuclear distribution of the full-length nsP2-KPNA1 interaction is similar to the distribution pattern observed for full-length VEE nsP2 when it is detected through fluorescent signals rather than indirect immunostaining (data not shown).
VEE nsP2 contains a CRM1-dependent NES.
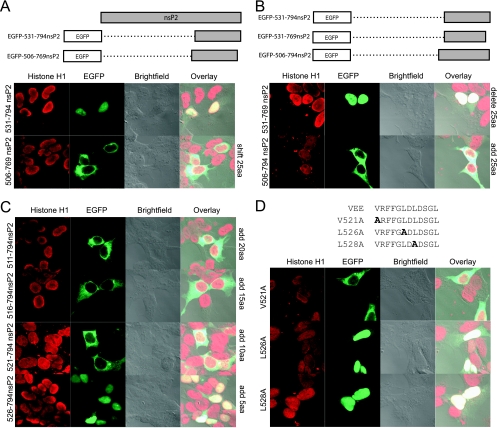
The results described above demonstrate that different regions of nsP2 vary in their subcellular localization when expressed separately, suggesting that VEE nsP2 contains multiple signals directing the localization of the protein during infection. While mapping the C-terminal NLS, we observed that simply shifting the EGFP-531-794nsP2 construct toward the N terminus by 25 residues to form EGFP-506-769nsP2 caused a complete loss of nuclear localization (Fig. 7A). That is, the addition of the endogenous 25 residues just upstream of residue 531 coupled with the deletion of 25 C-terminal residues yielded a protein localized predominantly to the cytoplasm rather than the nucleus. This suggested either that there was an NLS in the deleted 25 residues at the C terminus or that an NES was present in the N-terminal addition.
FIG. 7.
Identification of an NES in nsP2. (A) Confocal microscopy images show that shifting the C-terminal one-third of the nsP2 construct by 25 aa towards the N terminus resulted in an entirely cytoplasmic distribution of the fusion protein. (B) To test for the presence of an NLS in the C-terminal 25 aa, a fusion protein that had the original N-terminal end of the C-terminal third of nsP2 but had a 25-aa deletion at the C terminus was constructed. To test for the presence of an NES in the N-terminal 25-aa addition, an EGFP fusion protein was constructed in which the endogenous upstream 25 aa of nsP2 were added to the original C-terminal third of nsP2. (C) To map the domain required for cytoplasmic localization, the 25 residues present in the N-terminal addition were added in 5-aa increments. (D) The functionally critical residues of the NES were identified by mutating the indicated residues to alanine residues.
To test for the presence of an NLS in the C-terminal 25 residues, we deleted the C-terminal 25 aa from the original construct to form EGFP-531-769nsP2. Removal of the C-terminal 25 residues did not alter the ability of the protein to localize to the nucleus, demonstrating that there was not a required NLS at the extreme C terminus (Fig. 7B). To determine the possible presence of an NES in the upstream 25 residues, a protein was constructed such that the natural 25 residues just upstream of the original N terminus of the C-terminal one-third of nsP2 were added to form EGFP-506-794nsP2. The 25-residue addition rendered the fusion protein completely cytoplasmic, thus indicating that the 25 residues (residues 506 to 531) contained an NES, as they directed an otherwise nuclear protein fragment to the cytoplasm (Fig. 7B). To more carefully map the region within the 25-aa addition that conferred cytoplasmic localization, the residues within that region were sequentially added to the original nsP2 sequence in 5-aa increments. The addition of at least 10 aa was required to render the protein entirely cytoplasmic (Fig. 7C).
The canonical NES is composed of a short leucine-rich region, but other hydrophobic amino acids, such as valine, can be substituted for leucine residues (26, 41). Therefore, alanine residues were substituted for the valine residue and each of the leucine residues (V521A, L526A, and L528A) contained within the 10-aa segment 521-531. Examination of these mutants revealed that L526 and L528 were required for function of the NES, while V521 was not necessary. Sequence analysis of the identified NES across 10 representative alphaviruses revealed that the presence of a hydrophobic residue at VEE position 526 is conserved and that the leucine residue at VEE position 528 is highly conserved (7, 13, 27, 30, 37, 54, 57, 62, 64).
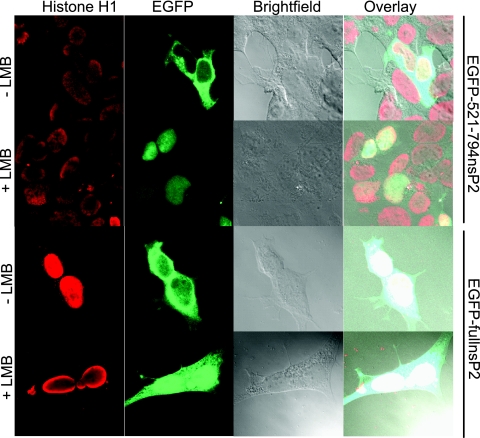
The export of a variety of proteins that contain a leucine-rich NES is mediated by the recognition and binding of the NES by the transport protein CRM1 (also called exportin1) (41). LMB is a small molecule that is able to directly bind CRM1 and consequently block its ability to export proteins (25). To determine if nsP2 export was mediated by CRM1, cells were treated with LMB 24 h after transfection with EGFP-521-794nsP2, and the localization of the fluorescent protein was examined by confocal microscopy. The cytoplasmic localization of EGFP-521-794nsP2 was sensitive to LMB, as LMB treatment resulted in the protein being confined entirely to the nucleus (Fig. 8). Hence, we have identified an NES within nsP2 that functions via the CRM1 transport pathway. To examine the sensitivity of the full-length protein to LMB treatment, we performed a similar experiment using EGFP-fullnsP2, which contains the full-length VEE nsP2 fused to EGFP. While LMB treatment caused the full-length nsP2 fusion protein to accumulate in the nucleus, it was not entirely confined there (Fig. 8). Thus, it is possible that nsP2 contains alternative export signals that function independent of the CRM1 pathway or that all of the nsP2 within a cell does not enter the nucleus within the time examined.
FIG. 8.
Function of the nsP2 NES is dependent on the CRM1 pathway. The sensitivity of the identified NES to LMB was examined. Twenty-four hours after transfection with EGFP-521-794nsP2 or EGFP-fullnsP2, cultures were treated with 20 ng/ml of LMB. At 6 h posttreatment, cells were fixed, stained for histone H1, and examined by confocal microscopy. Nuclear accumulation of the fusion proteins illustrates that the nuclear export of nsP2 is sensitive to LMB treatment.
Effect of disruption of the NLS or NES in nsP2 on viability.
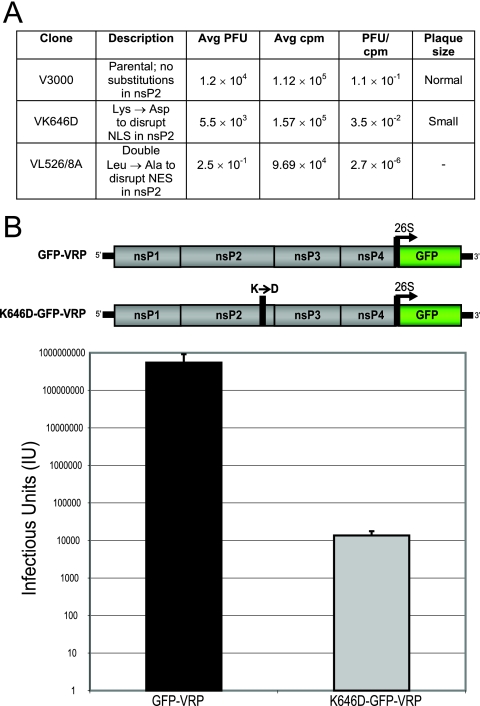
To examine the biological relevance of regions within nsP2 which are critical for the appropriate intracellular distribution of the protein, two nsP2 mutants were constructed independently in the context of the full-length VEE infectious clone (V3000). First, the VK646D mutant contained the K646D substitution in nsP2, which diminished the ability of nsP2 to accumulate in the nucleus. Second, the VL526/528A mutant contained a double leucine-to-alanine substitution, at nsP2 residues 526 and 528, which inhibits the nuclear export of nsP2. To determine the effects of these nsP2 mutations on viability, the mutants were compared to the parental strain through an RNA specific infectivity assay, which examines the ability of full-length viral RNA to produce virus. The VK646D mutant yielded reduced titers and dramatically reduced plaque sizes, while the VL526/528A mutant entirely abrogated the formation of plaques (Fig. 9A). This suggests that the ability of nsP2 to reach the nucleus improves the viability of VEE, while the ability of nsP2 to exit the nucleus is requisite for VEE viability.
FIG. 9.
Examination of the consequences of disruption of the nsP2 NLS and NES. (A) RNA specific infectivities were compared between the full-length VEE (V3000) and two mutants derived from it, one of which contained a mutation to disrupt an NLS within nsP2 (VK646D) and the other of which contained a double mutation to disrupt an NES within nsP2 (VL526/528A). Data shown are representative of one experiment out of three. Average numbers were calculated for quadruplicate samples. (B) GFP-VRP were packaged from a replicon genome expressing GFP, and K646D-GFP-VRP were packaged from a replicon genome expressing GFP and containing the K646D mutation within nsP2 (top). Infectious units were calculated by counting green fluorescent cells yielded from infecting BHK-21 cells with serially diluted preparations of VRP. Data shown represent 10 individual VRP packaging experiments (bottom).
To further examine the consequences of the K646D substitution on the function of full-length nsP2, a replicon genome expressing GFP but containing the nsP2 K646D mutation was constructed. When K646D-GFP-VRP were synthesized, the titers of VRP packaged were reproducibly reduced 10,000-fold compared to those of GFP-VRP, which express GFP but contain unaltered nsP2 (Fig. 9B; see image of nsP2 showing localization in Fig. 4C). The reduced titer of VRP when the K646D substitution is present, along with the small-plaque phenotype of the VK646D mutant, indicates that nuclear localization of nsP2 is needed for optimal alphavirus replication.
DISCUSSION
Although the alphavirus protein nsP2 already has multiple critical functions ascribed to it, some of its functions remain elusive. We sought to determine the subcellular localization of nsP2 from the New World alphavirus VEE during infection as a first step in elucidating some of these functions. Similar to reports of nsP2 from Old World alphaviruses (1, 20, 35, 42, 50, 51), VEE nsP2 localizes to both the cytoplasm and the nucleus in mammalian cells. Interestingly, VEE nsP2 contains signals that direct both its entry into and exit from the nucleus. We found that VEE nsP2 associates with karyopherin-α1, a cellular transport protein capable of translocating the protein into the nucleus. Additionally, we found that VEE nsP2 residues L526 and L528 are necessary for the function of an NES. Blocking CRM1-mediated nuclear export hindered the export of nsP2, suggesting that nsP2 is exported via the CRM1 pathway. Finally, disruption of the NLS and NES independently hindered the normal function of nsP2. Taken together, these data suggest that during the course of infection, there is a cycling of nsP2 in and out of the nucleus that contributes to optimal viral function.
To elucidate the signals that govern the subcellular localization of nsP2, we fused sequential thirds of nsP2 to the C terminus of EGFP. The distribution of each fusion protein fragment was dramatically different, which suggested that the intact nsP2 protein contains multiple signals directing its transport into and out of the nucleus. We have been unable to identify any such transport signal in the N-terminal third of nsP2, which led to a cytoplasmic and nuclear distribution of the fusion protein indistinguishable from the distribution of EGFP alone. Studies that deleted 85 residues contained in the N-terminal third of SFV nsP2 demonstrated that this region was necessary but not sufficient for localization of SFV nsP2 to the nucleus. The N terminus of VEE nsP2 does not seem to be required for nuclear localization of nsP2, although many proteins that localize to both the cytoplasm and the nucleus contain redundant signals directing their subcellular localization, which complicates their identification. Alternatively, incorrect folding of the fusion protein may result in a misrepresentation of the true conformation of the protein, consequently resulting in its incorrect subcellular compartmentalization.
The C-terminal third of VEE nsP2 was able to completely confine EGFP to the nucleus, demonstrating that this region contained an NLS. This VEE nsP2 fragment contains the region homologous to the defined NLS in SFV nsP2. The SFV nsP2 NLS is considered to be P648RRRV, a monopartite NLS which contains three positively charged arginine residues and in which mutation of the central arginine residue renders the protein cytoplasmic (50). Whereas other alphaviruses conserve the positive charge of at least two residues in the canonical NLS, VEE has only a single positively charged residue, the central lysine in the VEE sequence PGKMV. Mutation of the positively charged lysine to a negatively charged aspartic acid diminished but did not obliterate the ability of nsP2 to localize to the nucleus, suggesting that the sequence contributes to the nuclear localization of nsP2 but does not account for it entirely. Although the NLS in alphavirus nsP2 has some features of a classic NLS, fusing the SFV nsP2 NLS to cytoplasmic proteins fails to deliver them to the nucleus (50). Taken together, these data suggest that the alphavirus nsP2 NLS is relatively weak and that the environment in which it is contained is critical to its function.
Fusing the middle third of nsP2 to EGFP rendered the protein completely cytoplasmic. This nsP2 fragment contained residues L526 and L528, which we identified as being critical for the function of an NES. The addition of the 10-aa sequence VRFFGL526DL528DS to the completely nucleus-localizing EGFP-531-794nsP2 construct rendered the protein cytoplasmic, and we identified the leucine residues at VEE nsP2 positions 526 and 528 as necessary for the function of the NES. The VEE nsP2 NES appears to be potent, as it was able to completely localize an otherwise entirely nuclear protein fragment to the cytoplasm. An examination of the analogous sequence in other alphaviruses revealed that L528 is well conserved but L526 is not; however, at position 526 of other alphaviruses, a hydrophobic residue is conserved (M, F, or V), which is tolerated within NESs (26, 41). At VEE nsP2 position 532, there is a leucine residue that is conserved among the alphaviruses and likely functions as part of the NES. However, L532 was not included in EGFP-266-530nsP2, an entirely cytoplasmic protein fragment, suggesting that L532 is not required for NES function or that there is NES redundancy in that portion of nsP2.
We found that VEE nsP2 interacts in vivo with karyopherin-α1, a transport protein that recognizes and directly binds to NLSs. Karyopherin-α concurrently binds importin-β1, which allows the complex to traverse the nuclear membrane. Once inside the nucleus, the NLS-bearing cargo protein is released. Moreover, we found that the function of the VEE nsP2 NES is at least partially blocked by LMB, an inhibitor of the export transport protein CRM1. Analogous to nuclear import, CRM1 recognizes and directly binds a leucine-rich NES and transports cargo across the nuclear membrane, where it is released into the cytoplasm. Taken together, our data suggest a model in which VEE nsP2 cycles between the nucleus and the cytoplasm, being transported into the nucleus via karyopherin-α1, released, and later exported out of the nucleus via CRM1, where it is released again into the cytoplasm.
Nuclear transport of nsP2 appears to be critical for alphaviruses to realize their full replicative potential. The K646D mutation in VEE nsP2 not only disrupted the nuclear localization of nsP2 but also diminished the specific infectivity of the infectious genome and the ability to package VRP. Likewise, the neurovirulence of SFV is compromised in a mutant in which nsP2 nuclear localization is inhibited (14). Evidence of active export of nsP2 from the nucleus suggests that cycling of nsP2 between the nucleus and cytoplasm is also a key element of nsP2 function. This activity is critical, as a mutation which compromised the nsP2 NES function was lethal.
Hypothetically, the cycling of nsP2 through the nucleus may act at several levels, which are not mutually exclusive, to influence viral replication and the host cell response. First, aside from being an essential component of the replication complex in the cytoplasm, nsP2 may also have a specific nuclear function, such as impeding a normal host activity that occurs in the nucleus. Second, the bidirectional nuclear transport of nsP2 may establish an optimal distribution of nsP2 in both the cytoplasm and the nucleus for it to function efficiently in both compartments. Third, the process of transporting nsP2 into and out of the nucleus could impede the routine movement of essential cellular components through the nuclear pore, in either direction, by monopolizing critical cellular transport systems and consequently precluding normal transport of cellular factors. For example, cycling of VEE nsP2 may be analogous to the transport of the Ebola virus protein VP24 to the nucleus by karyopherin-α1, which precludes STAT1 from utilizing karyopherin-α1 and translocating into the nucleus, thus in turn likely blocking the host interferon response (46). Alternatively, because all of the karyopherin-α transport proteins must bind an importin-β1 protein to transport cargo into the nucleus, it is possible that the use of karyopherin-α1 by nsP2 also depletes the available population of importin-β1 molecules and affects the nuclear transport of proteins that rely on other karyopherin-α proteins. Likewise, nsP2 may occupy sufficient CRM1 to inhibit normal nuclear export. Further studies regarding the biochemistry of active nsP2 nuclear cycling will illuminate its function in virus replication and its effect on the host cell.
Acknowledgments
Confocal microscopy was conducted at the Microscopy Services Laboratory, a part of the Department of Pathology and Laboratory Medicine at UNC-CH. We are grateful to R. C. Bagnell for his expertise and enthusiasm in teaching the art of microscopy. We acknowledge the generosity of AlphaVax, Inc., for providing the VEE nsP2 antibody. We thank J. A. Rivera-Perez for sharing EGFP fusion protein reagents with us. We are grateful to P. Berglund, P. Palese, and S. Michnick for generously providing SFV4 nsP2, FLAG-tagged karyopherin-α plasmids, and the split YFP system, respectively. We appreciate the assistance of M. Frieman, who was instrumental in the acquisition of reagents. We are grateful to K. Young and R. S. Shabman for technical advice and assistance.
This work was supported by NIH grant AI51990.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1991. Solubilization and immunoprecipitation of alphavirus replication complexes. J. Virol. 65:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrington, J. C., and W. G. Dougherty. 1987. Small nuclear inclusion protein encoded by a plant potyvirus genome is a protease. J. Virol. 61:2540-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christophe, D., C. Christophe-Hobertus, and B. Pichon. 2000. Nuclear targeting of proteins: how many different signals? Cell Signal. 12:337-341. [DOI] [PubMed] [Google Scholar]
- 4.Corbett, A. H., and P. A. Silver. 1997. Nucleocytoplasmic transport of macromolecules. Microbiol. Mol. Biol. Rev. 61:193-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 6.Davis, N. L., K. W. Brown, and R. E. Johnston. 1996. A viral vaccine vector that expresses foreign genes in lymph nodes and protects against mucosal challenge. J. Virol. 70:3781-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, N. L., N. Powell, G. F. Greenwald, L. V. Willis, B. J. Johnson, J. F. Smith, and R. E. Johnston. 1991. Attenuating mutations in the E2 glycoprotein gene of Venezuelan equine encephalitis virus: construction of single and multiple mutants in a full-length cDNA clone. Virology 183:20-31. [DOI] [PubMed] [Google Scholar]
- 8.Davis, N. L., L. V. Willis, J. F. Smith, and R. E. Johnston. 1989. In vitro synthesis of infectious Venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology 171:189-204. [DOI] [PubMed] [Google Scholar]
- 9.Dé, I., S. G. Sawicki, and D. L. Sawicki. 1996. Sindbis virus RNA-negative mutants that fail to convert from minus-strand to plus-strand synthesis: role of the nsP2 protein. J. Virol. 70:2706-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot, R. J., W. R. Hardy, Y. Shirako, and J. H. Strauss. 1990. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 9:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dryga, S. A., O. A. Dryga, and S. Schlesinger. 1997. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology 228:74-83. [DOI] [PubMed] [Google Scholar]
- 12.Erwin, C., and D. T. Brown. 1983. Requirement of cell nucleus for Sindbis virus replication in cultured Aedes albopictus cells. J. Virol. 45:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faragher, S. G., A. D. Meek, C. M. Rice, and L. Dalgarno. 1988. Genome sequences of a mouse-avirulent and a mouse-virulent strain of Ross River virus. Virology 163:509-526. [DOI] [PubMed] [Google Scholar]
- 14.Fazakerley, J. K., A. Boyd, M. L. Mikkola, and L. Kaariainen. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontoura, B. M., P. A. Faria, and D. R. Nussenzveig. 2005. Viral interactions with the nuclear transport machinery: discovering and disrupting pathways. IUBMB Life 57:65-72. [DOI] [PubMed] [Google Scholar]
- 16.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Pragai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garmashova, N., R. Gorchakov, E. Volkova, S. Paessler, E. Frolova, and I. Frolov. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 81:2472-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez de Cedron, M., N. Ehsani, M. L. Mikkola, J. A. Garcia, and L. Kaariainen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448:19-22. [DOI] [PubMed] [Google Scholar]
- 20.Gorchakov, R., E. Frolova, and I. Frolov. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlich, D. 1997. Nuclear protein import. Curr. Opin. Cell Biol. 9:412-419. [DOI] [PubMed] [Google Scholar]
- 22.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiscox, J. A. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamrud, K. I., M. Custer, J. M. Dudek, G. Owens, K. D. Alterson, J. S. Lee, J. L. Groebner, and J. F. Smith. 2007. Alphavirus replicon approach to promoterless analysis of IRES elements. Virology 360:376-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 26.la Cour, T., R. Gupta, K. Rapacki, K. Skriver, F. M. Poulsen, and S. Brunak. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 31:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanciotti, R. S., M. L. Ludwig, E. B. Rwaguma, J. J. Lutwama, T. M. Kram, N. Karabatsos, B. C. Cropp, and B. R. Miller. 1998. Emergence of epidemic O'nyong-nyong fever in Uganda after a 35-year absence: genetic characterization of the virus. Virology 252:258-268. [DOI] [PubMed] [Google Scholar]
- 28.Lange, A., R. E. Mills, C. J. Lange, M. Stewart, S. E. Devine, and A. H. Corbett. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langenberg, W. G., and L. Zhang. 1997. Immunocytology shows the presence of tobacco etch virus P3 protein in nuclear inclusions. J. Struct. Biol. 118:243-247. [DOI] [PubMed] [Google Scholar]
- 30.Lee, E., C. Stocks, P. Lobigs, A. Hislop, J. Straub, I. Marshall, R. Weir, and L. Dalgarno. 1997. Nucleotide sequence of the Barmah Forest virus genome. Virology 227:509-514. [DOI] [PubMed] [Google Scholar]
- 31.Lemm, J. A., A. Bergqvist, C. M. Read, and C. M. Rice. 1998. Template-dependent initiation of Sindbis virus RNA replication in vitro. J. Virol. 72:6546-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, G., and C. M. Rice. 1993. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J. Virol. 67:5062-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (New York) 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 34.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, M. L., and D. T. Brown. 1993. The distribution of Sindbis virus proteins in mosquito cells as determined by immunofluorescence and immunoelectron microscopy. J. Gen. Virol. 74:293-298. [DOI] [PubMed] [Google Scholar]
- 36.Montgomery, S. A., P. Berglund, C. W. Beard, and R. E. Johnston. 2006. Ribosomal protein S6 associates with alphavirus nonstructural protein 2 and mediates expression from alphavirus messages. J. Virol. 80:7729-7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myles, K. M., D. J. Pierro, and K. E. Olson. 2003. Deletions in the putative cell receptor-binding domain of Sindbis virus strain MRE16 E2 glycoprotein reduce midgut infectivity in Aedes aegypti. J. Virol. 77:8872-8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 39.Nyfeler, B., S. W. Michnick, and H. P. Hauri. 2005. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. USA 102:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohno, M., M. Fornerod, and I. W. Mattaj. 1998. Nucleocytoplasmic transport: the last 200 nanometers. Cell 92:327-336. [DOI] [PubMed] [Google Scholar]
- 41.Pemberton, L. F., and B. M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187-198. [DOI] [PubMed] [Google Scholar]
- 42.Peranen, J., M. Rikkonen, P. Liljestrom, and L. Kaariainen. 1990. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 64:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perri, S., D. A. Driver, J. P. Gardner, S. Sherrill, B. A. Belli, T. W. Dubensky, Jr., and J. M. Polo. 2000. Replicon vectors derived from Sindbis virus and Semliki Forest virus that establish persistent replication in host cells. J. Virol. 74:9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239:389-401. [DOI] [PubMed] [Google Scholar]
- 45.Ranki, M., I. Ulmanen, and L. Kaariainen. 1979. Semliki Forest virus-specific nonstructural protein is associated with ribosomes. FEBS Lett. 108:299-302. [DOI] [PubMed] [Google Scholar]
- 46.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice, C. M. 1992. Examples of expression systems based on animal RNA viruses: alphaviruses and influenza virus. Curr. Opin. Biotechnol. 3:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rikkonen, M. 1996. Functional significance of the nuclear-targeting and NTP-binding motifs of Semliki Forest virus nonstructural protein nsP2. Virology 218:352-361. [DOI] [PubMed] [Google Scholar]
- 49.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 68:5804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rikkonen, M., J. Peranen, and L. Kaariainen. 1992. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 189:462-473. [DOI] [PubMed] [Google Scholar]
- 51.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. Nuclear targeting of Semliki Forest virus nsP2. Arch. Virol. 9(Suppl.):369-377. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez, M. S., C. Dargemont, and F. Stutz. 2004. Nuclear export of RNA. Biol. Cell 96:639-655. [DOI] [PubMed] [Google Scholar]
- 53.Russo, A. T., M. A. White, and S. J. Watowich. 2006. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 14:1449-1458. [DOI] [PubMed] [Google Scholar]
- 54.Salonen, A., L. Vasiljeva, A. Merits, J. Magden, E. Jokitalo, and L. Kaariainen. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 77:1691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawicki, D. L., and S. G. Sawicki. 1998. Role of the nonstructural polyproteins in alphavirus RNA synthesis. Adv. Exp. Med. Biol. 440:187-198. [DOI] [PubMed] [Google Scholar]
- 56.Sawicki, D. L., and S. G. Sawicki. 1993. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J. Virol. 67:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuffenecker, I., I. Iteman, A. Michault, S. Murri, L. Frangeul, M. C. Vaney, R. Lavenir, N. Pardigon, J. M. Reynes, F. Pettinelli, L. Biscornet, L. Diancourt, S. Michel, S. Duquerroy, G. Guigon, M. P. Frenkiel, A. C. Brehin, N. Cubito, P. Despres, F. Kunst, F. A. Rey, H. Zeller, and S. Brisse. 2006. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3:e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw, M. L., W. B. Cardenas, D. Zamarin, P. Palese, and C. F. Basler. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and Toll-like receptor 3-triggered signaling pathways. J. Virol. 79:6078-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suopanki, J., D. L. Sawicki, S. G. Sawicki, and L. Kaariainen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Gen. Virol. 79:309-319. [DOI] [PubMed] [Google Scholar]
- 62.Uryvaev, L. V., V. E. Volckhov, V. P. Iuferov, E. I. Samokhvalov, A. Lebedev, P. F. Safronov, and S. V. Netesov. 1994. Primary structure of proteins of the nsP2 and nsP3 polymerase complex confirm the recombinant nature of western encephalitis virus. Dokl. Akad. Nauk SSSR 335:813-818. [PubMed] [Google Scholar]
- 63.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kaariainen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 64.Wang, E., S. Paessler, P. V. Aguilar, A. S. Carrara, H. Ni, I. P. Greene, and S. C. Weaver. 2006. Reverse transcription-PCR-enzyme-linked immunosorbent assay for rapid detection and differentiation of alphavirus infections. J. Clin. Microbiol. 44:4000-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, P., P. Palese, and R. E. O'Neill. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71:1850-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Y. F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 68:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiss, B., R. Rosenthal, and S. Schlesinger. 1980. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J. Virol. 33:463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]