Abstract
The henipaviruses, Nipah virus (NiV) and Hendra virus (HeV), are lethal emerging paramyxoviruses. EphrinB2 and ephrinB3 have been identified as receptors for henipavirus entry. NiV and HeV share similar cellular tropisms and likely use an identical receptor set, although a quantitative comparison of receptor usage by NiV and HeV has not been reported. Here we show that (i) soluble NiV attachment protein G (sNiV-G) bound to cell surface-expressed ephrinB3 with a 30-fold higher affinity than that of sHeV-G, (ii) NiV envelope pseudotyped reporter virus (NiVpp) entered ephrinB3-expressing cells much more efficiently than did HeV pseudotyped particles (HeVpp), and (iii) NiVpp but not HeVpp entry was inhibited efficiently by soluble ephrinB3. These data underscore the finding that NiV uses ephrinB3 more efficiently than does HeV. Henipavirus G chimeric protein analysis implicated residue 507 in the G ectodomain in efficient ephrinB3 usage. Curiously, alternative versions of published HeV-G sequences show variations at residue 507 that can clearly affect ephrinB3 but not ephrinB2 usage. We further defined surrounding mutations (W504A and E505A) that diminished ephrinB3-dependent binding and viral entry without compromising ephrinB2 receptor usage and another mutation (E533Q) that abrogated both ephrinB2 and -B3 usage. Our results suggest that ephrinB2 and -B3 binding determinants on henipavirus G are distinct and dissociable. Global expression analysis showed that ephrinB3, but not ephrinB2, is expressed in the brain stem. Thus, ephrinB3-mediated viral entry and pathology may underlie the severe brain stem neuronal dysfunction seen in fatal Nipah viral encephalitis. Characterizing the determinants of ephrinB2 versus -B3 usage will further our understanding of henipavirus pathogenesis.
Nipah virus (NiV) and Hendra virus (HeV) are closely related zoonotic viruses comprising the newly defined Henipavirus genus of the Paramyxoviridae family (reviewed in references 16 and 47). HeV emerged in eastern Australia in 1994, causing severe respiratory disease in horses and humans (36, 41). Four years later, NiV emerged in peninsular Malaysia, resulting in predominantly nonlethal respiratory disease in pigs yet causing over 100 human deaths, mainly due to acute encephalitis (12, 13). Both of these outbreaks are believed to have occurred from zoonotic transmission between bats of the Pteropus species, the presumptive natural host (21, 49), and their amplifying animal host before subsequent human infections arose. However, in more recent NiV outbreaks in Bangladesh, NiV may have been transmitted directly from bats to humans, resulting in mortality rates of >70% (26, 34). Furthermore, there is epidemiological evidence for human-to-human transmission in the latter outbreaks (2). NiV and HeV are biosafety level 4 pathogens and are designated category C priority agents in the NIAID Biodefense Research Agenda.
Henipaviruses have a broad host range, including dogs, cats, guinea pigs, and hamsters in addition to the previously mentioned humans, horses, pigs, and bats. Recently, ephrinB2 was identified as the cellular receptor for both HeV and NiV (6, 37). Ephrins are ligands for the Eph family of receptor tyrosine kinases, and the Eph-Ephrin signaling axis directs cell-to-cell attraction and repulsion cues important for developmental processes, such as angiogenesis and axonal guidance (10, 39, 40). Ephrin ligands are evolutionarily conserved from nematodes to vertebrates (40). In fact, publicly available genomic drafts of the horse, pig, dog, and cat genomes (www.ncbi.nlm.nih.gov/Genomes) suggest that their ephrinB2 genes are extremely homologous to the human gene (96 to 98%), providing strong evidence that ephrinB2 serves as the cross-species receptor.
EphrinB3, a homolog of ephrinB2, was also recently described as an alternative receptor for NiV (38). The identification of these ephrin receptors has not only provided an explanation for the wide host range of henipaviruses but also shed light on several pathological features of the diseases caused by these viruses. EphrinB2 is specifically expressed on endothelial cells, neurons, and the smooth muscle cells surrounding arterioles (18, 42), a distribution pattern remarkably concordant with the known cellular tropism of NiV and HeV (25, 48). EphrinB3 is expressed in the central nervous system, in distinct regions such as the corpus callosum (31) and spinal cord (3), where ephrinB2 is lacking but where NiV-induced pathology has been described (32, 33). A more comprehensive global expression study from the Allen Brain Institute (www.brainatlas.org) indicates that ephrinB3, but not ephrinB2, is expressed in the several regions of the brain stem where lethal NiV disease is manifested (19, 28). Therefore, understanding the determinants of ephrinB2 versus ephrinB3 receptor usage by henipaviruses may shed light on the severe neurological dysfunction that occurs during fatal Nipah viral encephalitis.
Viral entry and membrane fusion of henipaviruses are mediated through the concerted action of two envelope glycoproteins, the attachment protein (G) and the fusion protein (F). Before the discovery of the ephrin receptors, functional analysis of F- and G-mediated fusion was used to show that NiV and HeV have identical patterns of fusion in permissive and nonpermissive cell types, which suggested that they share the same receptor set (9). However, no study to date has been able to quantitatively compare the efficiencies of fusion or viral entry between NiV and HeV. Although NiV and HeV may share the same receptor set, the efficiencies of receptor usage between the henipaviruses may be different. HeV has been responsible for a total of four human cases, with three patients developing respiratory infection, while NiV has caused several hundred human cases, with the majority of patients developing neurological disease. A comparison between NiV and HeV use of human ephrin receptors can potentially explain their variant pathological profiles or even their efficiencies of transmission to humans. In this study, we performed a quantitative comparison of ephrinB2 versus ephrinB3 usage, using cell surface binding experiments and pseudotyped viral entry assays. In addition, we also identified residues involved in distinct ephrinB3 usage that are dissociable from ephrinB2 usage.
MATERIALS AND METHODS
Cells and culture conditions.
CHOpgsA745 is a mutant cell line derived from Chinese hamster ovary (CHO) cells that lack the endogenous expression of heparin sulfate proteoglycans (17). CHOpgsA745 cells were specifically chosen as a strict negative cell line, as they were previously shown to have no endogenous ephrinB1, -B2, or -B3 and are uninfectable by live or pseudotyped NiV (NiVpp) (38). Furthermore, the use of cells deficient in cell surface heparin sulfates, which can serve as nonspecific attachment factors for many enveloped viruses, avoids a potentially confounding factor. CHOpgsA745 cells were maintained in Dulbecco's modified Eagle's medium-F12 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Omega Scientific, Tarzana, CA). Human ephrin B-class proteins were transfected into CHOpgsA745 cells and then selected for stable expression with 1.0 mg/ml of G418 to drive plasmid expression through neomycin resistance. Once selected, the cells were single cell cloned to ensure a homogenous level of ephrin expression. Since the EphB3 receptor binds to all ephrin B-class ligands with similar affinities (0.27 to 1.8 nM for ephrinB1, 0.28 to 0.78 nM for ephrinB2, and 1.5 nM for ephrinB3) (5), we used saturating amounts of soluble EphB3-Fc (R&D Systems, Minneapolis, MN) to determine the levels of ephrinB1 to -B3 expression in our stable cell lines by flow cytometry. Stable cells expressing similar amounts of ephrinB2 and ephrinB3 (mean florescence intensities [MFIs], 43.5 and 48.9, respectively) were selected to compare henipavirus binding and entry between these two receptors. EphrinB1-expressing cells had slightly higher levels of EphB3-Fc binding (MFI, 54.6).
Plasmids and reagents.
Soluble Fc fusion proteins mouse ephrinB2-Fc, human ephrinB3-Fc, and EphB3-Fc were purchased from R&D Systems. Human ephrinB2 and ephrinB3 plasmids were purchased from Genecopoeia (Germantown, MD), and human ephrinB1 was obtained from OpenBiosystems (Huntsville, AL). Each ephrin open reading frame was subcloned into the pcDNA3.1 vector (Invitrogen) under the control of a cytomegalovirus (CMV) promoter. NiV envelope nucleotide sequences of NiV-F and NiV-G were codon optimized and synthesized by Geneart (Germany) (GenBank accession numbers AY816748 and AY816745, respectively), based on previously published sequences (23). Similarly, HeV-G (AY816746) and HeV-F (AY816747) were synthesized in the same manner, based on previously published sequences (51). NiV-G and HeV-G were tagged at the C terminus with hemagglutinin (HA), while NiV-F and HeV-F were tagged with AU-1 at the C terminus.
Construction of soluble, chimeric, and mutant henipavirus G glycoproteins.
Soluble G (sG) proteins were made using the ectodomain of NiV-G (amino acid residues [aa] 71 to 602) or HeV-G (aa 71 to 604) cloned in frame with a C-terminal HA tag and placed under the control of a CMV promoter. Since NiV-G and HeV-G are type II transmembrane proteins, the kappa light chain signal sequence was placed at the N terminus of all sG-HA constructs to promote secretion into the supernatant. Chimeric HeV-G and NiV-G constructs were made using an overlap PCR strategy and subcloned into the pcDNA3.1 (Invitrogen) expression vector. Point mutations in both full-length and sG constructs were created using a QuikChange (Stratagene, La Jolla, CA) site-directed mutagenesis kit. All chimeric and mutant G constructs were confirmed by sequencing.
Production, concentration, and measurement of sG constructs.
sG DNA plasmids were transfected into 293T cells by using Lipofectamine 2000 (Invitrogen), and serum-free supernatants were collected at 2 and 4 days posttransfection. Proteins were then concentrated using Centricon (Millipore, Billerica, MA) centrifugation filtration units with a 10-kDa cutoff. sG-HA proteins alongside HA protein standards were detected by Western blotting using a horseradish peroxidase-conjugated anti-HA monoclonal antibody (Novus Biologicals, Littleton, CO) and an ECL Plus chemiluminescence detection system (GE Healthcare, Piscataway, NJ). Concentrations of sG-HA proteins were determined based on the intensities of serial dilutions of sG-HA proteins measured against an HA protein standard. The HA protein standard was an HeV-G-HA-Fc dually tagged protein, purified and quantified using a sandwich Fc-specific enzyme-linked immunosorbent assay and purified human immunoglobulin G (IgG) to generate a standard curve (38).
Cell surface binding assays.
EphrinB2 and -B3-expressing CHOpgsA745 cells (CHO-B2 and CHO-B3 cells, respectively) were made as described above. Increasing amounts of sG-HA were incubated with CHO-B2 or CHO-B3 cells for 1 h on ice. The cells were then washed with buffer (1% bovine serum in phosphate-buffered saline [PBS]) and incubated with mouse monoclonal anti-HA antibodies (Covance, Berkeley, CA) for 30 min on ice. Again, cells were washed with buffer and incubated with R-phycoerythrin-conjugated anti-mouse IgG antibodies (Invitrogen) for 30 min on ice. The cells were washed again and fixed with 2% paraformaldehyde, and the data were collected using a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The data were analyzed using FCS Express V2 (DeNovo Software, Thornhill, Ontario, Canada). The cell surface Kd values were obtained via linear regression, using GraphPad Prism software (San Diego, CA), by normalizing the highest MFI obtained to 100%.
Pseudotyped virus infection assay.
NiVpp and HeVpp were made from the VSV-ΔG-Luc virus, a recombinant vesicular stomatitis virus (VSV) derived from a full-length cDNA clone of the VSV Indiana serotype in which the G envelope protein has been replaced with Renilla luciferase (Luc; kindly provided by Andrea Bertolotti-Ciarlet and Robert Doms). 293T cells were transfected with NiV and HeV envelopes F and G and subsequently infected with VSV-ΔG-Luc virus, itself pseudotyped with VSV-G. The resulting F/G pseudotyped viruses were collected at 24 h postinfection. Viral particles were then pelleted via ultracentrifugation in 20% sucrose, and the resuspended pseudotyped viruses were used to infect the CHOpgsA745 stable cell lines after titration on the appropriate receptor-expressing cell lines, as indicated. Briefly, the cells were seeded overnight in a 48-well plate and then infected for 1.5 h at 37°C with NiV- and HeV-pseudotyped VSV-ΔG-Luc diluted in 1% fetal bovine serum in PBS. The inoculum was removed, and cells were washed with PBS, which was then replaced by culture medium. The next day, the cells were lysed, the lysates were mixed with Renilla Luc substrate (Promega, Madison WI), and a luminescence reading was performed on a Veritas luminometer (Turner BioSystems, Sunnyvale, CA). Each infection was performed in triplicate.
Preparation of anti-NiV-G polyclonal and monoclonal antibodies.
New Zealand White rabbits were genetically immunized four times (2 weeks apart) with a mixture of soluble and full-length NiV-G expression plasmids, using a proprietary electroporation protocol (Aldevron, Fargo, ND). After the last immunization, spleen cells were used for the production of rabbit hybridomas (Epitomics Inc., Burlingame, CA). Hybridomas were screened with an enzyme-linked immunosorbent assay using biotinylated NiV-G-transfected Vero cell lysate. Anti-NiV sera from the final bleed (polyclonal antibody 806) and two hybridoma clones (monoclonal antibodies 26 and 45) specifically recognized NiV-G and/or HeV-G by flow cytometry and in viral inhibition assays (1a).
Expression and conformation of NiV-G.
Cell surface expression and conformational integrity of the full-length wild-type and mutant NiV-G proteins were assayed by flow cytometry. Briefly, 293T cells were transfected with equal concentrations of plasmids encoding the indicated G proteins, and the following four antibodies were used to monitor expression levels: mouse anti-HA-tag monoclonal antibody (anti-HA; Covance), rabbit anti-NiV-G specific antiserum (806), and two distinct rabbit anti-NiV-G monoclonal antibodies, 26 and 45. While anti-HA and 806 antisera detected NiV-G both by flow cytometry and in Western blots under reducing and denaturing conditions (conformationally independent epitopes), the monoclonal anti-G antibodies, i.e., 26 and 45, detected NiV-G only by flow cytometry, not in a denatured form, e.g., on a Western blot after denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), suggesting that they recognize conformationally dependent epitopes. In addition, monoclonal antibodies 26 and 45 recognize nonoverlapping epitopes (H. C. Aguilar et al., unpublished data). All values were normalized to the MFIs of the wild-type proteins.
RESULTS
Construction, expression, and receptor binding analysis of henipavirus sG glycoproteins.
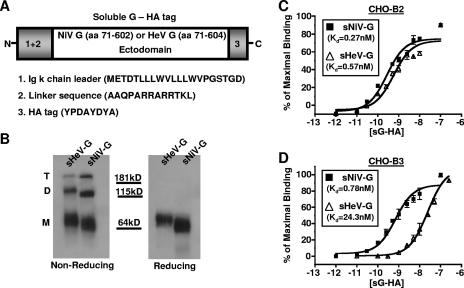
In order to compare ephrin B-class receptor usage between the henipaviruses, we first constructed similar sG glycoproteins for both NiV and HeV. Since the henipavirus G glycoprotein is a type II membrane protein, we substituted an immunoglobulin κ chain leader signal sequence for the N-terminal cytoplasmic and transmembrane domain sequence to generate our sG constructs (Fig. 1A). For ease of detection, the soluble NiV-G (sNiV-G) and HeV-G (sHeV-G) proteins were tagged with an HA tag at the C terminus.
FIG. 1.
Construction, expression, and receptor binding analysis of sNiV-G and sHeV-G. (A) NiV-G and HeV-G were codon optimized and synthesized as described in Materials and Methods. The ectodomain of NiV-G (amino acid residues 71 to 602) was C-terminally tagged with an HA tag and placed under the control of a CMV promoter (sNiV-G). A similar construct was made using the HeV-G ectodomain (amino acid residues 71 to 604) (sHeV-G). Since G is a type II transmembrane protein, the kappa light chain signal sequence was placed at the N termini of both sNiV-G and sHeV-G to promote secretion of soluble protein into the supernatant. A linker sequence was also added between the kappa leader and the start of the G ectodomain. (B) Various oligomeric forms that likely correspond to monomeric (M), dimeric (D), and tetrameric (T) species of sG-HA were seen after the proteins were separated by reducing and nonreducing SDS-PAGE and visualized by anti-HA Western blot analysis. (C and D) EphrinB2 and -B3 stably transfected CHOpgsA745 cells (CHO-B2 and CHO-B3 cells, respectively) were used to measure sNiV-G (filled squares) and sHeV-G (open triangles) cell surface binding. Increasing concentrations of sG-HA (10−12 to 10−7 M) were added to CHO-B2 or CHO-B3 cells, and binding was assessed by flow cytometry using anti-HA monoclonal antibodies and R-phycoerythrin-conjugated anti-mouse IgG. Regression curves were generated based on values displayed as percentages of maximal binding, with the saturation value set to 100%. Each data point is an average of triplicates ± standard deviation (SD).
The sG constructs were produced, concentrated, and analyzed by reducing or nonreducing SDS-PAGE (Fig. 1B). Under reducing conditions, both sNiV-G and sHeV-G ran as a single diffuse band with an apparent molecular mass of ∼64 kDa. Under nonreducing conditions, several bands were present for each construct, likely representing the different oligomeric states of G. Since paramyxovirus attachment proteins are known to exist as tetrameric structures on the viral envelope (27), the differently sized bands on a nonreducing gel most likely represent tetrameric (T), dimeric (D), and monomeric (M) species of sNiV-G or sHeV-G. Moreover, the SDS- and heat-stable oligomers in the nonreducing gel may represent disulfide-linked species since they were completely monomeric under reduced conditions (Fig. 1B). Other paramyxovirus attachment proteins, such as that of Newcastle disease virus, are thought to exist as tetramers consisting of disulfide-linked dimers (7, 35). The properties of our soluble henipavirus G constructs are thus far consistent with known features of paramyxovirus attachment proteins and similar to those described for other sHeV-G constructs (8).
We then used sNiV-G and sHeV-G to bind cell surface-expressed ephrin B-class ligands. CHO-pgsA745 (CHO) cells were stably transfected with human ephrinB1 (CHO-B1), ephrinB2 (CHO-B2), or ephrinB3 (CHO-B3) and single cell cloned to obtain cells that expressed equivalent amounts of ephrinB ligands, as determined by EphB3-Fc binding (see Materials and Methods) (38).Titration analysis revealed that while both sNiV-G and sHeV-G bound to ephrinB2-expressing cells with similar affinities (Kd = 0.27 nM and 0.57 nM, respectively) (Fig. 1C), they demonstrated a marked difference in binding to cell surface-expressed ephrinB3. Indeed, sNiV-G bound CHO-B3 cells with a Kd of 0.78 nM, in contrast to that of 24.3 nM with sHeV-G (Fig. 1D). Parental CHO and CHO-B1 cells were both negative for binding of sNiV-G and sHeV-G (38; data not shown). Cumulatively, our data show that while sNiV-G and sHeV-G bound human ephrinB2 with similar affinities, sNiV-G bound ephrinB3 with an ∼30-fold higher affinity (lower Kd) than that of sHeV-G.
HeV uses ephrinB3 less efficiently than does NiV.
Next, we sought to determine if the differences in ephrinB2 and -B3 binding translated into a differential efficiency of viral entry via these receptors. Paramyxovirus entry requires the concerted action of both the F and G envelope glycoproteins. Thus, we first produced homotypic NiVpp and HeVpp reporter viruses, using recombinant VSV expressing Luc, a system previously described to mimic live NiV entry (38). We infected CHO, CHO-B1, CHO-B2, and CHO-B3 cells with equivalent amounts of NiVpp and HeVpp, as titrated on CHO-B2 cells. We chose this infection strategy based on our binding studies (Fig. 1C) and on data from other groups who reported similar ephrinB2 binding between HeV and NiV (6). Figure 2A shows that the amount of NiVpp that gave similar infection efficiencies to those of HeVpp in CHO-B2 cells entered ephrinB3-expressing cells sevenfold more efficiently than did HeVpp (Fig. 2A). Minimal entry was seen in CHO or CHO-B1 cells.
FIG. 2.
HeV uses ephrinB3 less efficiently than does NiV. (A and B) Homotypic envelope-mediated infections were performed using NiV-G and NiV-F (NiVpp) or HeV-G and HeV-F (HeVpp) glycoproteins pseudotyped into a VSV-ΔG-Luc reporter virus. Heterotypic envelope-mediated infections were performed using NiV-G and HeV-F (NG/HF) or HeV-G and NiV-F (HG/NF) pseudotyped into the VSV reporter virus. Equivalent amounts of viral particles (titrated to give equivalent Luc activities on CHO-B2 cells) were used to infect parental CHOpgsA745 cells (CHO) or CHO-pgsA745 cells stably expressing human ephrinB1, -B2, and -B3 (CHO-B1, CHO-B2, and CHO-B3 cells, respectively). CHO-B2 and CHO-B3 cells expressed ephrinB2 and -B3, respectively, at similar levels (see Materials and Methods). Entry of homotypic and heterotypic viruses was measured by quantifying Renilla Luc activity according to the manufacturer's directions. Relative light units (RLU) were acquired and quantified on a luminometer. Data shown are averages for three independent experiments ± SD. (C and D) Dilutions of homotypic (NiVpp and HeVpp) and heterotypic (NG/HF and HG/NF) reporter viruses were made to span 2 log and subsequently used for viral entry into CHO-B2 and CHO-B3 cells. The amount of pseudotyped virus entry was measured in RLU. Data shown are averages for three independent experiments ± SD. Error bars are too small to be seen on the log scale presented for the RLU data.
To confirm that the less efficient usage of ephrinB3 seen with HeVpp was due to the G glycoprotein, we decided to produce heterotypic particles containing NiV-G and HeV-F (NG/HF) or HeV-G and NiV-F (HG/NF). Heterotypic complementation between HeV and NiV envelope glycoproteins was previously demonstrated in the context of a fusion assay (9). Figure 2B shows that when the heterotypic particles were titrated on CHO-B2 cells, HeV-G-containing HG/NF particles utilized ephrinB3 less efficiently than did NG/HF particles, essentially giving the same phenotype as homotypic particles. To confirm that the observed difference in ephrinB3 receptor usage between NiV and HeV particles was within the linear range of our infection assay, various dilutions of homotypic (NiVpp and HeVpp) and heterotypic (NG/HF and HG/NF) reporter viruses were used to infect CHO-B2 and CHO-B3 cells. The difference between NiV and HeV homotypic viral entry in CHO-B3 cells remained linear over the span of 100-fold viral dilutions (Fig. 2C). Similar results were seen with heterotypic particles, where HeV-G-containing HG/NF particles entered, on average, ninefold less efficiently than NiV-G-containing NG/HF particles at any given dilution within 2 log (Fig. 2D). Overall, these results suggest that the HeV G protein, rather than the F protein, is responsible for deficient ephrinB3 receptor usage.
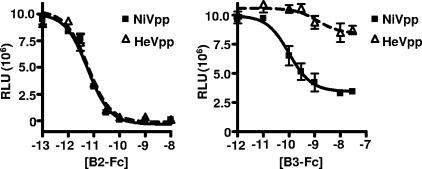
Soluble ephrinB3 inhibits HeV entry less efficiently than NiV entry.
To further verify the differential usage of ephrinB2 and -B3 between NiV and HeV, we infected CHO-B2 cells with our reporter NiV and HeV particles in the presence of increasing concentrations of soluble Fc fusion proteins of ephrinB2 and -B3 (B2-Fc and B3-Fc, respectively) (Fig. 3). Under these conditions, NiVpp and HeVpp were equally inhibited by B2-Fc, consistent with their similar ability to use ephrinB2 as an entry receptor. However, soluble ephrinB3 (B3-Fc) was clearly not as effective at inhibiting HeVpp entry as it was at inhibiting NiVpp entry in CHO-B2 cells. For example, while 1 nM of B3-Fc inhibited NiVpp entry almost 60%, it had little or no effect on HeVpp entry, suggesting that soluble human ephrinB3 binds to the NiV envelope more efficiently and prevents cell surface ephrinB2 engagement compared to that of HeV.
FIG. 3.
Soluble ephrin-B3 inhibits HeVpp less efficiently than NiVpp. NiV-F/G (NiVpp) and HeV-F/G (HeVpp) VSV-ΔG-Luc pseudotyped viruses were used to infect CHO-B2 cells in the presence of increasing amounts of soluble ephrinB2-Fc (10−12 to 10−8 M) and ephrinB3-Fc (10−13 to 10−7 M) fusion proteins (B2-Fc and B3-Fc, respectively). Entry was measured as described in the legend to Fig. 2A. Inhibition curves were generated via GraphPad Prism. Data are the averages for three independent experiments ± SD.
The determinant of ephrinB3 usage maps to a single amino acid in the G protein.
Our data thus far indicate that the reduced efficiency of ephrinB3 usage by HeV maps to the G protein and is likely due to HeV-G's lower affinity for ephrinB3 binding. HeV-G and NiV-G share 83% homology at the protein level, and most of the structurally important residues, such as those predicted to maintain the beta-propeller sheets, appear to be conserved (45). Thus, we constructed soluble chimeric G proteins between NiV and HeV and sought to determine the minimal domain in NiV-G that can restore NiV-G's level of ephrinB3 binding in the context of HeV-G. Figure 4A shows the initial chimeras constructed, which fused increasing numbers of N-terminal residues from NiV-G's extracellular domain with the remaining homologous C terminus of HeV-G. For example, the sNH183 chimera fused the proposed NiV-G stalk domain residues (aa 72 to 183) with the HeV-G globular domain (aa 184 to 604). Equal concentrations of sHeV-G, sNiV-G, and the indicated chimeric constructs were then used for binding experiments with cell surface-expressed ephrinB2 or -B3 (Fig. 4B). None of the chimeric constructs could restore ephrinB3 binding to the level seen with sNiV-G, even though they all bound equally well to ephrinB2, suggesting that these chimeras were also conformationally intact. sNH494, which consisted mostly of NiV-G residues fused to only the last 110 C-terminal residues of HeV-G, was just as deficient in ephrinB3 binding as sHeV-G. This suggests that the ephrinB3 binding determinants in NiV-G reside within this C-terminal domain. We therefore decided to make the reverse chimeric construct, which included mostly HeV-G residues but contained 80 residues from NiV-G (aa 494 to 573 [HN494-573]) (Fig. 4C).
FIG. 4.
Determinant of ephrinB3 usage maps to a single amino acid in the G protein. (A) Soluble chimeric G proteins were made by N-terminally fusing the indicated segments from the ectodomain of sNiV-G to the corresponding remaining C-terminal segment from sHeV-G. (B) Equal amounts of sHeV-G, sNiV-G, and the chimeric G proteins were incubated with CHO-B2 or CHO-B3 cells. The amount of cell surface binding was measured by flow cytometry and calculated as the MFI. (C) Full-length chimeric HeV-G proteins were made by replacing residues 494 to 573 (HN494-573), 512 to 573 (HN494-573), or 494 to 512 (HN494-512) with homologous NiV-G residues. (D) HeV-G, NiV-G, and the full-length chimeric G proteins were used to make a VSV-Luc reporter virus, which was used to infect CHO-B2 or CHO-B3 cells. The amount of entry was measured in RLU, based on the amount of Luc activity present in the cells 24 h after infection. (E) Single amino acid substitutions were made in full-length HeV-G at residues 498 (H-V498T), 502 (H-V502I), and 507 (H-S507V) to complement the NiV-G residues at the equivalent positions. (F) The H-V498T, H-V502I, and H-S507V mutants were used to make pseudotyped VSV, which was used, along with NiVpp and HeVpp, to infect CHO-B2 or CHO-B3 cells. All data points in panels B, D, and F are averages of triplicates ± SD.
Since infection efficiency is a more relevant indicator of receptor usage, a full-length version of this construct (HN494-573) was made for use in pseudotyped viral infections (Fig. 4C). Indeed, HN494-573 was found to gain ephrinB3-dependent viral entry at levels comparable to that of NiV (Fig. 4D). The 79 NiV residues within aa 494 to 573 were further divided into smaller regions (HN494-512 and HN512-573), which were then reconstructed in the context of full-length HeV-G. Viral entry experiments using these new constructs revealed that ephrinB3-dependent entry resided within a 19-aa stretch within the C-terminal domain of NiV-G (aa 494 to 512) (Fig. 4D). Only three amino acids differed between NiV-G and HeV-G within aa 494 to 512, and thus, single amino acids in HeV-G were converted to the corresponding NiV-G residues at positions 498, 502, and 507 (H-V498T, H-V502I, and H-S507V, respectively) (Fig. 4E). Full-length clones in the context of a viral entry assay indicated that a single amino acid change in HeV-G, i.e., conversion of residue 507 from serine to valine (H-S507V), was enough to confer ephrinB3 usage back to NiV levels (Fig. 4F).
A serine-to-threonine change at residue 507 in HeV-G confers ephrinB3 binding and viral entry abilities comparable to those of NiV.
The remarkable finding described above prodded us to more carefully examine the HeV-G sequences available in public databases. Curiously, we found that two previously published HeV-G sequences displayed variations at position 507 (23, 51). In fact, the only difference in the globular domain between these sequences was a serine-to-threonine change at aa 507. While the current HeV-G sequence in GenBank (accession no. NP_047112 [“version 2,” updated on 1 April 2003]) contains a threonine at position 507, it was originally entered as a sequence containing a serine at 507 when it was reported by Yu et al. (51) (GenBank sequence “version 1”). The reasons for this change are not entirely clear (see Discussion). The HeV-G sequences we have used until now contain a serine at position 507, as our codon-optimized sequence was made prior to 2003 (GenBank accession no. AY816746). Notably, residue 507 remains a valine in all available NiV-G sequences, suggesting that residue 507 is important for receptor interactions. To investigate the effect of a threonine at position 507 in the updated HeV-G sequence, we constructed both soluble and full-length HeV-G proteins with a threonine at residue 507 (sH-507T and H-507T, respectively) and renamed our original soluble and full-length HeV-G constructs, containing a serine at aa 507, with the designations sH-507S and H-507S, respectively.
sH-507T, from the updated sequence (“version 2”), bound cell surface-expressed ephrinB2 with a similar affinity to those of sNiV-G and sH-507S, our original HeV-G sequence (Fig. 5A). Remarkably, sH-507T bound to cell surface ephrinB3 almost as well as NiV-G did; it no longer exhibited the decrease in ephrinB3 binding seen with sH-507S (“version 1” [our original HeV-G sequence]). The full-length H-507T protein, along with NiV-G and the H-507S protein, were then used to produce pseudotyped viruses for use in viral entry assays. All three G proteins were equally incorporated into viral particles (Fig. 5B), and all three pseudotyped viruses infected CHO-B2 cells at similar levels, but now, the H-507T virus was able to infect CHO-B3 cells almost as well as NiVpp (<2-fold decrease) (Fig. 5C). As expected, the H-507S virus, which has the HeV-G sequence we used for all experiments up to now (Fig. 1 to 4), still had reduced ephrinB3-dependent entry compared to NiVpp (almost 10-fold decrease). Since both valine (from NiV-G sequences) and threonine (from the updated HeV-G sequence), but not serine (from the original HeV-G sequence), at residue 507 conferred efficient ephrinB3 receptor usage, it appears that the amino acid structure similarities between valine and threonine (which have terminal hydrophobic methyl side chains) are more important than polar similarities between threonine and serine.
FIG. 5.
A mutant with a serine-to-threonine change at residue 507 in HeV-G gains ephrinB3 binding and viral entry abilities comparable to those of NiV. (A) sNiV-G and sHeV-G with residue T507 (sH-507T) or S507 (sH-507S), used at 2 nM, 0.4 nM, and 0.1 nM concentrations, were allowed to bind independently to CHO-B2 or CHO-B3 cells. The amount of binding was assessed by flow cytometry using anti-HA monoclonal antibodies and R-phycoerythrin-conjugated anti-mouse IgG. Data are the averages of triplicates ± SD. (B) Henipavirus envelope glycoproteins, namely, NiV-F/G (NiVpp) or HeV-F/G with residue 507T or 507S (H-507T or H-507S, respectively), were pseudotyped onto VSV reporter viruses. NiVpp, H-507T, and H-507S virus particles were run in a reducing SDS-PAGE gel and detected by anti-HA (αHA) Western blotting to detect G. Note that HeV-G always runs slightly higher on the gel than NiV-G. (C) Fivefold serial dilutions were made with NiVpp, H-507T, and H-507S VSV reporter viruses and subsequently used to infect CHO-B2 and CHO-B3 cells at similar concentrations, based on the amount of G incorporated onto the viral particles (B). Luc reporter activity was measured in RLU. Data are the averages for three independent experiments ± SD.
Residues near aa 507 in NiV-G are involved in distinct ephrinB3 binding.
Since subtle changes at position 507 can significantly impact ephrinB3 receptor usage, our data suggest that residues around this region may be critical for envelope-receptor interactions. Guillaume et al. (20) recently implicated several residues, including residues 504 and 505, as part of the putative receptor binding domain in NiV-G. However, no biochemical characterization was provided to indicate whether mutation of these residues actually effected ephrinB2 or -B3 binding. Since these residues were near position 507, we constructed soluble and full-length NiV-Gs to include mutations W504A, E505A, V507S, and E533Q (in another putative receptor binding residue [implicated by Guillaume et al.], though slightly more distant from position 507) in order to characterize their effects on ephrinB2 and -B3 receptor usage. Soluble W504A, E505A, and V507S mutants bound to CHO-B2 cells as well as wild-type sNiV-G did, while the soluble E533Q mutant no longer bound ephrinB2 to any significant degree (Fig. 6A). Interestingly, all mutations significantly reduced ephrinB3 binding, with the most drastic decreases seen with the charged residue mutations E505A and E533Q. The reduction in ephrinB3 binding was likely not due to gross conformational changes, as (i) most of the mutants, except for the E533Q mutant, still bound ephrinB2; and (ii) probing with two conformational monoclonal antibodies (26 and 45) indicated that they had the same ratio of reactivity as two other conformationally independent antibodies (the anti-HA antibody and polyclonal 806 anti-NiV-G antibody) (Fig. 6B).
FIG. 6.
Residues near amino acid 507 in NiV-G are involved in distinct ephrinB3 binding. (A) Flow cytometry was used to assess the binding of cell surface-expressed ephrinB2 and -B3 with wild-type sNiV-G or sNiV-G with a mutation at W504A (sN-W504A), E505A (sN-E505A), V507S (sN-V507S), or E533Q (sN-E533Q). The amounts of binding of various concentrations (2 nM, 0.4 nM, and 0.1 nM) of wild-type or mutant sNiV-G to CHO-B2 and CHO-B3 cells are shown as MFI values. Data are the averages of triplicates ± SD. (B) Full-length wild-type NiV-G or mutant G proteins (W504A, E505A, V507S, and E533Q) were expressed in 293T cells and used for binding analysis with two nonconformational (anti-HA and 806) and two conformational (26 and 45) antibodies. All values were normalized to the MFI of the wild-type protein, set at 100% (data shown are averages of triplicates ± SD). (C) NiV envelope glycoproteins F and wild-type (WT) G (NiVpp) or NiV-F and mutant G (N-W504A, N-E505A, N-V507S, and N-E533Q) were pseudotyped onto VSV-ΔG-Luc reporter viruses. Wild-type NiV and mutant virus particles were run in a reducing SDS-PAGE gel and detected by anti-HA (αHA) Western blotting to detect G. (D) Dilutions (1×, 5×, and 25×) of wild-type NiV and mutant VSV reporter viruses were used to infect CHO-B2 and CHO-B3 cells at similar concentrations, based on the amount of G incorporated onto the viral particles (C). Data are the averages for three independent experiments ± SD.
Full-length constructs of the aforementioned mutations were then used in pseudotyped viral entry assays after ensuring that the various G mutants were incorporated into the virions in roughly equal amounts (Fig. 6C). The pattern of viral entry into CHO-B2 and CHO-B3 cells mimicked soluble G binding, where the W504A, E505A, and V507S mutants entered ephrinB2-expressing cells as well as wild-type NiV did but ephrinB3-dependent entry was reduced compared to that of NiV (Fig. 6D). E533Q mutant pseudotyped virus particles were essentially not competent for either ephrinB2 or -B3-mediated entry. These results suggest that residues near position 507 are also important for distinct ephrinB3 binding, while residue 533 has overlapping ephrinB2 and -B3 binding properties.
DISCUSSION
Here we show that both ephrinB2 and -B3 can serve as functional receptors for NiV and HeV. While NiV and HeV can use ephrinB2 equivalently well, we initially found that HeV (i.e., H-507S) uses ephrinB3 much less efficiently than does NiV. Mapping studies indicated that the valine 507 residue in NiV-G can confer efficient ephrinB3 usage when placed in the context of HeV-G. Upon careful inspection, we found that two alternative published HeV-G sequences exist, differing by one amino acid at residue 507 in the globular domain (23, 51) (GenBank accession no. AF01749.2 and AF01749). Further analysis indicated that the HeV sequence with a threonine at residue 507 (H-507T) in the attachment protein has comparable ephrinB2 and -B3 usage to that of NiV, while a serine at residue 507 can significantly reduce HeV and NiV usage of ephrinB3 without compromising their ability to use ephrinB2 efficiently.
The frequency at which these two alternative HeV-G sequences exist in natural viral isolates is currently unknown. Perhaps these particular viral isolates reported in GenBank were sequenced from a mixed population of viruses. RNA viruses are prone to high mutation rates due to the lack of proofreading ability of their RNA-dependent RNA polymerases. Thus, RNA viruses exist as quasispecies, which can be an intrinsic property of the virus that determines its pathogenicity (44). In order to determine which HeV-G sequence is the more prominent “wild-type” sequence, more HeV isolates need to be sequenced directly from infected sources or from early-passage stocks. Indeed, multiple horse, bat, and human isolates appear to have been reported (15), although their sequences have not been made public.
With regards to NiV, only minor changes have been reported between pig, bat, and human NiV isolates from the Malaysian outbreak, but more dramatic changes were seen in viral isolates from the Malaysian and Bangladeshi outbreaks (1, 14, 22, 23). Since our results indicate that even a slight conservative change in the putative receptor binding domain, such as that between serine and threonine, can significantly affect receptor usage, it would be interesting to determine if the Malaysian, Bangladeshi, and various animal NiV isolates differ in their efficiencies of ephrinB2 versus -B3 usage. Strikingly, a serine-to-threonine change in the receptor binding site of the severe acute respiratory syndrome (SARS) coronavirus spike protein has also been implicated in its ability to bind efficiently to the human ACE2 receptor (30). Indeed, structural evidence indicates that the terminal methyl group of the critical threonine fills a hydrophobic cavity in the ACE2 receptor which stabilizes the envelope-receptor interaction (29). As with the SARS viral isolates, we hypothesize that a serine-to-threonine change in the receptor binding region of the henipavirus attachment protein can alter the pathogenicity of the virus via its ability to affect ephrinB3 usage (see below and Fig. 8).
FIG. 8.
EphrinB2 and -B3 are coexpressed in the cerebral cortex, but ephrinB3 is distinctly expressed in the brain stem. (A and B) A three-dimensional model of the adult mouse brain was colored differently to represent the three major brain regions, i.e., the cerebral cortex (light blue), the cerebellum (green), and the brain stem (purple). The left hemisphere of the mouse brain was viewed from either a dorsal plane (A, C, and E) or a sagittal plane (B, D, and F). EphrinB2 expression in the cerebral cortex (C) or the brain stem (D) is indicated by orange and yellow dots. EphrinB3 expression in the cerebral cortex (E) or the brain stem (F) is displayed as red dots. All images were created using Brain Explorer v1.3, the Allen Brain Atlas (28) gene expression viewer program.
Guillaume et al. (20) described a reduction in cell-cell membrane fusion in Vero cells and ephrinB2-transfected CHO cells when they explored some of the same NiV-G mutants that we used. They did not explore our position 507 mutants, but the proximity of residue 507 to the putative receptor binding site identified by their group prompted us to carefully examine their mutations in the context of ephrinB2 and -B3 usage. We found that residues 504 and 505, in addition to residue 507, reduce ephrinB3 receptor usage without compromising ephrinB2 usage. Also, the E533Q mutation was the most potent fusion-nullifying mutation in both of our studies, although we now attribute that phenotype to a lack of both ephrinB2 and -B3 binding. Therefore, we confirm and extend the findings of Guillaume et al. (20) by attributing the underlying phenotype of their fusogenic mutants to a specific defect in ephrinB2 and/or ephrinB3 receptor usage. In addition, a recent study by Bishop et al. (4) also identified residues important for henipavirus receptor binding. These residues (G439, K443, G449, K465, and D468) are clustered in beta-sheet 4 strands 1 to 3 (β4S1-3), which are actually not predicted to be on the surface of the globular domain by either their own modeled structure (46) or the one published by Guillaume et al. (20). The authors themselves raised the possibility that the identified residues form a discontinuous epitope that affects receptor binding via indirect means.
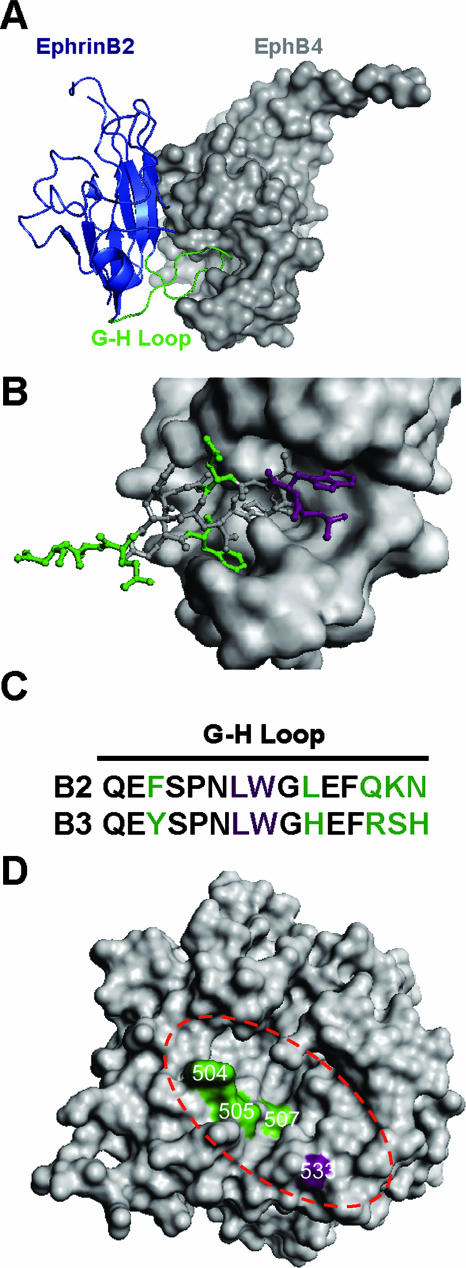
We previously characterized two overlapping residues (Leu-Trp) at the tip of the solvent-exposed G-H loop of ephrinB2 and -B3 that are critical determinants of NiV binding and entry (38). The cocrystal structure of ephrinB2 in complex with EphB4 (an endogenous ephrinB2 receptor) (11) indicates that the G-H loop of ephrinB2 inserts into a hydrophobic canyon of EphB4 (Fig. 7A). On closer inspection, several residues in the G-H loop of ephrinB2, on either side of the L-W tip, also make substantial contacts with the binding cleft of EphB4 (Fig. 7B). Since the G-H loop is an important region of contact with EphB4, EphB2 (24), and NiV-G, we hypothesize that residues in the G-H loop that are different between ephrinB2 and -B3 may interact with the distinct ephrinB3 binding determinants on NiV-G. Therefore, to correlate our previous findings (38) with current data, we propose that the tip of the G-H loop containing the L-W residues in ephrinB2 and -B3 interacts with residues at or around position 533 in NiV-G, since a mutation at this residue affected both ephrinB2 and -B3 binding. As a consequence, the differing residues in ephrinB2 and -B3 immediately surrounding the L-W tip of the G-H loop (Fig. 7C) are positioned to interact with distinct ephrinB3 binding determinants (residues 504, 505, and 507) in NiV-G. Interestingly, when the putative ephrinB2 and -B3 binding determinants were mapped onto a published model of NiV-G (20), we noted that the receptor binding face of NiV-G also has a cleft or canyon, where residue 533 is positioned at one end of the canyon and residues 504, 505, and 507 line the “walls” of the canyon, not unlike the ephrinB2-interacting canyon seen in EphB4.
FIG. 7.
NiV-G model and proposed site of ephrinB2 and -B3 interaction in comparison to the crystal structure of the ephrinB2-EphB4 complex. (A) The crystal structure of an ephrinB2 monomer (blue cartoon representation) in complex with its cognate receptor, EphB4 (surface representation), indicates that the G-H loop of ephrinB2 (green) inserts into a hydrophobic canyon of EphB4 (11). (B) Detailed view of the G-H loop of ephrinB2 (stick representation) that makes contact with the binding cleft of EphB4 (surface representation). (C) Alignment of the G-H loop residues of ephrinB2 and -B3. Color-coded residues correspond to those indicated in panel B. The purple residues are critical for ephrinB2 and -B3 binding to NiV-G, and the green residues indicate the differences between ephrinB2 and -B3 in the G-H loop. (D) Structural model of the NiV-G protein globular domain described by Guillaume et al. (20), displayed as a top view with surface representation. Green residues (W504, E505, and V507) localize to the top surface of the NiV-G model and represent residues involved in distinct ephrinB3 binding. The purple residue (E533) has overlapping ephrinB2 and -B3 binding properties. The dashed red oval highlights a predicted binding cleft or canyon in the NiV-G model. All images were created using PyMol v0.99 (DeLano Scientific LLC, Palo Alto, CA).
Finally, we suggest that ephrinB3-mediated entry may actually be the ultimate cause of death in acute Nipah viral encephalitis. High-resolution expression mapping data using the Allen Brain Atlas (28) indicate that while ephrinB2 and -B3 are coexpressed in overlapping regions of the mouse cerebrum (Fig. 8A, C, and E; compare panels C and E), ephrinB3, but not ephrinB2, is expressed in the brain stem (Fig. 8B, D, and F; compare panels D and F). Tissue gene expression studies with humans have also found that ephrinB3, but not ephrinB2, is expressed in the pons region of the brain stem (43). These findings underscore the biological relevance of ephrinB3-mediated usage, as severe brain stem dysfunction is the major clinical defining feature of fatal Nipah viral encephalitis (19). In addition, histological data from fatal cases of NiV infection found abundant NiV antigen staining in the pons region of the brain stem (48), while magnetic resonance images of NiV-infected patients also found lesions in the pons in three of eight persons examined (33). Thus, the ability of NiV or HeV to use ephrinB3 efficiently may be a key pathogenic determinant in Nipah viral encephalitis. Indeed, we hypothesize that mutations that enhance ephrinB3 usage may result in a more encephalopathic virus.
More data are required in several henipavirus research areas to determine if alterations in receptor usage by NiV or HeV may account for differences seen in transmission efficiency to humans, tropism, and/or severity of the disease they cause. For example, it was noted that NiV caused more severe infection in the respiratory epithelium than did HeV in a feline model of henipavirus infection, suggesting a concrete example to examine if differential receptor usage contributes to the variant pathological profiles observed (25). In addition, the use of a recently described NiV rescue system (50) to generate a virus that can use ephrinB2, but not ephrinB3, to infect an animal model that recapitulates NiV neuronal disease may help to dissect the role of ephrinB3 in fatal Nipah viral encephalitis. A more comprehensive delineation of henipavirus receptor interactions will thus aid our understanding of the pathology underlying this emergent infectious disease.
Acknowledgments
We thank Yao Wang for helpful comments and a critical reading of the manuscript. We also thank Su-Yang Liu for technical assistance, Andy Grock for thoughtful discussions, and Zeynep Akyol Ataman for creation of the protein structure images.
This work was supported by NIH grants (AI059051, AI060694, AI 070495, and AI069317) to B.L., by an NIH NRSA grant (GM07185), and by a UCLA dissertation year fellowship to O.A.N. We also acknowledge support to the UCLA AIDS Institute and CFAR flow cytometry core funded through NIH grants (CA16042 and AI28697).
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.AbuBakar, S., L. Y. Chang, A. R. Ali, S. H. Sharifah, K. Yusoff, and Z. Zamrod. 2004. Isolation and molecular identification of Nipah virus from pigs. Emerg. Infect. Dis. 10:2228-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Aguilar, H. C., O. A. Negrete, K. A. Matreyek, D. Y. Choi, and B. Lee. 2006. Annu. Meet. Am. Soc. Virol., abstr. P19-4. American Society for Virology, Madison, WI.
- 2.Anonymous. 2004. Person-to-person transmission of Nipah virus during outbreak in Faridpur District, 2004. Health Sci. Bull. 2:5-9. [Google Scholar]
- 3.Benson, M. D., M. I. Romero, M. E. Lush, Q. R. Lu, M. Henkemeyer, and L. F. Parada. 2005. Ephrin-B3 is a myelin-based inhibitor of neurite outgrowth. Proc. Natl. Acad. Sci. USA 102:10694-10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. A., T. S. Stantchev, A. C. Hickey, D. Khetawat, K. N. Bossart, V. Kerasnoperov, P. Gill, Y. R. Feng, L. Wang, B. Eaton, L. Wang, and C. C. Broder. 2007. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J. Virol. 81:5893-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blits-Huizinga, C. T., C. M. Nelersa, A. Malhotra, and D. J. Liebl. 2004. Ephrins and their receptors: binding versus biology. IUBMB Life 56:257-265. [DOI] [PubMed] [Google Scholar]
- 6.Bonaparte, M. I., A. S. Dimitrov, K. N. Bossart, G. Crameri, B. A. Mungall, K. A. Bishop, V. Choudhry, D. S. Dimitrov, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 102:10652-10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bossart, K. N., and C. C. Broder. 2007. Paramyxovirus entry, p. 1-20. In S. Pohlmann and G. Simmons (ed.), Viral entry into host cells. Landes Bioscience, Austin, TX.
- 8.Bossart, K. N., G. Crameri, A. S. Dimitrov, B. A. Mungall, Y. R. Feng, J. R. Patch, A. Choudhary, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 79:6690-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossart, K. N., L. F. Wang, M. N. Flora, K. B. Chua, S. K. Lam, B. T. Eaton, and C. C. Broder. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 76:11186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, N., D. M. Brantley, and J. Chen. 2002. The ephrins and Eph receptors in angiogenesis. Cytokine Growth Factor Rev. 13:75-85. [DOI] [PubMed] [Google Scholar]
- 11.Chrencik, J. E., A. Brooun, M. L. Kraus, M. I. Recht, A. R. Kolatkar, G. W. Han, J. M. Seifert, H. Widmer, M. Auer, and P. Kuhn. 2006. Structural and biophysical characterization of the EphB4-ephrinB2 protein-protein interaction and receptor specificity. J. Biol. Chem. 281:28185-28192. [DOI] [PubMed] [Google Scholar]
- 12.Chua, K. B. 2003. Nipah virus outbreak in Malaysia. J. Clin. Virol. 26:265-275. [DOI] [PubMed] [Google Scholar]
- 13.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 14.Chua, K. B., C. L. Koh, P. S. Hooi, K. F. Wee, J. H. Khong, B. H. Chua, Y. P. Chan, M. E. Lim, and S. K. Lam. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4:145-151. [DOI] [PubMed] [Google Scholar]
- 15.Eaton, B., J. Mackenzie, and L. Wang. 2007. Henipaviruses, p. 1587-1600. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 16.Eaton, B. T., C. C. Broder, D. Middleton, and L. F. Wang. 2006. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 4:23-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esko, J. D., T. E. Stewart, and W. H. Taylor. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 82:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gale, N. W., P. Baluk, L. Pan, M. Kwan, J. Holash, T. M. DeChiara, D. M. McDonald, and G. D. Yancopoulos. 2001. Ephrin-B2 selectively marks arterial vessels and neovascularization sites in the adult, with expression in both endothelial and smooth-muscle cells. Dev. Biol. 230:151-160. [DOI] [PubMed] [Google Scholar]
- 19.Goh, K. J., C. T. Tan, N. K. Chew, P. S. Tan, A. Kamarulzaman, S. A. Sarji, K. T. Wong, B. J. Abdullah, K. B. Chua, and S. K. Lam. 2000. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N. Engl. J. Med. 342:1229-1235. [DOI] [PubMed] [Google Scholar]
- 20.Guillaume, V., H. Aslan, M. Ainouze, M. Guerbois, T. F. Wild, R. Buckland, and J. P. Langedijk. 2006. Evidence of a potential receptor-binding site on the Nipah virus G protein (NiV-G): identification of globular head residues with a role in fusion promotion and their localization on an NiV-G structural model. J. Virol. 80:7546-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halpin, K., P. L. Young, H. E. Field, and J. S. Mackenzie. 2000. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 81:1927-1932. [DOI] [PubMed] [Google Scholar]
- 22.Harcourt, B. H., L. Lowe, A. Tamin, X. Liu, B. Bankamp, N. Bowden, P. E. Rollin, J. A. Comer, T. G. Ksiazek, M. J. Hossain, E. S. Gurley, R. F. Breiman, W. J. Bellini, and P. A. Rota. 2005. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg. Infect. Dis. 11:1594-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harcourt, B. H., A. Tamin, T. G. Ksiazek, P. E. Rollin, L. J. Anderson, W. J. Bellini, and P. A. Rota. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334-349. [DOI] [PubMed] [Google Scholar]
- 24.Himanen, J. P., K. R. Rajashankar, M. Lackmann, C. A. Cowan, M. Henkemeyer, and D. B. Nikolov. 2001. Crystal structure of an Eph receptor-ephrin complex. Nature 414:933-938. [DOI] [PubMed] [Google Scholar]
- 25.Hooper, P., S. Zaki, P. Daniels, and D. Middleton. 2001. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 3:315-322. [DOI] [PubMed] [Google Scholar]
- 26.Hsu, V. P., M. J. Hossain, U. D. Parashar, M. M. Ali, T. G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee, and R. F. Breiman. 2004. Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10:2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lein, E. S., M. J. Hawrylycz, N. Ao, M. Ayres, A. Bensinger, A. Bernard, A. F. Boe, M. S. Boguski, K. S. Brockway, E. J. Byrnes, L. Chen, T. M. Chen, M. C. Chin, J. Chong, B. E. Crook, A. Czaplinska, C. N. Dang, S. Datta, N. R. Dee, A. L. Desaki, T. Desta, E. Diep, T. A. Dolbeare, M. J. Donelan, H. W. Dong, J. G. Dougherty, B. J. Duncan, A. J. Ebbert, G. Eichele, L. K. Estin, C. Faber, B. A. Facer, R. Fields, S. R. Fischer, T. P. Fliss, C. Frensley, S. N. Gates, K. J. Glattfelder, K. R. Halverson, M. R. Hart, J. G. Hohmann, M. P. Howell, D. P. Jeung, R. A. Johnson, P. T. Karr, R. Kawal, J. M. Kidney, R. H. Knapik, C. L. Kuan, J. H. Lake, A. R. Laramee, K. D. Larsen, C. Lau, T. A. Lemon, A. J. Liang, Y. Liu, L. T. Luong, J. Michaels, J. J. Morgan, R. J. Morgan, M. T. Mortrud, N. F. Mosqueda, L. L. Ng, R. Ng, G. J. Orta, C. C. Overly, T. H. Pak, S. E. Parry, S. D. Pathak, O. C. Pearson, R. B. Puchalski, Z. L. Riley, H. R. Rockett, S. A. Rowland, J. J. Royall, M. J. Ruiz, N. R. Sarno, K. Schaffnit, N. V. Shapovalova, T. Sivisay, C. R. Slaughterbeck, S. C. Smith, K. A. Smith, B. I. Smith, A. J. Sodt, N. N. Stewart, K. R. Stumpf, S. M. Sunkin, M. Sutram, A. Tam, C. D. Teemer, C. Thaller, C. L. Thompson, L. R. Varnam, A. Visel, R. M. Whitlock, P. E. Wohnoutka, C. K. Wolkey, V. Y. Wong, M. Wood, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168-176. [DOI] [PubMed] [Google Scholar]
- 29.Li, F., W. Li, M. Farzan, and S. C. Harrison. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864-1868. [DOI] [PubMed] [Google Scholar]
- 30.Li, W., C. Zhang, J. Sui, J. H. Kuhn, M. J. Moore, S. Luo, S. K. Wong, I. C. Huang, K. Xu, N. Vasilieva, A. Murakami, Y. He, W. A. Marasco, Y. Guan, H. Choe, and M. Farzan. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebl, D. J., C. J. Morris, M. Henkemeyer, and L. F. Parada. 2003. mRNA expression of ephrins and Eph receptor tyrosine kinases in the neonatal and adult mouse central nervous system. J. Neurosci. Res. 71:7-22. [DOI] [PubMed] [Google Scholar]
- 32.Lim, C. C., W. L. Lee, Y. S. Leo, K. E. Lee, K. P. Chan, A. E. Ling, H. Oh, A. P. Auchus, N. I. Paton, F. Hui, and P. A. Tambyah. 2003. Late clinical and magnetic resonance imaging follow up of Nipah virus infection. J. Neurol. Neurosurg. Psych. 74:131-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim, C. C., Y. Y. Sitoh, F. Hui, K. E. Lee, B. S. Ang, E. Lim, W. E. Lim, H. M. Oh, P. A. Tambyah, J. S. Wong, C. B. Tan, and T. S. Chee. 2000. Nipah viral encephalitis or Japanese encephalitis? MR findings in a new zoonotic disease. Am. J. Neuroradiol. 21:455-461. [PMC free article] [PubMed] [Google Scholar]
- 34.Luby, S. P., M. Rahman, M. J. Hossain, L. S. Blum, M. M. Husain, E. Gurley, R. Khan, B. N. Ahmed, S. Rahman, N. Nahar, E. Kenah, J. A. Comer, and T. G. Ksiazek. 2006. Foodborne transmission of Nipah virus, Bangladesh. Emerg. Infect. Dis. 12:1888-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinnes, L. W., and T. G. Morrison. 1994. The role of the individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology 200:470-483. [DOI] [PubMed] [Google Scholar]
- 36.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell, et al. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94-97. [DOI] [PubMed] [Google Scholar]
- 37.Negrete, O. A., E. L. Levroney, H. C. Aguilar, A. Bertolotti-Ciarlet, R. Nazarian, S. Tajyar, and B. Lee. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401-405. [DOI] [PubMed] [Google Scholar]
- 38.Negrete, O. A., M. C. Wolf, H. C. Aguilar, S. Enterlein, W. Wang, E. Muhlberger, S. V. Su, A. Bertolotti-Ciarlet, R. Flick, and B. Lee. 2006. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer, A., and R. Klein. 2003. Multiple roles of ephrins in morphogenesis, neuronal networking, and brain function. Genes Dev. 17:1429-1450. [DOI] [PubMed] [Google Scholar]
- 40.Poliakov, A., M. Cotrina, and D. G. Wilkinson. 2004. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev. Cell 7:465-480. [DOI] [PubMed] [Google Scholar]
- 41.Selvey, L. A., R. M. Wells, J. G. McCormack, A. J. Ansford, K. Murray, R. J. Rogers, P. S. Lavercombe, P. Selleck, and J. W. Sheridan. 1995. Infection of humans and horses by a newly described morbillivirus. Med. J. Aust. 162:642-645. [DOI] [PubMed] [Google Scholar]
- 42.Shin, D., G. Garcia-Cardena, S. Hayashi, S. Gerety, T. Asahara, G. Stavrakis, J. Isner, J. Folkman, M. A. Gimbrone, Jr., and D. J. Anderson. 2001. Expression of ephrinB2 identifies a stable genetic difference between arterial and venous vascular smooth muscle as well as endothelial cells, and marks subsets of microvessels at sites of adult neovascularization. Dev. Biol. 230:139-150. [DOI] [PubMed] [Google Scholar]
- 43.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vignuzzi, M., J. K. Stone, J. J. Arnold, C. E. Cameron, and R. Andino. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]
- 46.White, J. R., V. Boyd, G. S. Crameri, C. J. Duch, R. K. van Laar, L. F. Wang, and B. T. Eaton. 2005. Location of, immunogenicity of and relationships between neutralization epitopes on the attachment protein (G) of Hendra virus. J. Gen. Virol. 86:2839-2848. [DOI] [PubMed] [Google Scholar]
- 47.Wolf, M. C., O. A. Negrete, and B. Lee. 2007. Pathobiology of henipavirus entry: insights into therapeutics strategies. Future Virol. 2:267-282. [Google Scholar]
- 48.Wong, K. T., W. J. Shieh, S. Kumar, K. Norain, W. Abdullah, J. Guarner, C. S. Goldsmith, K. B. Chua, S. K. Lam, C. T. Tan, K. J. Goh, H. T. Chong, R. Jusoh, P. E. Rollin, T. G. Ksiazek, and S. R. Zaki. 2002. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am. J. Pathol. 161:2153-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yob, J. M., H. Field, A. M. Rashdi, C. Morrissy, B. van der Heide, P. Rota, A. bin Adzhar, J. White, P. Daniels, A. Jamaluddin, and T. Ksiazek. 2001. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoneda, M., V. Guillaume, F. Ikeda, Y. Sakuma, H. Sato, T. F. Wild, and C. Kai. 2006. Establishment of a Nipah virus rescue system. Proc. Natl. Acad. Sci. USA 103:16508-16513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, M., E. Hansson, J. P. Langedijk, B. T. Eaton, and L. F. Wang. 1998. The attachment protein of Hendra virus has high structural similarity but limited primary sequence homology compared with viruses in the genus Paramyxovirus. Virology 251:227-233. [DOI] [PubMed] [Google Scholar]