Abstract
Interferons (IFNs) play a major role in the control of hepatitis B virus (HBV), whether as endogenous cytokines limiting the spread of the virus during the acute phase of the infection or as drugs for the treatment of its chronic phase. However, the mechanism by which IFNs inhibit HBV replication has so far remained elusive. Here, we show that type I and II IFN treatment of human hepatocytes induces the production of APOBEC3G (A3G) and, to a lesser extent, that of APOBEC3F (A3F) and APOBEC3B (A3B) but not that of two other cytidine deaminases also endowed with anti-HBV activity, activation-induced cytidine deaminase (AID), and APOBEC1. Most importantly, we reveal that blocking A3B, A3F, and A3G by combining RNA interference and the virion infectivity factor (Vif) protein of human immunodeficiency virus does not abrogate the inhibitory effect of IFNs on HBV. We conclude that these cytidine deaminases are not essential effectors of IFN in its action against this pathogen.
More than 350 millions individuals worldwide suffer from chronic hepatitis B virus (HBV) infection (66), a condition that evolves towards liver insufficiency and hepatocellular carcinoma in approximately 15 to 40% of cases (31). However, the majority of de novo HBV infections, except for those occurring during the perinatal period, are cleared during the acute phase. This appears to result largely from noncytopathic host defense mechanisms involving cytokines, most notably interferons (IFNs) (2, 21). A key role for IFN in the biology of HBV infection is further illustrated by studies in animal models of the disease. For instance, HBV-specific T cells from acutely infected chimpanzees produce high levels of gamma IFN (IFN-γ) coincident with viral clearance (23, 37). Also, the induction of alpha/beta IFN (IFN-α/β) or the production of IFN-γ by transferred HBV-specific cytotoxic T lymphocytes inhibits HBV replication in the liver of HBV transgenic mice (9, 19, 20, 35). Finally, IFN-α is the standard treatment for chronic hepatitis B (17). Although type I and II IFNs are the most important immune regulators of HBV replication, the downstream effectors mediating the inhibitory action of IFN on HBV replication have not yet been identified.
HBV replication is blocked by human APOBEC3G (A3G) and APOBEC3F (A3F) (43, 44, 57, 58), two closely related editing enzymes initially recognized for their ability to inhibit retroviruses (24, 29, 32, 49, 70) and as the targets of the virion infectivity factor (Vif) protein of human immunodeficiency virus (HIV) (34, 50, 68). APOBEC3B (A3B), a Vif-resistant member of the same family, exhibits modest levels of activity against HIV (3). Notwithstanding, the full-length protein APOBEC3BL (A3BL) has recently been found to be as potent as A3G in inhibiting HBV replication, in contrast to A3BS, which results from alternative splicing of the same transcript (6).
A3G, A3F, and A3B belong to a family of polynucleotide cytidine deaminases that also comprises activation-induced cytidine deaminase (AID), APOBEC1 (A1), APOBEC2 (A2), and the products of the APOBEC3A (A3A) through the APOBEC3H gene cluster. AID is essential for class switch recombination and somatic hypermutation of the immunoglobulin locus in pre-B lymphocytes and thus for the generation of antibody diversity (25), while A1 edits the apolipoprotein B mRNA in the guts, thereby regulating cholesterol metabolism (61). By comparison, little is known about the physiological functions of the other family members. However, apart from that for A2 and A3A, sequence analysis of the genes encoding these enzymes reveals that they rapidly accumulated nonsynonymous changes during primate evolution, suggesting that their products were involved in molecular conflicts and thus possibly evolved as a host defense against pathogens (46, 71). This hypothesis was confirmed for most APOBEC3 proteins, which were independently found to restrict the replication of various exogenous and endogenous retroelements, as well as the adeno-associated virus (4, 12, 14, 67). Like A3G, A3F, A3B, and A3DE can inhibit the replication of both Vif-defective (ΔVif) HIV type 1 (HIV-1) and ΔVif simian immunodeficiency virus (14, 67, 72), while A3C can block ΔVif simian immunodeficiency virus but not ΔVif HIV-1 (67). A3A restricts the parvovirus adeno-associated virus as well as autonomous non-long terminal repeat (LTR) retroelements, such as the human long interspersed nucleotide element 1 (L1) (5, 12, 36). L1 retrotransposition is also inhibited by A3B, A3C, and A3F (5, 12, 36, 52). Interestingly, A3G does not affect L1 retrotransposition. Nevertheless, A3G impairs L1-mediated Alu retrotransposition, along with A3A, A3B, and, to some extent, A3C (5, 13, 26).
Cytidine deaminases block HBV infection not by lethal editing of nascent reverse transcripts (38, 43, 57, 58), as for retroviruses (24, 29, 32, 70), but rather by impairing the accumulation of pregenomic RNA-containing viral capsids in which HBV reverse transcription takes place (57, 58). Remarkably, this is similar to the decreased steady-state levels of pregenomic RNA-containing capsids which are observed upon exposure of HBV-infected cells to IFN (63, 64). This suggests that IFNs and cytidine deaminases might affect HBV replication through the same pathway.
A first step to unveil the in vivo impact of antiviral cytidine deaminases on HBV replication was provided by the recent demonstration that the expression of A3G and, to a smaller degree, that of A3F and A3B is up-regulated in hepatocytes upon IFN treatment (6, 28, 45, 56). Likewise, it has been shown that alpha IFN (IFN-α) treatment induces the expression of A3G in macrophages, albeit not in T lymphocytes, the main target of HIV in the body. Notably, the IFN-α-mediated inhibition of HIV-1 in macrophages results at least in part from this induction of A3G, to levels or in a form that allows escape from Vif countering (41). These data led us to ask whether the induction of antiviral cytidine deaminases by cytokines similarly contribute to the IFN-induced clearance of HBV, as IFN-α and IFN-γ are both found in high amounts in the livers of acutely infected individuals.
For this, we first explored the range of cytidine deaminases active on HBV and their expression and cytokine inducibility in cells of hepatic origin. We then assessed their relevance for the anti-HBV effect of IFNs. Our results indicate that antiviral cytidine deaminases are not the main mediators of the action of IFNs on this pathogen.
MATERIALS AND METHODS
Constructs, virus production, infections, and titrations.
The plasmids expressing the hemagglutinin (HA)-tagged form of A3G (49) and A3A (3) were a kind gift from M. Malim (King's College, London, United Kingdom). The human A3B and A3F cDNAs were amplified from activated peripheral blood lymphocytes. We used the forward (cem196, 5′-AGATTAAGCTTGGCTGAACATGAATCCACAGATCAG-3′) and reverse (cem197, 5′-TTACTTCTAGAGTTTCCCTGATTCTGGAGAATGG-3′) primers for A3B and the forward (cem157, 5′-AGATTAAGCTTCCAAGGATGAAGCCTCACTTCAG-3′) and reverse (cem156, 5′-TTACTTCTAGACTCGAGAATCTCCTGCAGCTTGC-3′) primers for A3F. These cDNAs were then introduced in the HindIII and XbaI sites of the pCMV4/CEM15HA plasmid (49), replacing the human A3G cDNA. The resulting proteins correspond to the NP_004891 and NP_660341 NCBI entries, respectively. Note that A3B bears the known T146K variation. The cDNAs for A3C (pcDNA3.1-APOBEC3C-V5-6XHis), A2 (pcDNA3.1-APOBEC2-V5-6XHis), A1 (pcDNA3.1-APOBEC1-V5-6XHis), and AID (pcDNA3.1-AID-V5-6XHis) from B. Majita Peterlin and Yong-Hui Zheng (72) were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. All cDNAs for APOBEC family members were inserted into the same expression vector (pCMV4-HA). HBV was encoded by the payw1.2 construct (47), kindly provided by R. J. Schneider (New York University), and contains a 1.2 copy of the HBV genome (subtype ayw) from which the pregenomic and subgenomic RNAs are expressed under the control of the endogenous viral promoters. The murine stem cell virus-based pNG/A3G-HA retroviral vector and the corresponding empty vector pNG95 (49) were kindly provided by M. Malim (King's College, London, United Kingdom). The pNG/A3B-HA vector was constructed by replacing the A3G-HA cassette from the pNG/A3G-HA plasmid by the A3B-HA cassette generated by PCR using pCMV4/A3B-HA as the template and cem272/cem273 as primers (5′-AGACTAGTCGATAAGCTTGGCTGAACATGAATC-3′ and 5′-TTATCAATTGGACGCGTCCCGGGTCACTGAGCAGCGTAATC-3′, respectively). Murine leukemia virus-based particles were used to establish stable cell lines expressing either A3G-HA, A3B-HA, or the control vector and were produced by transient transfection of 293T cells with calcium phosphate using the pNG transfer vectors together with the packaging construct pCIG3 NB (kindly provided by O. Danos, Généthon-CNRS, Evry, France), the envelope plasmid pMD2G (http://tronolab.epfl.ch/page58122.html), and a Vif-expressing plasmid in the case of the pNG/A3G-HA vector. The simultaneous expression of HXB2 Vif and green fluorescent protein (GFP) was obtained through the use of a T2A self-cleaving peptide (55). For this, a cassette coding for this T2A peptide followed by a myc-tagged version of Vif was appended to the GFP 3′ end in the pRRLsin.cPPT.PGK.GFP.WPRE lentivector plasmid (16), yielding the pRRLsin.cPPT.PGK.GFP-2A-Vif-myc plasmid. The same construct encoding the C133S Vif mutant was used as a control vector and was generated using the QuikChange site-directed mutagenesis kit (Stratagene) and the forward (Vif35, 5′-GGACACATAGTTAGCCCTAGATCTGAATATCAAGCAGGAC-3′) and reverse (Vif36, 5′-GTCCTGCTTGATATTCAGATCTAGGGCTAACTATGTGTCC-3′) primers. Lentiviral particles were produced by transient transfection of 293T cells with calcium phosphate by use of the pRRL transfer vectors, the second-generation packaging construct pCMV-ΔR8.91, and the envelope plasmid pMD2G (see http://tronolab.epfl.ch/page58122.html for details). For the infectivity assay, ΔVif HIV-1 particles were produced by transient transfection of HepG2-H1.3 cells with a proviral clone (60) by use of JetPEI-Gal (Polyplus Transfection). Virus-containing supernatants were collected, and infections and titrations on target cells were performed as previously described (33). Briefly, virion release was scored by monitoring the reverse transcriptase (RT) enzymatic activity in the producer cell supernatant (1). Infectious titer was determined in a single-round infectivity assay by applying supernatant on HeLa-CD4-LTRLacZ indicator cells (11). Virion infectivity was derived by dividing the infectious titer by the amount of physical particles as determined with RT activity.
Generation of HepG2-H1.3.
HepG2-H1.3 cells were generated by stably transfecting human hepatoma HepG2 cells with the pTH1.3 plasmid containing a 1.3-fold-overlength HBV genome (51) and the pSV2neo plasmid (Clontech) conferring neomycin resistance.
DNA dot blot analysis of progeny HBV DNA.
HBV particles concentrated from 20 ml of cell culture medium at 4°C using centrifugal filter devices (Centricon Plus-70, Biomax 100,000-Da membrane; Millipore Corp., Bedford, MA) were sedimented through a cesium chloride step gradient (density, 1.15 to 1.4 g/ml) to separate unenveloped capsids from enveloped HBV virions, typically sedimenting at 1.2 g/ml (51). Gradient fractions were collected from the bottom. HBV DNA was detected by dot blot hybridization with a 32P-labeled HBV DNA probe.
Cell culture and transfections.
293T, P4.2, and Huh7 hepatoma cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS), 10 mM HEPES, 2 mM glutamine, and antibiotics (100 IU/ml penicillin, 100 mg/ml streptomycin). For the antiviral activity assay, six-well plates of Huh7 cells at 80% confluence were cotransfected with the payw1.2 plasmid and either a vector encoding one of the cytidine deaminases or an empty vector using FUGENE 6 (Roche) as recommended by the manufacturer. Inclusion of a GFP-expressing plasmid provided an internal control for transfection efficiency in each experiment. Where indicated, 20 μM 3TC (lamivudine; Moravek Biochemicals Inc.) was added to the culture at the time of cell washes after transfection. Cells were harvested 3 days after transfection for fluorescence-activated cell sorter analysis, protein analysis, and core-associated HBV DNA extraction. HepG2 hepatoma cells were maintained in Iscove's modified Dulbecco's medium containing 8% FCS, 2 mM glutamine, and antibiotics. HepG2-H1.3 cells were maintained in DMEM complemented with 10% FCS, 2 mM glutamine, antibiotics, 1% nonessential amino acids, and 200 μg/ml G418. Primary hepatocytes were freshly isolated from three different donors and seeded in a 24-multiwell plate (BD Bioscience). Donors included a 28-year-old male who tested negative for HIV, human T-cell leukemia virus, HBV, and HCV but positive for cytomegalovirus, a 44-year-old male who tested negative by the above-mentioned serology testing procedure, and a 4-year-old female with a history of Crigler-Najjar type I disease (hepatocytes from this last patient were kindly provided by T.H. Nguyen, University of Geneva). Upon reception, primary hepatocytes were maintained in serum-free and phenol red-free DMEM-F-12 medium to which only antibiotics were added. For cytidine deaminase induction experiments, cells were treated as indicated with 1,000 U/ml of human recombinant IFN-γ (Peprotech, Calbiochem), 10,000 U/ml of human recombinant IFN-α (Peprotech, Calbiochem), and 100 ng/ml of tumor necrosis factor alpha (TNF-α) (Peprotech), interleukin-6 (IL-6), IL-8 (Sigma), or IL-15 (Endogen). Primary hepatocytes were seeded in 24-well plates, treated, and harvested 24 h later for RNA extraction. Cell lines were treated each 2 days with the cytokines and harvested after 6 days of treatment for cellular RNA, protein, and core-associated HBV DNA extractions.
Protein analyses.
Cells were lysed with radioimmunoprecipitation assay buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate in phosphate-buffered saline). Lysates were precleared (13,000-rpm tabletop spin) and subjected to standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis. HA-tagged cytidine deaminases were detected with the mouse HA-specific monoclonal 3F10-peroxidase-conjugated antibody (Roche Applied Science). PCNA was used as a protein loading control and was detected with the monoclonal anti-PCNA antibody (Oncogene Research Products, Boston, MA). In order to visualize the IFN-induced endogenous A3G protein, we used a rabbit polyclonal antibody that specifically reacts with the C-terminal 16 amino acids of the enzyme (53). The expression of HIV-1 Vif was monitored using a rabbit HIV-1HXB2Vif antiserum (18). These antibodies were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Warner C. Greene and Dana Gabuzda, respectively.
Core-associated HBV DNA purification and analysis.
The purification of cytoplasmic core-associated HBV DNA was performed as previously described (58). Briefly, cells were disrupted in lysis buffer (100 mM Tris-HCl [pH 8.0], 0.2% NP-40), and the lysate was purified from nuclei and insoluble material by centrifugation. The resulting supernatant was treated with 200 μg/ml of DNase I and 100 μg/ml of RNase A for 2 h at 37°C. After removal of digested nucleic acids, the supernatant was incubated with 200 μg/ml of proteinase K for 1 h at 55°C. DNA was recovered through phenol-chloroform extraction and ethanol precipitation, resuspended in TE, pH 8, and finally treated with 100 ng/μl of RNase A for 30 min at 37°C. Purified HBV DNA was then subjected to quantification using quantitative real-time PCR with HBV primers and probes specific for the polymerase and pre-S1 regions (PB.HBV1, 5′-AAGGTAGGAGCTGGAGCATTCG; PB.HBV2, 5′-AGGCGGATTTGCTGGCAAAG; PB.HBVFAM1, 5′-6-carboxyfluorescein [FAM]-AGCCCTCAGGCTCAGGGCATAC-6-carboxytetramethylrhodame [TAMRA]) and Southern blot analysis.
RNA isolation and quantitative real-time RT-PCR analysis.
Total RNA was isolated using an RNeasy Plus kit (QIAGEN) according to the manufacturer's instructions. RNA was then converted into double-stranded cDNA by the Superscript II RT (Invitrogen) with random hexamers (Promega) as primers. For each sample, a control reaction lacking the RT was included. To compare the levels of expression of cytidine deaminases in different cell lines or in cells treated or not with cytokines, diluted cDNA was analyzed using primers and probes specific for A3G (cem162, 5′-ATGCAACCAGGCTCCACATAA; cem163, 5′-GGAATCACGTCCAGGAAGCA; FAM3G.2, 5′-FAM-CTTGAAGGCCGCCATGCAGAGC-TAMRA), A3F (cem177, 5′-CCTATGGTCGGAACGAAAGC; cem178, 5′-GCATGACAATGGGTCTCAGG; and FAM3F.1, 5′-FAM-TCCACCTGGTTTCGGAAGACGCC-TAMRA), A3B (cem172, 5′-CTATGGTCGGAGCTACACTTG; cem173, 5′-ACATTTCTGCGTGGTACTGAG; and FAM3B.1, 5′-FAM-AAATACACCTGGCCTCGAAAGACCC-TAMRA), A1 (huA1f, 5′-CCACTCTGAGGAGAAGAATCGAA; huA1r, 5′-GACAGGCCTCTTTACGAAGTTCTCT; and huA1.FAM, 5′-FAM-CATAGAAGACGTCAAACTCCCAGG-TAMRA), AID (huAIDf, 5′-TCCTTTTCACTGGACTTTGGTTATC; huAIDr, 5′-TGTAGCGGAGGAAGAGCAATTC; and huAID.FAM, 5′-FAM-TCGCAATAAGAACGGCTGCCACGT-TAMRA), and three of the four following housekeeping genes: those for TATA-box-binding protein (TBP) (huTBPf, 5′-GCCCGAAACGCCGAATATA; huTBPr, 5′-CGTGGCTCTCTTATCCTCATGA; and huTBP.PBE, 5′-FAM-CCGCAGCAAACCGCTTGGGABHQ), transferrin receptor (TFRC) (huTFRCf, 5′-CATTTGTGAGGGATCTGAACCA; huTFRCr, 5′-CGAGCAGAATACAGCCACTGTAA; and huTFRC.PBE, 5′-FAM-CAGGCCCATTTCCTTTATGTCTGCTCTGTABHQ), beta-2-microglobulin (β2MG) (huβ2MGf, 5′-TGCTCGCGCTACTCTCTCTTT; huβ2MGr, 5′-TCTGCTGGATGACGTGAGTAAAC; and huβ2MG.PBE, 5′-FAM-CTGGAGGCTATCCAGCGTACTCCAAAGATTBHQ), and eukaryotic elongation factor 1 alpha (eEF1A) (eEF1Af, 5′-AGCAAAAATGACCCACCAATG; eEF1Ar, 5′-GGCCTGGATGGTTCAGGATA; and eEF1A.PBE, 5′-FAM-CACCTGAGCAGTGAAGCCAGCTGCTTBHQ). Levels of expression of A3G and A3F in H9 and SupT1 human lymphoid cells were used as positive and negative controls, respectively. H9 cells express high levels of these proteins (49) and are thus nonpermissive for the replication of ΔVif HIV-1, whereas SupT1 cells fail to produce A3G or A3F and thus fully support ΔVif HIV replication. Total RNA from Huh7 cells transfected with A3B-, AID-, or A1-expressing constructs was used as a control for the amplification of these RNAs. To avoid amplification of potentially contaminating genomic DNA, all the primers were designed on exon junctions. For the standardization, the two most stable housekeeping genes out of three tested were selected by submitting the data to the GeNorm software (59). For each sample, a normalization factor was obtained by calculating the geometric mean of the values of the selected housekeeping genes and subsequently used to normalize the relative amounts of the RNAs of interest. The expression levels of the different APOBEC transcripts could be directly compared, since the values were also normalized for the real efficacy of the primers, which was calculated for each target sequence by determining the slope of the respective amplification curves.
RNA interference.
The following sense and antisense oligonucleotides were used for the cloning of small hairpin RNA (shRNA)-encoding sequences in lentiviral vectors: for shA3B, 5′-GATCCCCGGATGTATCGAGACACATTTTCAAGAGAAATGTGTCTCGATACATCCTTTTTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAGGATGTATCGAGACACATTTCTCTTGAAAATGTGTCTCGATACATCCGGG-3′ (antisense); and for shCtrl, 5′-GATCTCCCCGCATCGTGCACAGGAGTATTTCAAGAGAATACTCCTGTGCACGATGCTTTTTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAGCATCGTGCACAGGAGTATTCTCTTGAAATACTCCTGTGCACGATGCGGGGA-3′ (antisense). shCtrl contained seven mismatches compared to the A3G, A3F, and A3B mRNA sequences. The oligonucleotides were first annealed and ligated into the BglII and HindIII sites of the pSUPER plasmid (8). BamHI-SalI fragments of pSUPER which correspond to the cassette containing the H1 RNA polymerase III promoter and the hairpin sequence were then cloned into the BamHI and SalI sites of the pRDI292 lentivector providing puromycin resistance (7). Retroviral particles were produced by transient transfection of 293T cells with calcium phosphate by use of the pRDI transfer vector, the second-generation packaging construct psPAX2, and the envelope plasmid pMD2G (see http://tronolab.epfl.ch/page58122.html for details). Transduced HepG2-H1.3 cells were selected using 2 μg/ml puromycin.
RESULTS
A3G, A3F, A3B, AID, and A1 can inhibit HBV.
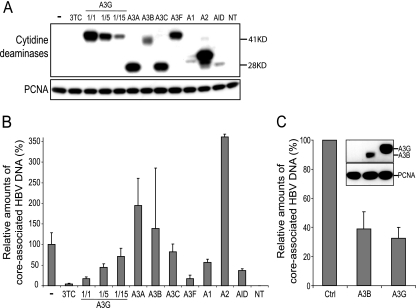
A3G, A3F, and A3BL were shown to inhibit HBV replication (6, 43, 44, 57, 58). We first tested whether other human polynucleotide cytidine deaminases can act on this virus. For this, we cotransfected Huh7 human hepatoma cells with an HBV-producing plasmid and either a control vector or vectors encoding HA-tagged forms of these enzymes. Transfection rates were similar, as confirmed by the inclusion of a GFP-expressing plasmid. Intracellular levels of tagged cytidine deaminases induced by this procedure varied, with A2, A3A, A3C, and A3G being the most abundant and A1 and AID exhibiting the lowest levels (Fig. 1A). Amounts of core-associated HBV DNA were measured by quantitative real-time PCR as previously described (58) (Fig. 1B). Among the well-expressed cytidine deaminases, A3G and A3F exerted a strong inhibition, almost as pronounced as that induced by the RT antagonist 3TC. Serial dilutions of the A3G plasmid confirmed that this protein acted in a dose-dependent manner. A2 and A3A were without effect on HBV, while A3C had only a minimal influence. In contrast, the lowly expressed A1 and AID consistently reduced levels of core-associated HBV DNA. Indeed, transfection of AID and A1 led to protein levels that were below those of the 15-fold-diluted A3G yet were twice as active. With the transiently expressed A3B, results were very variable from one experiment to the other, precluding any conclusion (Fig. 1B). To solve this issue, we generated HBV-producing hepatoma cells stably expressing the HA-tagged versions of A3G or A3B. In this context, expression of A3G and A3B reproducibly and similarly restricted HBV replication despite a lower expression of the A3B protein (Fig. 1C).
FIG. 1.
A3G, A3F, A3B, AID, and A1 can inhibit HBV. (A) Western blot analysis of cytoplasmic extracts from HBV-transfected Huh7 cells in the presence of either 3TC or the indicated HA-tagged cytidine deaminase, including dilutions of HA-A3G. The protein expression levels at day 3 posttransfection were estimated using an anti-HA antibody, while PCNA served as protein loading control (NT, nontransfected). Molecular masses in kDa are given to the right. (B) Quantitative real-time PCR analysis of core-associated HBV DNA purified from HBV-transfected Huh7 cells in the presence of either 3TC or the indicated cytidine deaminase at day 3 posttransfection. This is one representative experiment out of three. Values represent means ± standard deviations of two independent duplicates and are expressed relative to the amounts of core-associated HBV DNA produced in HBV-transfected control cells, which were given the arbitrary value of 100%. The antiviral activity of A3B was assessed overall in five independent experiments, which consistently triggered highly variable results. (C) Quantitative real-time PCR analysis of core-associated HBV DNA purified from HBV-producing cells stably expressing the HA-tagged A3B or A3G. Values are expressed relative to the amounts of core-associated HBV DNA in HBV-producing cells transduced with the control vector (Ctrl), which were given the arbitrary value of 100%, and represent means ± standard deviations of two independent experiments, one of which was performed in duplicate. (Inset) Protein levels of the HA-tagged A3B and A3G in the cell lines were assessed by Western blotting using an anti-HA antibody and PCNA as the protein loading control.
Baseline levels of expression of cytidine deaminases in human hepatocytes.
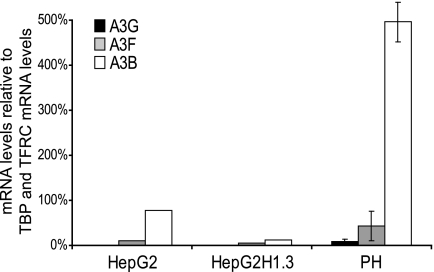
We then examined the baseline levels of expression of the cytidine deaminases endowed with anti-HBV activity in human hepatic cells by use of quantitative real-time RT-PCR. As expected, neither transcripts of B lymphocyte-specific AID nor those of the gut-specific A1 were detected in any of the analyzed cells (data not shown). Expression of A3G was undetectable in various immortalized human hepatocytic cell lines, including the immortalized line IHH10.3 (39), the hepatoma lines Huh7 and HepG2, and its HBV-producing HepG2-H1.3 derivative (see Fig. 4A), while A3F was lowly expressed in these cells, reaching at most 20% of the TBP and TFRC RNA levels, which were used as internal controls (Fig. 2 and data not shown). In primary human hepatocytes, levels of A3G and A3F RNA represented 10% and 40%, respectively, of the TBP and TFRC RNA levels. A3B expression was higher in most tested cells, notably reaching 500% of the TBP and TFRC RNA levels in primary hepatocytes (Fig. 2 and data not shown). Interestingly, expression levels of the three antiviral cytidine deaminases were particularly low in HepG2-H1.3 cells (Fig. 2), a condition which perhaps facilitated the establishment of this constitutively HBV-producing derivative of HepG2 hepatoma cells.
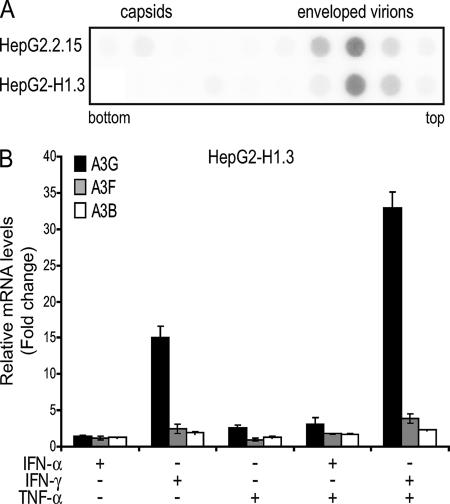
FIG. 4.
Infectious HBV particle production and IFN-mediated induction of antiviral cytidine deaminases in HepG2-H1.3 cells. (A) Secretion of HBV virions from HBV-producing hepatoma cell lines. HBV was concentrated from cell culture medium of HepG2.2.15 and HepG2-H1.3, and obtained virus stocks were sedimented through a CsCl step gradient (1.2 to 1.4 g/ml). The different gradient fractions from bottom to top were analyzed by dot blot analysis using a 32P-labeled HBV DNA probe. (B) A3G and, to a small extent, A3F and A3B are induced in HepG2-H1.3 cells upon IFN-γ treatment. Levels of A3G, A3F, and A3B gene expression were assessed by quantitative real-time RT-PCR 24 h after treatment with the indicated cytokines. Quantities were normalized using TBP and TFRC and expressed as change (n-fold) relative to the baseline levels obtained from untreated HepG2-H1.3 cells. Values represent means ± standard deviations for independent duplicates.
FIG. 2.
Baseline levels of A3G, A3F, and A3B in human hepatocytes. Levels of A3G, A3F, and A3B gene expression in various hepatic cells were evaluated by quantitative real-time RT-PCR and expressed relative to the levels of the two normalization genes, the TBP and TFRC genes. Amounts in primary hepatocytes represent means ± standard deviations of values obtained from three donors. PH, primary hepatocytes.
A3G, A3F, and A3B are induced upon cytokine treatment of hepatocytes.
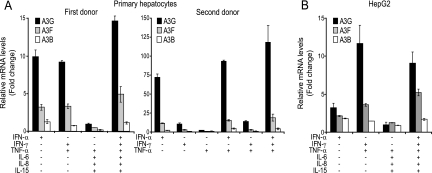
It was recently demonstrated that IFN-α treatment up-regulates A3G, A3F, A3B, and A3C in primary human hepatocytes as well as in hepatocarcinoma cell lines (6, 28, 45, 56). However, it is still not known whether the expression of cytidine deaminases endowed with anti-HBV activity is up-regulated by treatment with other cytokines, notably those known to be induced during acute HBV infection. We thus treated hepatic cells with IFN-α, IFN-γ, TNF-α, IL-6, IL-8, and IL-15 alone or in various combinations and measured the levels of A3G, A3F, A3B, A1, and AID mRNA 24 h later by quantitative real-time RT-PCR. A3G was induced about 10-fold in primary hepatocytes from a first donor upon treatment with either IFN-α or IFN-γ combined with TNF-α and up to 15-fold when all cytokines were used together (Fig. 3A, left). A3F exhibited the same pattern, albeit with lower levels of induction, while A3B was less than 1.5 times up-regulated. In contrast, A1 and AID mRNA remained below the threshold of detection in all conditions tested (data not shown). Similar results were obtained with primary hepatocytes from a second donor, although in this case the induction of antiviral cytidine deaminases was very strong upon IFN-α stimulation, with A3G reaching levels six times higher than those observed upon IFN-γ treatment (Fig. 3A). There thus appear to be donor-dependent variations in the regulation of APOBEC proteins. Likewise, A3G, A3F, and, to a small degree, A3B were induced by IFN-γ plus TNF-α in hepatoma cell lines, including Huh7 and HepG2 cells (Fig. 3B and data not shown). Of note is the fact that IFN-α alone, the therapeutic agent used to treat chronic HBV infection, significantly up-regulated A3G expression exclusively in primary hepatocytes (Fig. 3).
FIG. 3.
A3G, A3F, and A3B are induced in primary hepatocytes (A) and HepG2 cells (B) upon cytokine treatment. Levels of A3G, A3F, and A3B gene expression were assessed by quantitative real-time RT-PCR 24 h after treatment with the indicated cytokines. Quantities were normalized using two of the following housekeeping genes: β2MG, TBP, TFRC, and eEF1A. Normalized quantities were expressed as change (n-fold) relative to the baseline levels obtained from untreated hepatocytes. Values represent means ± standard deviations for independent triplicates.
Inhibition of HBV replication by IFNs is independent of antiviral cytidine deaminases.
IFN-γ and IFN-α are demonstrated inhibitors of HBV replication both in vitro and in vivo (9, 10, 15, 22, 35, 40, 42, 54), and both cytokines induce the expression of cytidine deaminases capable of blocking this virus (Fig. 3). We thus investigated a possible link between these two observations by asking whether IFNs conserve their inhibitory effect on HBV when A3G, A3F, and A3B are blocked. To address this point, we used HBV-producing HepG2-H1.3 cells. These cells release enveloped HBV virions at levels comparable to the previously characterized HepG2.2.15 cells (48), but in contrast to this other line they are sensitive to IFNs (D. Webb, F. Bohne, M. Hösel, and U. Protzer, unpublished data) (Fig. 4A and data not shown). Upon IFN-γ, IFN-α, and TNF-α treatment, HepG2-H1.3 cells exhibited patterns of A3G induction similar to those observed for other hepatoma cell lines, while A3F and A3B were both less than fourfold up-regulated (Fig. 4B).
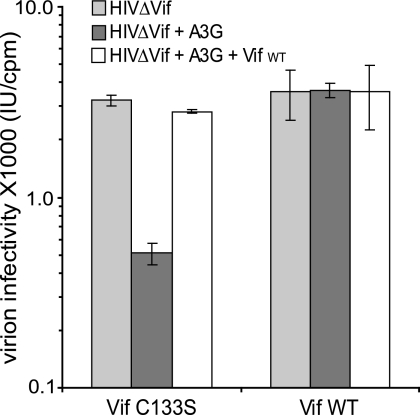
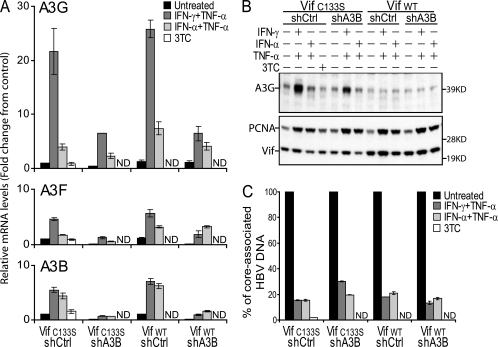
In order to suppress the consequences of the induction of A3F, A3G, and A3B by IFN, we used a combined protein- and RNA-based approach. We first took advantage of HIV-1 Vif, which counters A3G and A3F by triggering their proteasomal degradation (30, 50, 53). We previously demonstrated that Vif rescues HBV DNA production in cells overexpressing A3G and A3F (reference 58 and data not shown), which explained our approach. We thus transduced HepG2-H1.3 cells with lentiviral vectors expressing either wild-type Vif or the C133S Vif mutant, which still interacts with A3G but fails to induce its degradation (69). We verified the functionality of wild-type Vif in HepG2-H1.3 cells by demonstrating that it could counter the effect of A3G on ΔVif HIV-1, in contrast to the C133S Vif mutant (Fig. 5). However, Vif is inactive against A3B, and in spite of the low levels of induction of this enzyme in the IFN-treated HepG2-H1.3 cells, we further modified the two cell lines by transduction with lentiviral vectors expressing either a shRNA directed to A3B (shA3B) or a mismatched shRNA (shCtrl) as a negative control. Treatment with IFN-γ plus TNF-α induced A3G RNA expression in HepG2-H1.3 cells expressing shCtrl by 25-fold, while induction of A3F and A3B was markedly lower (Fig. 6A). Upon IFN-α-plus-TNF-α treatment, A3G and A3B were about fivefold up-regulated, while levels of A3F increased three times at most. In cells expressing shA3B, A3B expression was effectively suppressed, whether at baseline or upon cytokine treatment. Surprisingly, A3F and A3G transcripts were also down-regulated by shA3B, albeit to a lesser extent, in spite of one mismatch between their sequences and that of the shRNA (Fig. 6A). The increased transcription of A3G in cells expressing shCtrl and the C133S Vif mutant correlated with increased levels of the protein, as verified by Western blotting, which also confirmed that A3G was completely suppressed in the presence of wild-type Vif (Fig. 6B). Per the lack of antibodies recognizing endogenous A3F and A3B, we could not assess the protein levels of these cytidine deaminases, but the results of the RNA analyses demonstrated their very low levels of expression in wild-type Vif- and shA3B-expressing cells, even upon IFN treatment. Most importantly, neither Vif nor either of the two shRNAs affected baseline levels of HBV replication (data not shown).
FIG. 5.
ΔVif-HIV-1 infectivity is rescued in HepG2-H1.3 cells stably expressing the wild-type Vif. CD4+, HIV-1 LTR-LacZ-containing HeLa-derived P4.2 cells were infected with ΔVif HIV-1 exposed or not to A3G during production in HepG2-H1.3 cells expressing either the C133S mutant or the wild-type Vif (Vif WT), as indicated. Viral production in the presence of cotransfected A3G- and wild-type Vif-expressing vectors was used as a control. Values represent means ± standard deviations for independent duplicates. IU, infectious units.
FIG. 6.
A3G, A3F, and A3B are not required for the antiviral effect of IFNs against HBV. Doubly transduced HepG2-H1.3 cells were treated every 2 days with IFN-γ plus TNF-α, IFN-α plus TNF-α, or the RT inhibitor 3TC as indicated and collected after 6 days for total RNA, protein, and core-associated HBV DNA analyses. ND, not determined. (A) Levels of A3G, A3F, and A3B gene expression were assessed by quantitative real-time RT-PCR. Quantities were normalized using TFRC and TBP and expressed as change (n-fold) relative to the baseline levels obtained from untreated control HepG2-H1.3 cells (Vif C133S shCtrl). Values represent means ± standard deviations for independent duplicates. (B) Aliquots of the lysates were subjected to Western blot analysis using anti-A3G, anti-Vif, and anti-PCNA antibodies. Each lane is a pool of independent duplicates. Molecular masses in kDa are given to the right. (C) Core-associated HBV DNA was purified from the remaining lysate and analyzed by quantitative real-time PCR. Levels are expressed relative to the amounts of core-associated HBV DNA produced in the different untreated HepG2-H1.3 cell lines, which were given the arbitrary value of 100%. Values represent means ± standard deviations for independent duplicates. Data are from one independent experiment representative of two. WT, wild type.
IFN-γ combined with TNF-α induced an approximately 80% decrease in core-associated HBV DNA both in control C133S Vif- and shCtrl-expressing HepG2-H1.3 cells and in wild-type Vif- and shCtrl-expressing cells, demonstrating that A3G and A3F are not essential effectors of the IFN-γ-induced inhibition of HBV replication (Fig. 6C). Moreover, HBV was similarly suppressed by IFN-γ-plus-TNF-α treatment in wild-type Vif- and shA3B-expressing cells, in which all three antiviral cytidine deaminases are blocked. This was in agreement with our finding that when combined with TNF-α, IFN-α was as active as IFN-γ at blocking HBV, whereas only the latter regimen significantly up-regulates the expression of A3G, A3F, and A3B in these targets (Fig. 6). These results converge to indicate that the anti-HBV effect of IFNs does not stem from the induction of antiviral cytidine deaminases, at least in this system.
DISCUSSION
In this study, we first show that, in addition to A3G, A3F, and A3B, two other human polynucleotide cytidine deaminases, AID and A1, can inhibit HBV replication. Whether these two enzymes act by editing the viral genome or by some other mechanism was not investigated. Indeed, AID and A1 were not found to be expressed in hepatocytes or induced in these cells upon treatment with relevant cytokines, making it unlikely that they affect HBV replication in vivo.
In contrast, the expression of A3G, of A3F, and, to a small extent, of A3B was induced by type I and II IFNs in hepatic cells, particularly when IFN-γ was combined with TNF-α. This is interesting because these two cytokines have long been known to play a key role during the acute phase of HBV infection and act synergistically to curtail HBV replication in vitro (27). As suggested by the recent demonstration that IFN-α-induced inhibition of HIV in macrophages is partly mediated by APOBEC3 enzymes (41), a similar involvement of cytidine deaminases might have accounted for the IFN-induced block of HBV. Here, we demonstrate that in spite of the induction of A3G, A3F, and A3B in hepatoma cells exposed to IFNs, the cytokine-mediated inhibition of HBV replication is not affected when the expression of these cytidine deaminases is suppressed by combining RNA interference and the Vif protein of HIV-1. From these data, we conclude that there must be other effectors that account for the antiviral action of IFNs, at least in this system.
While demonstrating that A3G, A3F, and A3B are not required for the inhibition of HBV by IFNs in cell culture, these results do not imply that antiviral cytidine deaminases have no relevance for HBV biology and therapy. First, the A3G protein is weakly expressed in IFN-treated HepG2-H1.3 compared to H9 lymphoid cells. As cytidine deaminases act in a dose-dependent manner (58), they may have a greater impact on HBV replication upon stronger expression, which may occur in the infected liver. Thus, it is not excluded that antiviral cytidine deaminases act in concert with other yet-undefined IFN-induced effectors during the clearance of HBV in vivo. Some genes were previously shown to be specifically up-regulated during HBV clearance in both chimpanzees and IFN-treated mouse hepatocytes, and thus they represent potential mediators of the antiviral effect of IFNs against HBV (62, 65). However, it remains to be clarified whether these genes are induced in the hepatocytes or in the surrounding inflammatory cells in the liver.
To assess further the role of antiviral cytidine deaminases in the IFN-mediated clearance of HBV, it might be revealing to compare levels of A3G, A3F, and A3B in the livers of uninfected, acutely infected, and chronically infected individuals. Informative data might also be obtained by looking at possible interindividual differences in the sequences of the antiviral cytidine deaminase promoters, particularly with respect to IFN-responsive elements. As well, an evaluation of the sensitivity to A3G, A3F, and A3B of HBV strains isolated at various stages of the disease could be indicative.
Finally, IFN-α treatment rarely leads to the clearance of HBV and in addition triggers very strong systemic symptoms, and thus it is poorly tolerated. Therefore, the induction of antiviral cytidine deaminases in the liver, by either genetic or pharmacological means, can still be envisioned as a therapeutic strategy to treat chronically infected patients.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation and the Strauss Foundation to D.T. and from the Deutsche Forschungsgemeinschaft to U.P.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biermer, M., R. Puro, and R. J. Schneider. 2003. Tumor necrosis factor alpha inhibition of hepatitis B virus replication involves disruption of capsid integrity through activation of NF-κB. J. Virol. 77:4033-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonvin, M., F. Achermann, I. Greeve, D. Stroka, A. Keogh, D. Inderbitzin, D. Candinas, P. Sommer, S. Wain-Hobson, J. P. Vartanian, and J. Greeve. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43:1364-1374. [DOI] [PubMed] [Google Scholar]
- 7.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 8.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 9.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1998. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J. Virol. 72:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1997. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J. Virol. 71:3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480-485. [DOI] [PubMed] [Google Scholar]
- 13.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumortier, J., K. Schonig, H. Oberwinkler, R. Low, T. Giese, H. Bujard, P. Schirmacher, and U. Protzer. 2005. Liver-specific expression of interferon gamma following adenoviral gene transfer controls hepatitis B virus replication in mice. Gene Ther. 12:668-677. [DOI] [PubMed] [Google Scholar]
- 16.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 17.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 350:1118-1129. [DOI] [PubMed] [Google Scholar]
- 18.Goncalves, J., P. Jallepalli, and D. H. Gabuzda. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., P. Borrow, M. V. Hobbs, B. Matzke, I. Gresser, M. B. Oldstone, and F. V. Chisari. 1996. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc. Natl. Acad. Sci. USA 93:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guidotti, L. G., and F. V. Chisari. 1999. Cytokine-induced viral purging—role in viral pathogenesis. Curr. Opin. Microbiol. 2:388-391. [DOI] [PubMed] [Google Scholar]
- 21.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 22.Guidotti, L. G., T. Ishikawa, M. V. Hobbs, B. Matzke, R. Schreiber, and F. V. Chisari. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25-36. [DOI] [PubMed] [Google Scholar]
- 23.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 24.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 25.Honjo, T., M. Muramatsu, and S. Fagarasan. 2004. AID: how does it aid antibody diversity? Immunity 20:659-668. [DOI] [PubMed] [Google Scholar]
- 26.Hulme, A. E., H. P. Bogerd, B. R. Cullen, and J. V. Moran. 2007. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawanishi, Y., N. Hayashi, K. Katayama, K. Ueda, T. Takehara, E. Miyoshi, E. Mita, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Tumor necrosis factor-alpha and interferon-gamma inhibit synergistically viral replication in hepatitis B virus-replicating cells. J. Med. Virol. 47:272-277. [DOI] [PubMed] [Google Scholar]
- 28.Komohara, Y., H. Yano, S. Shichijo, K. Shimotohno, K. Itoh, and A. Yamada. 2006. High expression of APOBEC3G in patients infected with hepatitis C virus. J. Mol. Histol. 37:327-332. [DOI] [PubMed] [Google Scholar]
- 29.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 30.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lok, A. S. 2002. Chronic hepatitis B. N. Engl. J. Med. 346:1682-1683. [DOI] [PubMed] [Google Scholar]
- 32.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 33.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 34.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 35.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 37.Murray, J. M., S. F. Wieland, R. H. Purcell, and F. V. Chisari. 2005. Dynamics of hepatitis B virus clearance in chimpanzees. Proc. Natl. Acad. Sci. USA 102:17780-17785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen, D. H., S. Gummuluru, and J. Hu. 2007. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J. Virol. 81:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen, T. H., G. Mai, P. Villiger, J. Oberholzer, P. Salmon, P. Morel, L. Buhler, and D. Trono. 2005. Treatment of acetaminophen-induced acute liver failure in the mouse with conditionally immortalized human hepatocytes. J. Hepatol. 43:1031-1037. [DOI] [PubMed] [Google Scholar]
- 40.Pasquetto, V., S. F. Wieland, S. L. Uprichard, M. Tripodi, and F. V. Chisari. 2002. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J. Virol. 76:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng, G., K. J. Lei, W. Jin, T. Greenwell-Wild, and S. M. Wahl. 2006. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 203:41-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romero, R., and J. E. Lavine. 1996. Cytokine inhibition of the hepatitis B virus core promoter. Hepatology 23:17-23. [DOI] [PubMed] [Google Scholar]
- 43.Rosler, C., J. Kock, M. Kann, M. H. Malim, H. E. Blum, T. F. Baumert, and F. von Weizsacker. 2005. APOBEC-mediated interference with hepadnavirus production. Hepatology 42:301-309. [DOI] [PubMed] [Google Scholar]
- 44.Rosler, C., J. Kock, M. H. Malim, H. E. Blum, and F. von Weizsacker. 2004. Comment on “Inhibition of hepatitis B virus replication by APOBEC3G.” Science 305:1403. [DOI] [PubMed] [Google Scholar]
- 45.Sarkis, P. T., S. Ying, R. Xu, and X. F. Yu. 2006. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J. Immunol. 177:4530-4540. [DOI] [PubMed] [Google Scholar]
- 46.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scaglioni, P. P., M. Melegari, and J. R. Wands. 1997. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J. Virol. 71:345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sells, M. A., M. L. Chen, and G. Acs. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 84:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 50.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 51.Sprinzl, M. F., H. Oberwinkler, H. Schaller, and U. Protzer. 2001. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: crossing the species barrier. J. Virol. 75:5108-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 53.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 54.Suri, D., R. Schilling, A. R. Lopes, I. Mullerova, G. Colucci, R. Williams, and N. V. Naoumov. 2001. Non-cytolytic inhibition of hepatitis B virus replication in human hepatocytes. J. Hepatol. 35:790-797. [DOI] [PubMed] [Google Scholar]
- 55.Szymczak, A. L., C. J. Workman, Y. Wang, K. M. Vignali, S. Dilioglou, E. F. Vanin, and D. A. Vignali. 2004. Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat. Biotechnol. 22:589-594. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, Y., H. Marusawa, H. Seno, Y. Matsumoto, Y. Ueda, Y. Kodama, Y. Endo, J. Yamauchi, T. Matsumoto, A. Takaori-Kondo, I. Ikai, and T. Chiba. 2006. Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem. Biophys. Res. Commun. 341:314-319. [DOI] [PubMed] [Google Scholar]
- 57.Turelli, P., S. Jost, B. Mangeat, and D. Trono. 2004. Response to comment on “Inhibition of hepatitis B virus replication by APOBEC3G.” Science 305:1403. [DOI] [PubMed] [Google Scholar]
- 58.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 59.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 60.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19:207-216. [DOI] [PubMed] [Google Scholar]
- 62.Wieland, S., R. Thimme, R. H. Purcell, and F. V. Chisari. 2004. Genomic analysis of the host response to hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 101:6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wieland, S. F., A. Eustaquio, C. Whitten-Bauer, B. Boyd, and F. V. Chisari. 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc. Natl. Acad. Sci. USA 102:9913-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wieland, S. F., R. G. Vega, R. Muller, C. F. Evans, B. Hilbush, L. G. Guidotti, J. G. Sutcliffe, P. G. Schultz, and F. V. Chisari. 2003. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J. Virol. 77:1227-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.World Health Organization. October 2000. revision date. Hepatitis B. World Health Organization Fact Sheet 204. http://www.who.int/mediacentre/factsheets/fs204/en/index.html.
- 67.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 68.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 69.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, J., and D. M. Webb. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 13:1785-1791. [DOI] [PubMed] [Google Scholar]
- 72.Zheng, Y.-H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]