Abstract
An intact B-box 2 domain is essential for the antiretroviral activity of TRIM5α. We modeled the structure of the B-box 2 domain of TRIM5α based on the existing three-dimensional structure of the B-box 2 domain of human TRIM29. Using this model, we altered the residues predicted to be exposed on the surface of this globular structure. Most of the alanine substitutions in these residues exerted little effect on the antiretroviral activity of human TRIM5αhu or rhesus monkey TRIM5αrh. However, alteration of arginine 119 of TRIM5αhu or the corresponding arginine 121 of TRIM5αrh diminished the abilities of the proteins to restrict retroviral infection without affecting trimerization or recognition of the viral capsid. The abilities of these functionally defective TRIM5α proteins to accelerate the uncoating of the targeted retroviral capsid were abolished. Removal of the positively charged side chain from B-box 2 arginines 119/120/121 resulted in diminished proteasome-independent turnover of TRIM5α and the related restriction factor TRIMCyp. However, testing of an array of mutants revealed that the rapid turnover and retroviral restriction functions of this B-box 2 region are separable.
Proteins of the tripartite motif (TRIM) family contain RING, B-box, and coiled-coil domains and thus have also been referred to as RBCC proteins (25). TRIM proteins often self-associate and form aggregates called nuclear or cytoplasmic bodies (3, 25, 27). Although TRIM proteins have been implicated in transcriptional regulation, cell division, retroviral restriction, determination of cell polarity, and differentiation, the precise functions of most TRIM proteins remain to be determined (15, 19, 34).
TRIM5α was identified in a random genetic screen as the factor responsible for the early postentry block to human immunodeficiency virus type 1 (HIV-1) in Old World monkeys (30). TRIM5α is a cytoplasmic protein that is capable of restricting infection by different retroviruses in a species-dependent manner (13, 29). Variation among TRIM5α proteins in different primates accounts for the early postentry blocks to infection by particular retroviruses (5, 9, 12, 13, 37). For example, the TRIM5α proteins of several Old World monkey species block HIV-1 infection (8, 13, 18), whereas the TRIM5α proteins of New World monkeys block infection by simian immunodeficiency virus (29). TRIM5α from humans (TRIM5αhu) is not as potent in restricting HIV-1 infection as Old World monkey TRIM5α proteins, but TRIM5αhu potently restricts other retroviruses, e.g., N-tropic murine leukemia virus (N-MLV), equine infectious anemia virus (EIAV), and feline immunodeficiency virus (FIV) (8, 13, 21, 26, 37).
Variation in splicing of the TRIM5 primary transcript leads to the expression of TRIM5 isoforms, designated α, γ, and δ (25). The TRIM5α isoform contains, in addition to the RING, B-box 2, and coiled-coil domains, a carboxy-terminal B30.2(SPRY) domain. The B30.2(SPRY) domain is essential for the antiretroviral activity of TRIM5α (32). In some cases, the differences in the abilities of TRIM5α proteins from various primate species to restrict particular retroviruses are determined by sequences in the B30.2(SPRY) domain (20, 22, 28, 35). The B30.2 domain of rhesus monkey TRIM5α (TRIM5αrh) is required for specific recognition of the HIV-1 capsid (31). HIV-1 capsid recognition is also dependent on TRIM5αrh trimerization, which is mediated by the coiled-coil and adjacent linker 2 regions (4, 12, 16).
Disruption of the TRIM5αrh or TRIM5αhu B-box 2 domain by mutagenesis resulted in loss of retroviral restriction (11, 22, 24). Thus, it has been suggested that the B-box 2 TRIM5 mutants may lack an “effector” function important for restriction. Studies following the fate of the HIV-1 and N-MLV capsids in the cytoplasm of cells expressing TRIM5αrh or TRIM5αhu, respectively, indicate that the conversion of particulate capsids to soluble capsid proteins is accelerated by a restricting TRIM5α protein (24, 31). Thus, the B-box 2 domain may contribute to this process.
In this work, we modeled the structure of the B-box 2 domains of TRIM5αrh and TRIM5αhu based on the available B-box 2 domain structure from the TRIM29 protein (9). The modeled TRIM5 structure revealed that the B-box 2 domain is globular and highly charged on its surface. We tested the hypothesis that residues predicted to be on the surface of the B-box 2 domain contribute to the “effector” function of TRIM5α. These residues were individually altered, and the phenotypes of the mutants were characterized. We found that the positively charged arginine 121 residue on the surface of the B-box 2 domain of TRIM5αrh was important for the restriction of several retroviruses, including HIV, EIAV, and FIV. Moreover, the homologous residue, arginine 119, of TRIM5αhu was found to be essential for the restriction of N-MLV, EIAV, and FIV. Studies of the oligomerization, capsid-binding abilities, and effects of these mutant proteins on the infecting retroviral capsid suggested that this B-box 2 residue contributes to the ability of TRIM5α to accelerate the conversion of the particulate cytosolic capsid to soluble capsid proteins. The analysis of a larger panel of mutants involving residue 121 of TRIM5αrh or residue 119 of TRIM5αhu revealed that changing the positively charged side chain of this B-box 2 residue dramatically reduced TRIM5α turnover. The B-box 2-dependent turnover of TRIM5α was not inhibited by proteasome inhibitors. Some B-box 2 mutants of TRIM5αrh and TRIM5αhu were long-lived but exhibited potent antiretroviral activity. Thus, the rapid turnover and effector functions of this TRIM5α B-box 2 region are separable.
MATERIALS AND METHODS
Modeling the TRIM5α B-box 2 domain.
The structure of the human TRIM5α B-box 2 domain and the immediate flanking regions was modeled using the MODELLER (version 9.0) program (5) in Discovery Studio 1.7 (Accelrys Software, Inc., San Diego, CA). The structure of the TRIM29 B-Box 2 domain (Protein Data Bank file 2CSV) was used as the template for homology-based protein modeling (9). The B-box protein sequences from human TRIM5α and TRIM29 exhibit 50% sequence identity, and the TRIM5α model was built based on the sequence alignment. The model was further refined through loop and side chain refinement and energy minimization. The coordinates of zinc atoms were derived from the template.
Creation of cells stably expressing TRIM5α variants.
Retroviral vectors encoding wild-type or mutant TRIM5αhu, TRIM5αrh, or TRIMCyp proteins were created using the pLPCX vector. Recombinant viruses were produced in 293T cells by cotransfecting the pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus (VSV) G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells. Cf2Th canine thymocytes were transduced and selected in 5 μg/ml puromycin (Sigma).
Infection with viruses expressing GFP.
Recombinant HIV-1, N-MLV, and B-tropic murine leukemia virus (B-MLV) expressing green fluorescent protein (GFP) were prepared as described previously (19). For infections, 3 × 104 Cf2Th cells seeded in 24-well plates were incubated with virus for 24 h. The cells were washed and returned to culture for 48 h and then subjected to fluorescence-activated cell sorter analysis with a FACScan (Becton Dickinson). The FIV vector expressing GFP was obtained from System Biosciences (Mountain View, CA) and was prepared following the manufacturer's instructions. The EIAV vector expressing GFP was a gift from John Olsen (21). HIV-1, N-MLV, B-MLV, FIV, and EIAV viral stocks were titrated by serial dilution on Cf2Th cells to determine the concentrations of infectious viruses.
Protein analysis.
Cellular proteins were extracted with radioimmunoprecipitation assay buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 2 mg/ml aprotinin, 2 mg/ml leupeptin, 1 mg/ml pepstatin A, 100 mg/ml phenylmethylsulfonyl fluoride). The cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (10% acrylamide), followed by blotting them onto nitrocellulose membranes (Amersham Pharmacia Biotech). The detection of protein by Western blotting utilized monoclonal antibodies directed against the hemagglutinin (HA) epitope tags (Roche) and monoclonal antibodies to β-actin (Sigma). Detection of proteins was performed by enhanced chemiluminescence (NEN Life Sciences Products), using anti-mouse immunoglobulin (for β-actin) and anti-rat immunoglobulin (for HA) secondary antibodies (Amersham Pharmacia Biotech).
TRIM5α turnover.
The turnover rates of wild-type and mutant TRIM5α proteins were estimated as previously described (3). Briefly, cells expressing the TRIM5 proteins were incubated with medium containing 100 μg/ml cycloheximide, and the steady-state levels of TRIM5 protein were determined by Western blotting cell lysates for the HA epitope tags or, as a control, for β-actin, as described above. In some cases, TRIM5α proteins with HA epitope tags at both the N and C termini were studied. In some of the experiments, cycloheximide-treated cells were also incubated with the proteasome inhibitor MG115 (50 μM), MG132 (50 μM), or clasto-lactacystin (10 μM) before lysis and Western blotting as described above.
Localization of wild-type and mutant TRIM5α proteins.
The localization of TRIM5αrh, TRIM5αhu, or TRIMCyp variants in expressing cells was investigated as previously described (2). Briefly, cells were grown overnight on 12-mm-diameter coverslips and fixed in 3.9% paraformaldehyde (Sigma) in phosphate-buffered saline (PBS) (Cellgro) for 30 min. The cells were washed in PBS, incubated in 0.1 M glycine (Sigma) for 10 min, washed in PBS, and permeabilized with 0.05% saponin (Sigma) for 30 min. Samples were blocked with 10% donkey serum (Dako, Carpinteria, CA) for 30 min and incubated for 1 h with antibodies. The anti-HA fluorescein isothiocyanate-conjugated 3F10 antibody (Roche) was used to stain HA-tagged proteins. Subsequently, samples were mounted for fluorescence microscopy by using the ProLong Antifade Kit (Molecular Probes, Eugene, OR). Images were obtained with a Bio-Rad Radiance 2000 laser scanning confocal microscope with Nikon 60× 1.4 numerical aperture optics.
TRIM5α oligomerization.
Approximately 1 × 107 293T cells transfected with 5 μg of plasmids expressing TRIM5αrh or TRIM5αhu variants were lysed in immunoprecipitation buffer and cross-linked with different concentrations (0 to 10 mM) of glycolbis-succinimidylsuccinate for 30 min as previously described (11). The cross-linking reaction was stopped by adding 10 μl of 1 M Tris-HCl. After the cross-linking, samples were resuspended in 2× sample buffer and incubated at 37°C for 30 min. The samples were analyzed by SDS-PAGE and Western blotting with an anti-HA antibody (Roche).
HIV CA-NC expression and purification.
HIV-1 CA-NC protein was expressed, purified, and assembled as previously described (6, 7). The pET11a expression vector (Novagen) expressing the CA-NC protein of HIV-1 was used to transform Escherichia coli BL-21(DE3). CA-NC expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the culture reached an optical density of 0.6 at 600 nm. After 4 h of induction, the cells were harvested and resuspended in 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, 10 mM 2-mercaptoethanol, and protease inhibitors (Roche). Lysis was performed through sonication, and the debris was pelleted for 30 min at 35,000 × g. Nucleic acids were stripped from the solution by using 0.11 equivalent of 2 M (NH4)2SO4 and the same volume of 10% polyethylenimine. Nucleic acids were removed by stirring and centrifugation at 29,500 × g for 15 min. The protein was recovered by the addition of 0.35 equivalent of saturated (NH4)2SO4. The protein was centrifuged at 9,820 × g for 15 min and resuspended in 100 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, and 10 mM 2-mercaptoethanol. The protein was dialyzed against 50 mM NaCl, 20 mM Tris-HCl (pH 7.5), 1 μM ZnCl2, and 10 mM 2-mercaptoethanol.
In vitro assembly of CA-NC complexes.
HIV-1 CA-NC particles were assembled in vitro by diluting the CA-NC protein to a concentration of 0.3 mM in 50 mM Tris-HCl (pH 8.0), 0.5 M NaCl, and 2 mg/ml DNA oligo-(TG)50. The mixture was incubated at 4°C overnight and centrifuged at 8,600 × g for 5 min. The pellet was resuspended in assembly buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl) at a final protein concentration of 0.15 mM (6, 7).
Binding of TRIM5αrh variants to HIV-1 capsid complexes.
293T cells were transfected with plasmids expressing wild-type or mutant TRIM5αrh proteins. Forty-eight hours after transfection, cell lysates were prepared as follows. Previously washed cells were resuspended in hypotonic lysis buffer (10 mM Tris, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol). The cell suspension was frozen and thawed and incubated on ice for 10 min. Afterwards, the lysate was centrifuged at full speed in a refrigerated Eppendorf microcentrifuge (∼14,000 × g) for 5 min. The supernatant was supplemented with 1/10 volume of 10× PBS and then used in the binding assay. To test binding, 5 μl of CA-NC particles assembled in vitro was incubated with 200 μl of cell lysate at room temperature for 1 h. A fraction of this mixture was stored. The mixture was spun through a 70% sucrose cushion (70% sucrose, 1× PBS, and 0.5 mM dithiothreitol) at 100,000 × g in an SW55 rotor (Beckman) for 1 h at 4°C. After centrifugation, the supernatant was carefully removed and the pellet was resuspended in 1× SDS-PAGE loading buffer. The level of TRIM5αrh proteins was determined by Western blotting, as described above.
RESULTS
Modeling the B-box 2 domain of human and rhesus TRIM5α proteins.
The B-box domains of TRIM proteins contain conserved cysteine and histidine residues that have been shown in some instances to bind zinc atoms (1). Modification of particular cysteine residues predicted to bind zinc atoms in the B-box 2 domain of TRIM5αrh abrogates its anti-HIV-1 activity (11). To gain additional insights into the structure-function relationships of TRIM5α, we used the available structure of the B-box 2 domain of human TRIM29 to model this domain of TRIM5 (Fig. 1). Both human and rhesus monkey TRIM5α B-box 2 domains exhibit approximately 50% identity to the B-box 2 domain of human TRIM29, implying a high likelihood that the TRIM5 and TRIM29 B-box 2 domains are folded similarly (Fig. 1). The modeled B-box 2 domain of TRIM5α exhibits a globular structure, with the N-terminal linker (L1) that connects the B-box 2 domain to the RING domain and the C-terminal helical coiled coil projecting orthogonally from this globular structure (Fig. 1). The amphipathic character of the nascent coiled-coil helix suggests a probable orientation of the modeled B-box 2 domain with respect to the trimer axis. The globular B-box 2 domain presents a polar surface with prominent clusters of positively and negatively charged amino acids.
FIG. 1.
Modeling the B-box 2 domain of TRIM5αhu based on the TRIM29 B-box 2 structure. Modeling of the TRIM5αhu B-box 2 domain and flanking sequences was performed using the Modeler program (Accelrys Software, Inc.). Modeling was based on the TRIM29 B-box 2 structure (9). The Cα trace of the modeled TRIM5αhu B-box 2 domain is shown, with the side chains of lysine 111 and arginine 119. The beginnings of the L1 linker and the coiled coil are depicted in cyan and purple, respectively. The molecule is seen from the perspective of the putative trimer axis. The globular B-box 2 domain is colored green, with basic and acidic residues shown in blue and red, respectively. The alignment of the B-box 2 domains of TRIM5αrh and TRIM5αhu with human TRIM29 (TRIM29hu) is shown below. Identical residues are highlighted in yellow. Residues involved in coordinating zinc ions are colored red. Lysines 111/113 and arginines 119/121 of TRIM5αhu and TRIM5αrh, respectively, are colored blue.
The role in antiretroviral activity of TRIM5α residues predicted to be on the B-box 2 surface.
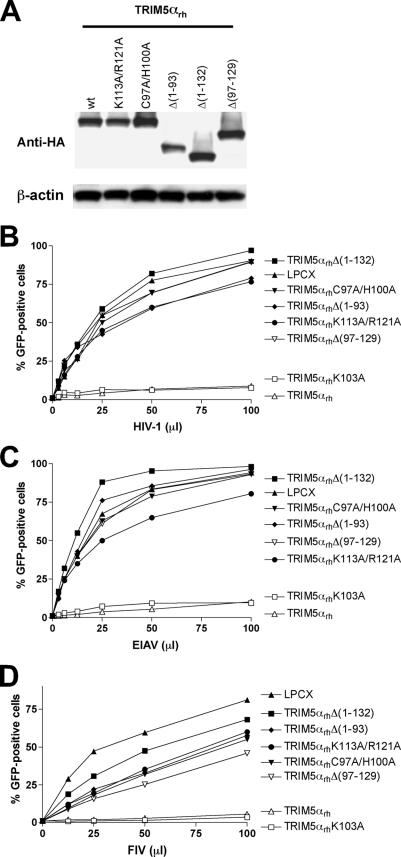
To understand the contribution of the predicted surface-exposed residues of the B-box 2 domain to the anti-HIV-1 activities of TRIM5 proteins, we changed these residues in the TRIM5αrh B-box 2 domain individually to alanine (Fig. 2A). Dog cells (Cf2Th) stably expressing the different mutants (Fig. 2B) were challenged with different amounts of recombinant HIV-1 expressing (HIV-1-GFP) (Fig. 2C and D). Deletion of the B-box 2 domain of TRIM5αrh [Δ(97-129)] completely eliminated its ability to restrict HIV-1 (Fig. 2C and D). By contrast, all of the TRIM5αrh proteins with single amino acid changes restricted HIV-1 infection. However, the K113A and R121A TRIM5αrh mutants inhibited HIV-1 infection less efficiently than the wild-type TRIM5αrh protein (Fig. 2C and D). Thus, replacement of individual TRIM5αrh residues predicted to be exposed on the surface of the B-box 2 domain with alanine exerted at most a modest effect on anti-HIV-1 activity.
FIG. 2.
TRIM5αrh and TRIM5αhu mutants and retroviral restriction. (A) The primary sequences of the B-box 2 domains from TRIM5αrh and TRIM5αhu are aligned. Residues depicted in red are predicted to be exposed on the surface of the B-box 2 domain. The asterisks indicate residues that were changed by site-directed mutagenesis. wt, wild type. (B) Cf2Th cells were transduced with the LPCX vector expressing HA-tagged wild-type and mutant TRIM5αrh proteins. Stable cell lines were selected with 5 μg/ml of puromycin. Cell lysates were analyzed by Western blotting using antibodies against HA (Anti-HA) and β-actin. (C and D) Cf2Th cells expressing the indicated wild-type and mutant TRIM5αrh proteins were incubated with HIV-1-GFP, and GFP-positive cells were measured by fluorescence-activated cell sorting. The results of two independent experiments were similar, and the results of one experiment are shown.
Lysine 113 and arginine 121 are predicted to be located on opposite sides of the upper surface of the TRIM5αrh B-box 2 domain (Fig. 1). To examine further the potential functional roles of these basic residues, we altered lysine 113 and arginine 121 simultaneously to alanine residues. The K113A/R121A TRIM5αrh protein was expressed at a level equivalent to that of the wild-type TRIM5αrh protein (Fig. 3A). The susceptibility of Cf2Th cells expressing the K113A/R121A mutant to infection by HIV-1, EIAV, and FIV was examined (Fig. 3B, C, and D). For comparison, the abilities of these three retroviruses to infect cells expressing wild-type TRIM5αrh or TRIM5αrh mutants with other alterations affecting the B-box 2 domain were examined in parallel. Cells expressing wild-type TRIM5αrh and the K103A mutant strongly resisted infection by HIV-1, EIAV, and FIV. As expected, TRIM5αrh variants with deletions [Δ(1-132) and Δ(97-129)] or disruption (C97A/H100A) of the B-box 2 domain did not inhibit infection by the three retroviruses. The antiretroviral activity of the K113A/R121A mutant was significantly attenuated, similar to that of the TRIM5αrh Δ(1-93) mutant, which lacks the RING and L1 regions. Thus, lysine 113 and arginine 121 contribute to the antiretroviral activity of TRIM5αrh.
FIG. 3.
Alteration of two basic residues in the TRIM5αrh B-box 2 domain. Cells were transduced with either the empty LPCX vector or a vector expressing wild-type (wt) TRIM5αrh or the following TRIM5αrh mutants: K113A/R121A, C97A/H100A, K103A, the mutant with RING deleted [Δ(1-93)], the mutant with the RING-B box deleted [Δ(1-132)], or the mutant with the B-box deleted [Δ(97-129)]. (A) Cell lysates were analyzed by Western blotting using antibodies against HA (Anti-HA) and β-actin. (B to D) Cf2Th cells expressing the different TRIM5αrh variants were challenged with HIV-1-GFP (B), EIAV-GFP (C), or FIV-GFP (D). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
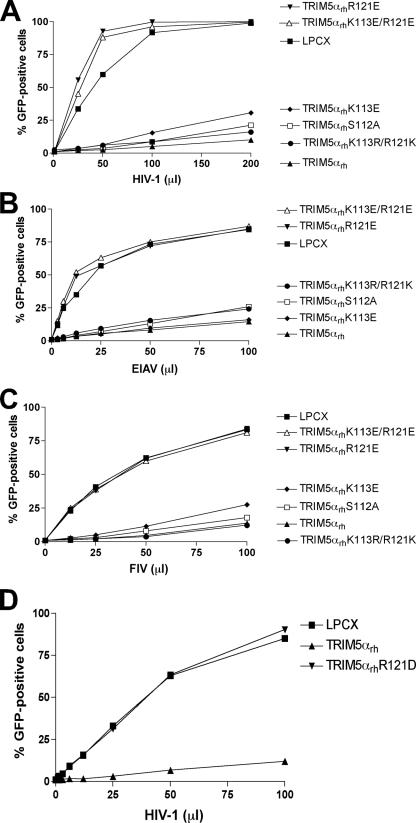
Residues 113 and 121 of TRIM5αrh were altered individually or in combination to other basic residues or to acidic residues. The substitution of negatively charged residues for arginine 121 completely eliminated the ability of TRIM5αrh to restrict HIV-1, EIAV, and FIV infections (Fig. 4A to D), despite efficient expression of these mutants (see below). Substitution of acidic residues at position 113 exerted only modest effects on the antiretroviral activity of TRIM5αrh. The simultaneous replacement of lysine 113 with arginine and arginine 121 with lysine only minimally affected the ability of TRIM5αrh to inhibit HIV-1, EIAV, and FIV infection (see K113R/R121K in Fig. 4A to C). Thus, the presence of a negatively charged residue at position 121 is particularly disruptive of the antiretroviral activity of TRIM5αrh.
FIG. 4.
Effects of altering the charge on residues 113 and 121 of the TRIM5αrh B-box 2 domain. Cf2Th cells expressing the indicated wild-type and mutant TRIM5αrh proteins were challenged with HIV-1-GFP (A and D), EIAV-GFP (B), or FIV-GFP (C). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
Roles of charged B-box 2 surface residues in the antiretroviral activity of human TRIM5α.
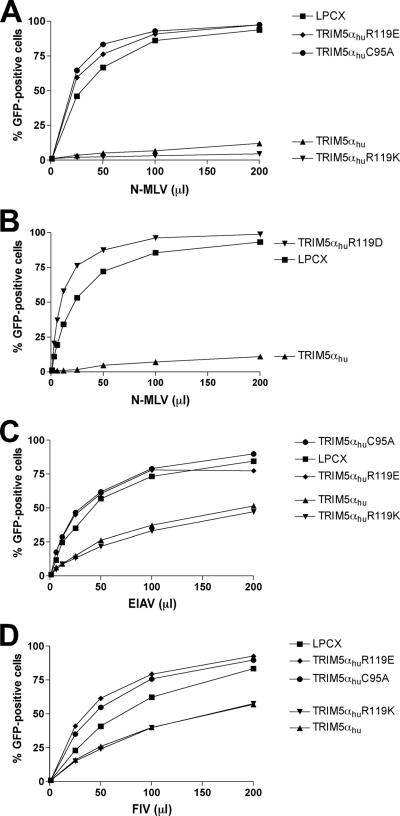
Arginine 121 is conserved in all primate TRIM5α proteins, whereas lysine 113 is conserved in Old World primate TRIM5α proteins (see below). To examine the contributions of these basic residues to the antiretroviral function of another TRIM5α protein, the equivalent residues, lysine 111 and arginine 119, of TRIM5αhu were altered. The abilities of the wild-type and mutant TRIM5αhu proteins to restrict infection of Cf2Th cells by N-MLV, EIAV, and FIV were examined. Alteration of arginine 119 of TRIM5αhu to alanine resulted in complete loss of the ability to restrict N-MLV and EIAV (Fig. 5A and C) and significant reduction in activity against FIV (Fig. 5D). Alanine substitution for lysine 111 in TRIM5αhu exerted less of an effect on N-MLV and EIAV inhibition (Fig. 5A and C). The TRIM5αhu K111A mutant exhibited partial restricting activity against FIV (Fig. 5D). As expected, none of the TRIM5αhu variants were active against B-MLV infection (Fig. 5B). Thus, the alteration of arginine 119 significantly affects the retrovirus-restricting ability of TRIM5αhu.
FIG. 5.
Effects of altering residues 111 and 119 of the TRIM5αhu B-box 2 domain. Cf2Th cells expressing the indicated wild-type and mutant TRIM5αhu proteins were challenged with N-MLV-GFP (A), B-MLV-GFP (B), EIAV-GFP (C), or FIV-GFP (D). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
To examine the role of charge in modulating the function of arginine 119 of TRIM5αhu, mutants were created with a basic substitution (R119K) or acidic substitutions (R119E and R119D) at this residue. The R119K mutant exhibited wild-type levels of activity against N-MLV, EIAV, and FIV infections (Fig. 6A to D). By contrast, the R119E and R119D mutants were devoid of detectable antiretroviral activity. The antiretroviral activities of the R119E and R119D mutants were no better than that of the C95A mutant, which has a disruption of the B-box 2 domain. Apparently, as was seen for TRIM5αrh, introduction of an acidic residue at this position in TRIM5αhu abolishes antiretroviral activity.
FIG. 6.
Effects of altering the charge on residues 111 and 119 of the TRIM5αhu B-box 2 domain. Cf2Th cells expressing the indicated wild-type and mutant TRIM5αhu proteins were challenged with N-MLV-GFP (A and B), EIAV-GFP (C), or FIV-GFP (D). GFP-positive cells were counted. Similar results were obtained in three independent experiments.
Trimerization and capsid binding of TRIM5α B-box 2 mutants.
To investigate the mechanistic basis of the attenuation of antiretroviral activity associated with changes in the B-box 2 residues, the mutants were compared with wild-type TRIM5α proteins for oligomerization and capsid-binding abilities. Both TRIM5αrh and TRIM5αhu form trimers (16). Cross-linking of lysates from cells expressing wild-type or mutant TRIM5α proteins suggested that all of the human and rhesus monkey TRIM5α mutants efficiently formed trimers (Fig. 7A and B and data not shown). These data are consistent with previous results demonstrating that the TRIM5α B-box 2 domain is not necessary for trimerization (12).
FIG. 7.
Oligomerization of TRIM5αrh and TRIM5αhu mutants. 293T cells were transiently transfected with plasmids expressing the indicated wild-type (wt) or mutant TRIM5α proteins with HA epitope tags. Cells expressing wild-type and mutant TRIM5αrh (A) or TRIM5αhu (B) proteins were lysed 48 h after transfection and cross-linked with the indicated concentrations of glycolbis-succinimidylsuccinate. The cell lysates were subsequently Western blotted with an anti-HA antibody. t, trimer; d, dimer.
The abilities of the TRIM5αrh mutants to bind HIV-1 CA-NC complexes assembled in vitro were examined. As shown in Fig. 8, deletion or amino acid alteration of the B-box 2 domain did not significantly compromise the ability of TRIM5αrh to bind HIV-1 capsid complexes. As expected (12, 31), deletion of the TRIM5αrh coiled coil [Δ(132-233)] disrupted its capsid-binding ability. Also as expected (14c, 31), TRIM5αhu associated less efficiently with HIV-1 capsid complexes than TRIM5αrh.
FIG. 8.
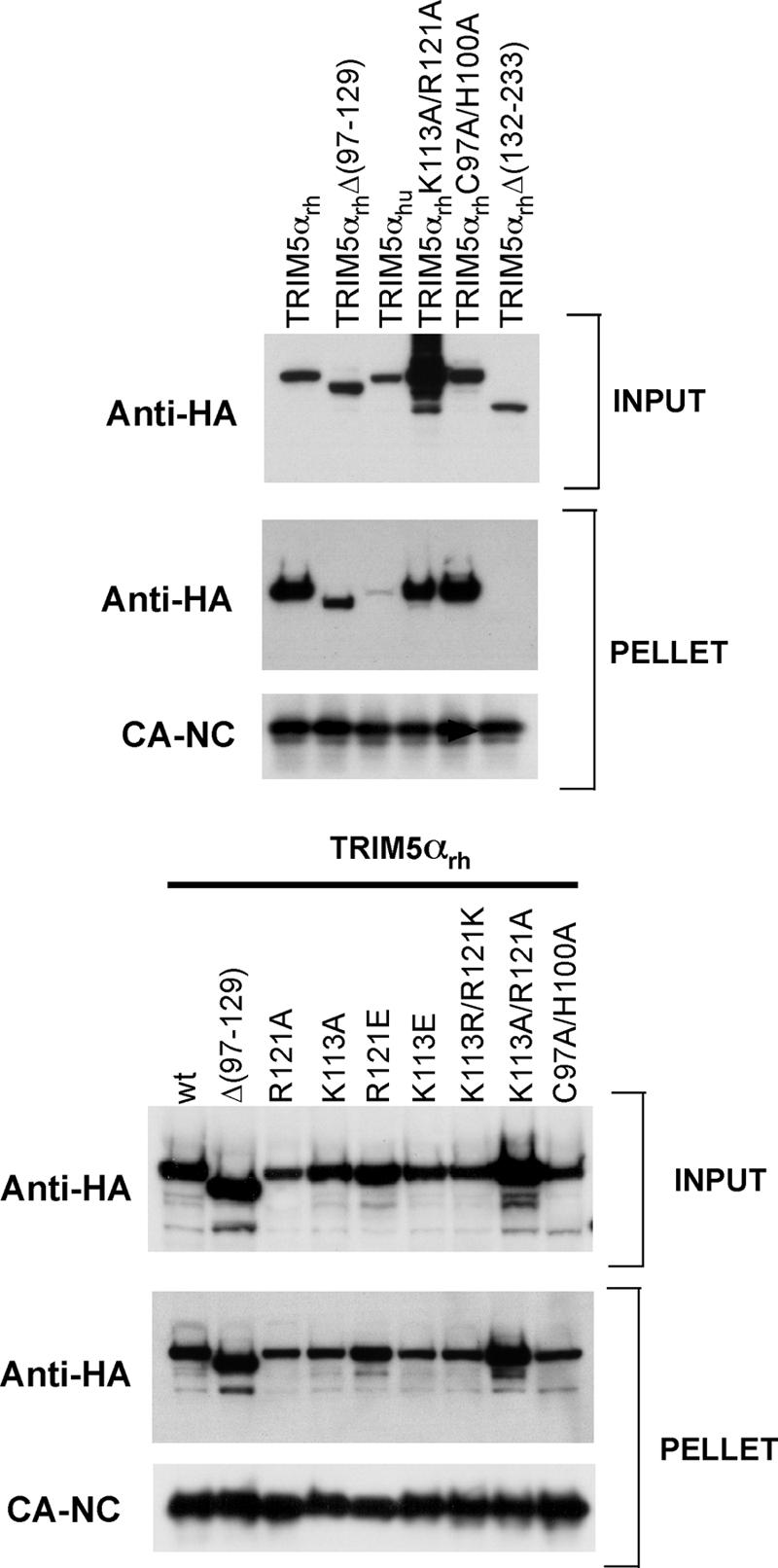
Binding of TRIM5αrh B-box 2 mutants to assembled HIV-1 capsids. 293T cells were transfected with plasmids expressing the indicated wild-type (wt) and mutant TRIM5αrh proteins tagged with HA epitopes. Thirty-six hours after transfection, the cells were lysed and the lysates were mixed with HIV-1 CA-NC complexes that had been assembled in vitro. The mixtures were applied to a 70% sucrose cushion. INPUT represents the mixtures analyzed by Western blotting before being applied to the 70% cushion. The INPUT mixtures were Western blotted for the HA tag. The PELLET from the 70% cushion was analyzed by Western blotting using antibodies against the HA tag and HIV-1 CA-NC protein.
These results suggest that the changes introduced into the TRIM5αrh B-box 2 domain allow trimerization and HIV-1 capsid binding.
Effects of B-box 2 changes in TRIM5α on the fate of the retroviral capsid in infected cells.
Previous studies (24, 31) suggested that restriction of HIV-1 and N-MLV infection is accompanied by an accelerated conversion of the cytosolic retroviral capsid from particulate to soluble forms. We wished to examine the effects of restriction-attenuating changes in the TRIM5α B-box 2 domain on the fate of the retroviral capsid. First, we examined the fate of the HIV-1 capsid in cells expressing the wild-type or R121E TRIM5αrh protein or in control cells transduced with the empty LPCX vector. No cytosolic HIV-1 capsid protein, in either soluble or particulate form, was detected in the control LPCX-transduced cells incubated with HIV-1 lacking envelope glycoproteins (Fig. 9A). By contrast, at 1 hour after infection of these cells with a VSV G glycoprotein-pseudotyped HIV-1, particulate and soluble capsid proteins were detected in the assay. This result demonstrates that the assay can distinguish retroviral capsid proteins that have entered the cytosol through the action of the VSV G glycoprotein from virions that are nonspecifically attached to or endocytosed into cells (24, 31). Compared with the results in the control LPCX cells, a significant decrease in the level of the particulate HIV-1 capsid was observed in the cytosol of TRIM5αrh-expressing cells. By contrast, particulate capsid was detected at this time point in the cytosol of cells expressing the TRIM5αrh R121E mutant.
FIG. 9.
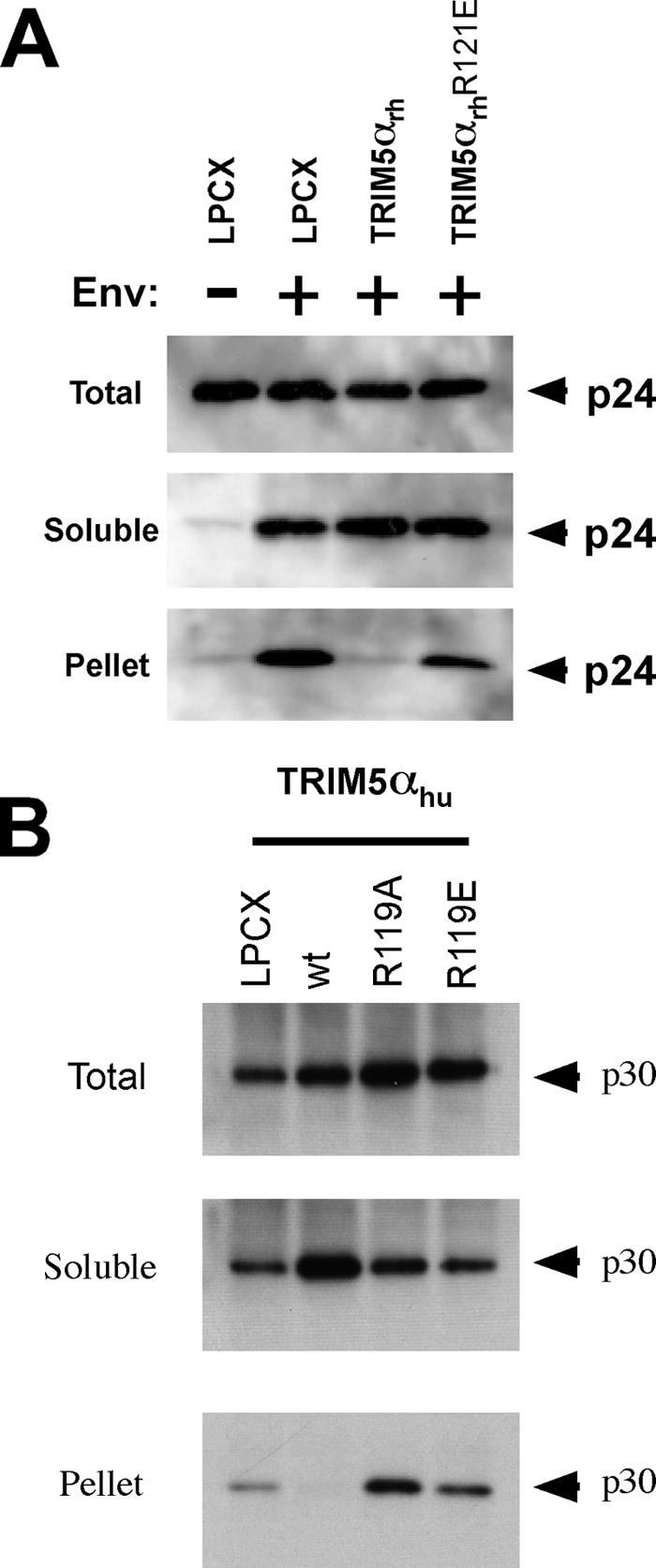
Fates of the retroviral capsid in cells expressing TRIM5 variants. (A) Cf2Th cells containing the LPCX empty vector or expressing wild-type TRIM5αrh or the R121E mutant TRIM5αrh protein were incubated with similar amounts of HIV-1-GFP at 4°C for 30 min. The cells were washed and returned to 37°C, and infection was allowed to proceed for 16 h. Cell extracts were fractionated on sucrose gradients, as described in Materials and Methods. Total, soluble and pellet fractions were analyzed by Western blotting using antibodies against the HIV-1 p24 capsid protein. (B) Cf2Th cells containing the LPCX empty vector or expressing the indicated wild-type (wt) and mutant TRIM5αhu proteins were incubated with similar amounts of N-MLV-GFP at 4°C for 30 min. The cells were washed and returned to 37°C, and infection was allowed to proceed for 4 h. Cell extracts were fractionated on sucrose gradients, as described in Materials and Methods. Total, soluble, and pellet fractions were analyzed by Western blotting using antibodies against the N-MLV p30 capsid protein.
The fate of the N-MLV capsid in the cytosol of cells expressing wild-type TRIM5αhu or the R119A and R119E TRIM5αhu mutants was compared with that seen in control cells transduced with the empty LPCX vector. Particulate capsid was detected in the LPCX-transduced cells and in the cells expressing the R119A and R119E TRIM5αhu mutants (Fig. 9B). By contrast, no detectable particulate capsid was observed in cells expressing wild-type TRIM5αhu. Consistent with previous observations (24, 31), an increase in soluble forms of the N-MLV capsid was observed in cells expressing the wild-type TRIM5αhu protein compared with the levels of soluble capsid proteins in the other cells. Thus, although the B-box 2 mutants are trimeric and retain the ability to bind retroviral capsids, they are deficient in promoting a decrease in the level of particulate cytosolic capsid and in restricting infection. Apparently, an “effector” function necessary for the last two processes has been compromised by the changes introduced into the TRIM5α B-box 2 domain.
Effects of B-box 2 changes on TRIM5α degradation.
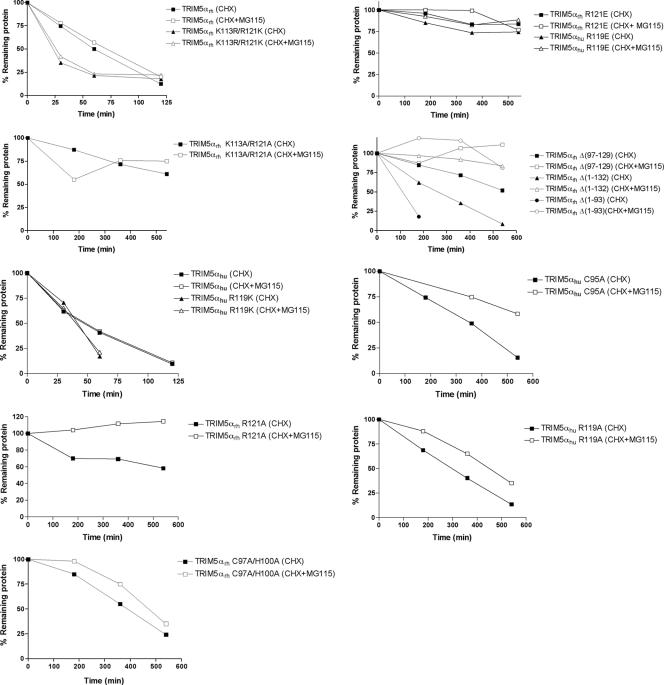
During the course of these studies, we noticed that the steady-state levels of some of the B-box 2 mutants of TRIM5αrh and TRIM5αhu were increased. Wild-type TRIM5α turns over with a half-life of approximately 50 to 70 min, and B-box 2 changes have been shown to affect TRIM5α turnover (3). As steady-state levels reflect rates of both synthesis and degradation, we examined whether the B-box 2 mutants exhibited changes in half-life compared with the wild-type TRIM5α protein. Cf2Th cells stably expressing the mutant proteins were incubated with cycloheximide for several hours, during which time the steady-state levels of the TRIM5α variants were determined by Western blotting. Levels of a control protein, β-actin, were measured in parallel and used as a normalization standard. Because the proteasome can contribute to the degradation of overexpressed TRIM5α (3), we also measured TRIM5α turnover in the presence of proteasome inhibitors. Pilot studies indicated that three different proteasome inhibitors, MG115, MG132, and clasto-lactacystin, all exerted similar effects on TRIM5α turnover (data not shown); thus, MG115 was used in the majority of the experiments.
As expected, the wild-type TRIM5αrh and TRIM5αhu proteins exhibited half-lives of 48 and 40 min, respectively. These half-lives were increased only modestly by incubation with the proteasome inhibitor MG115 (Fig. 10). The measured half-lives of these proteins were similar regardless of whether the HA epitope tag used for detection was at the N or C terminus (or on both termini) (data not shown). Thus, both TRIM5αrh and TRIM5αhu proteins are degraded in Cf2Th cells with a half-life of less than 1 hour by a process that is resistant to proteasome inhibitors.
FIG. 10.
Turnover of wild-type and mutant TRIM5α proteins. Cf2Th cells stably expressing the indicated TRIM5α proteins were treated with cycloheximide (CHX) or with cycloheximide and MG115 (CHX + MG115). At the indicated times after the initiation of treatment, the cells were lysed. The cell lysates were Western blotted using an antibody directed against the HA tags on the TRIM5 proteins. The intensities of the bands were estimated by densitometry and plotted versus time. The results of a typical experiment are shown, with similar results obtained in three separate experiments.
The effects of changes in the B-box 2 domain on TRIM5αrh and TRIM5αhu half-lives are shown in Fig. 10 and Table 1. Many of the B-box 2 changes involving arginine 121 of TRIM5αrh and arginine 119 of TRIM5αhu resulted in proteins with dramatic increases in half-life. Notably, replacement of these arginine residues with lysine did not increase the stability of TRIM5αrh or TRIM5αhu. We conclude that the presence of a basic residue at position 121 of TRIM5αrh or position 119 of TRIM5αhu is important for the rapid turnover of these proteins via a pathway resistant to proteasome inhibitors.
TABLE 1.
Half-lives, retrovirus-restricting activities, and cytoplasmic staining of TRIM variants
| TRIM variant | Half-life (min)a
|
Retrovirus- restricting abilityb | Cytoplasmic stainingc | |
|---|---|---|---|---|
| CHX | CHX + MG115 | |||
| Wild-type TRIM5αrh | 48 | 59 | ++++ | CB |
| TRIM5αrh K113R/R121K | 23 | 29 | +++ | CB |
| TRIM5αrh R121E | 1,541 | >10,000 | − | D |
| TRIM5αrh K113A/R121A | 764 | 1,464 | + | CB |
| TRIM5αrh R121A | 675 | >10,000 | +++ | CB |
| TRIM5αrh R121D | 173 | 714 | − | D |
| TRIM5αrh C97A/H100A | 336 | 470 | − | CB |
| TRIM5αrh Δ(1-93) | 150 | 1,783 | ++ | D |
| TRIM5αrh Δ(97-129) | 629 | >10,000 | − | D |
| TRIM5αrh Δ(1-132) | 203 | 3,072 | − | D |
| TRIM6-5 (CC-B30.2) | 4,933 | >10,000 | ++++ | ND |
| TRIM34-5 (CC-B30.2) | >10,000 | >10,000 | ++++ | ND |
| Wild-type TRIM5αhu | 40 | 42 | ++++ | CB |
| TRIM5αhu R119K | 29 | 31 | ++++ | CB |
| TRIM5αhu R119A | 231 | 439 | + | CB |
| TRIM5αhu R119D | 361 | 602 | − | D |
| TRIM5αhu R119E | 749 | 1,518 | − | D |
| TRIM5αhu C95A | 253 | 768 | − | CB |
| Wild-type TRIMCyp | 56 | 63 | ++++ | CB |
| TRIMCyp R120K | 58 | 61 | ++++ | CB |
| TRIMCyp R120D | 381 | 577 | ++++ | D |
| TRIMCyp R120E | 804 | 1,350 | ++++ | D |
| TRIMCyp R120A | 320 | 680 | ++++ | CB |
The half-life of the indicated TRIM protein stably expressed in Cf2Th cells was determined in the presence of cycloheximide (CHX) or cycloheximide and MG115 (CHX + MG115), as described in Materials and Methods. The half-lives were estimated from semilog plots of TRIM level versus time, in some cases extrapolating to make the estimate. The values shown are typical of those obtained in at least two experiments. The half-lives of the TRIM6-5(CC-B30.2) and TRIM34-5(L2-B30.2) proteins were determined in HeLa cells stably expressing these proteins; the half-life of the wild-type TRIM5αrh protein in these cells was 52 min in the presence of CHX and 115 min in the presence of CHX + MG115.
The retrovirus-restricting ability of each variant was estimated, in comparison to the value (++++) observed for the corresponding wild-type protein, using the data shown in Fig. 2 to 6 and 12. The minus sign indicates no detectable restricting ability for the appropriate susceptible virus, ++++ indicates activity comparable to that of the wild-type protein, and + to +++ indicate intermediate levels of potency.
The intracellular localization of the TRIM variant was determined. The intracellular localization of the TRIM5αrh C97A/H100A, Δ(1-93) and Δ(1-132) variants was described in reference 11. The TRIM5αrhΔ(1-93) protein is also found in the nuclei of expressing cells, where it is associated with nuclear bodies. CB, cytoplasmic bodies; D, diffuse cytoplasmic staining; ND, not determined.
With respect to HIV-1 restriction, the TRIM5αrh RING, L1, and B-box 2 domains can be functionally replaced by those of human TRIM6 or TRIM34 (14). Because TRIM6 and TRIM34 exhibit long half-lives compared with TRIM5α (14), we measured the half-lives of the chimeric proteins [Table 1, TRIM6-5(CC-B30.2) and TRIM34-5(CC-B30.2)]. Both chimeric proteins exhibited very long half-lives compared with that of wild-type TRIM5αrh.
Modest (approximately twofold) increases in the half-lives of most of the single-residue B-box 2 TRIM5α mutants were observed in the presence of proteasome inhibitors (Table 1), consistent with the observations made of the wild-type TRIM5αrh and TRIM5αhu proteins in Cf2Th cells. For a few mutants, like TRIM5αrh R121A and R121D, the effects of MG115 treatment on the half-life were somewhat greater. A TRIM5αrh protein with a deletion of the RING domain and L1 region [TRIM5αrh Δ(1-93)] or TRIM5αrh proteins with deletions encompassing the B-box 2 domain [TRIM5αrh Δ(1-132) and Δ(97-129)] exhibited longer half-lives than the wild-type TRIM5αrh protein. These half-lives were increased significantly (12- to 45-fold) by MG115 treatment. Thus, some TRIM5αrh mutants, particularly those with deletions encompassing the RING and B-box 2 domains, are turned over by proteasome-mediated pathways.
Effects of changes in arginine 120 in the TRIMCyp B-box 2 domain.
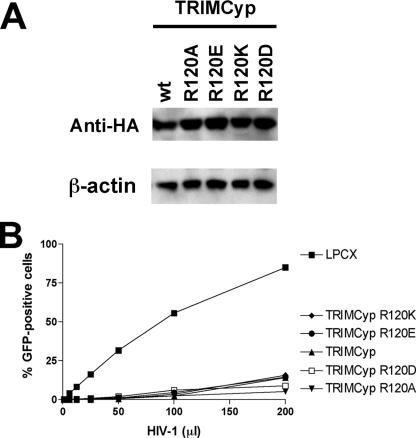
The B-box 2 arginine residue (arginine 121 in TRIM5αrh and arginine 119 in TRIM5αhu) is also conserved in the owl monkey restriction factor TRIMCyp as arginine 120. To examine the contribution of TRIMCyp arginine 120 to protein turnover and antiretroviral function, this residue was altered and the phenotypes of the mutants were examined. The steady-state levels of the TRIMCyp mutants stably expressed in Cf2Th cells are shown in Fig. 11A. The wild-type TRIMCyp protein and the TRIMCyp R120K mutant exhibited half-lives of about 1 hour (Table 1). By contrast, the R120D, R120E, and R120A mutants of TRIMCyp had half-lives greater than 5 h. Proteasome inhibition exerted only one- to twofold effects on the turnover of the wild-type and mutant TRIMCyp proteins. Thus, like TRIM5α, TRIMCyp is rapidly degraded by a process that is not inhibited by proteasome inhibitors; this process is disrupted by replacement of the TRIMCyp B-box 2 arginine 120 with amino acid residues that do not carry a positive charge.
FIG. 11.
Effects of B-box 2 arginine changes on TRIMCyp. (A) The steady-state expression levels of the indicated TRIMCyp variants in Cf2Th cells were determined by Western blotting with an anti-HA antibody. wt, wild type. (B) Cf2Th cells expressing the indicated TRIMCyp variants were incubated with HIV-1-GFP. GFP-positive cells were counted. The results of a typical experiment are shown. The experiment was repeated with similar results.
The abilities of the TRIMCyp B-box 2 mutants to restrict HIV-1 infection were examined by challenging Cf2Th cells expressing the wild-type and mutant TRIMCyp proteins with recombinant HIV-1-GFP. The TRIMCyp R120K, R120E, R120D, and R120A proteins inhibited HIV-1 infection as efficiently as the wild-type TRIMCyp protein (Fig. 11B). Thus, the phenotypes of the B-box 2 domain changes differ in the context of the TRIMCyp and TRIM5α proteins.
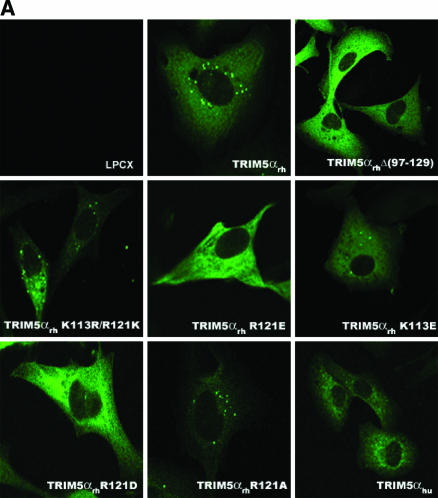
Cellular localization of TRIM5α B-box 2 mutants.
The intracellular localization of the wild-type TRIM5αrh, TRIM5αhu, and TRIMCyp proteins was compared with that of the B-box 2 mutants. The mutants in which arginines 119/120/121 in the B-box 2 domain were altered to an acidic amino acid residue demonstrated intense, diffuse staining throughout the cytoplasm of the cell (Fig. 12A and B). A similar pattern of staining was observed for TRIM5αrhΔ(97-129), in which the B-box 2 domain is deleted. The B-box 2 mutants in which arginines 119/120/121 were replaced by lysine or alanine residues exhibited faint, diffuse cytoplasmic staining punctuated by the presence of scattered cytoplasmic bodies, a pattern similar to that exhibited by the wild-type TRIM5α proteins.
FIG. 12.
Subcellular localization of TRIM5αrh and TRIM5αhu mutants. (A and B) Cf2Th cells stably expressing the indicated wild-type and mutant TRIM5α or TRIMCyp proteins were fixed and stained using a fluorescein isothiocyanate-conjugated anti-HA antibody as described in Materials and Methods. Representative confocal microscope images are shown.
DISCUSSION
The functions of most TRIM proteins are not known. In a few cases, TRIM proteins have been shown to recognize a protein target via their C-terminal domains; the binding of the TRIM protein results in ubiquitylation and proteasomal degradation of the target protein (10, 17, 33). The mechanism of retroviral restriction by TRIM5α is currently under investigation. TRIM5α variants that inhibit HIV-1 infection specifically recognize HIV-1 capsid complexes assembled in vitro (31). In the infected cell, TRIM5α promotes the premature conversion of particulate capsids into soluble capsid proteins (24, 31). Studies of TRIM5α variants with the RING domain deleted, cells expressing temperature-sensitive E1 ubiquitin ligase mutants, and the effects of proteasome inhibitors argue against a model of capsid ubiquitylation and proteasomal degradation (23, 31).
The work reported here provides insights into the structure-function relationships of the TRIM5 B-box 2 domain. We studied a panel of human and rhesus monkey TRIM5α B-box 2 mutants containing alterations of amino acid residues predicted to be surface exposed (Fig. 1). Most of the mutants phenotypically resembled the wild-type TRIM5α protein, indicating some degree of structural flexibility on the B-box 2 surface. On the other hand, some mutants were severely attenuated in the ability to restrict viral infection despite retaining the ability to trimerize and to recognize the relevant retroviral capsid. Thus, these mutants appear to lack an effector function necessary for retroviral restriction. An overlapping subset of B-box 2 mutants lost the ability to be rapidly degraded by a pathway insensitive to proteasome inhibitors. Some changes in the B-box 2 domain also influenced the subcellular localization of TRIM5α and TRIMCyp proteins.
A discrete TRIM5 B-box 2 region predicted to reside near the trimer axis was found to be important for retroviral restriction, formation of cytoplasmic bodies, and protein turnover. The structural requirements for these three processes differ subtly but involve arginines 119/120/121 in all three instances. All primate TRIM5 proteins, as well as owl monkey TRIMCyp, have arginine residues at these positions (Fig. 13). The rapid turnover of TRIM5α and TRIMCyp apparently requires a basic residue at these positions. The structural requirements for cytoplasmic-body formation and retroviral restriction appear to be less stringent. Substitution of acidic residues for arginines 119/120/121 or deletion of the B-box 2 domain results in a diffuse cytoplasmic staining pattern for TRIM5α and TRIMCyp, with lack of cytoplasmic-body formation. These acidic substitutions also attenuate retroviral restriction by rhesus monkey and human TRIM5α proteins, but not by TRIMCyp. Replacement of arginine 119 by alanine significantly impaired the retrovirus-restricting ability of TRIM5αhu, although cytoplasmic-body formation and rapid turnover were not affected. Retroviral restriction by TRIM5αrh, on the other hand, can tolerate the substitution of an alanine residue for arginine 121. Thus, TRIM5αrh R121A forms cytoplasmic bodies and retains potent restricting activity against HIV-1 but, unlike the wild-type protein, exhibits a long half-life. Likewise, replacement of the RING and/or B-box 2 domains of TRIM5αrh with those of the long-lived TRIM6, TRIM34, or TRIM21 proteins also resulted in stable proteins that exhibited potent antiretroviral activity (Table 1) (3, 14a). Thus, the functions of the TRIM5α B-box 2 domain related to rapid turnover and retroviral restriction are separable. These results seem incompatible with recently proposed (1a) models of restriction in which the TRIM5αrh protein and the capsid subunits associated with it are degraded through a proteasome-independent host cell pathway.
FIG. 13.
Sequence alignment of the B-box 2 domain sequences from different TRIM5 proteins. The B-box 2 domain sequences of the TRIM5 relatives TRIM6, TRIM34, and TRIM22 are also shown for comparison. Residues predicted to coordinate the two zinc ions in the B-box 2 domain are depicted in red. Residues depicted in black correspond to the consensus sequence for this set of proteins, whereas residues in blue differ from the consensus sequence. The arginine (119/120/121) studied in this work is conserved in all TRIM5 proteins (black box). In contrast, lysine (111/113) is conserved only in the TRIM5 proteins of hominoids and Old World monkeys.
The pathway by which TRIM5α and TRIMCyp are rapidly turned over is not understood. Despite testing multiple proteasome inhibitors, we were unable to eliminate the rapid turnover of the wild-type TRIM5α and most of the TRIM5α single-amino-acid mutants. Apparently, a nonproteasome protein degradation pathway interacts with the amino-terminal TRIM5α domains and results in rapid turnover. The role of this rapid turnover in the natural function of TRIM5α requires further investigation.
Our results suggest that the TRIM5 B-box 2 domain contributes to cytoplasmic-body formation but cannot do so when an acidic residue is present at positions 119/120/121. The formation of cytoplasmic bodies is not sufficient for potent retroviral restriction, as evidenced by the weak retroviral restriction mediated by the TRIM5αhu R119A and TRIM5αrh K113A/R121A mutants, both of which are found in cytoplasmic bodies. The necessity of cytoplasmic-body formation for retroviral restriction by TRIM5α cannot be ruled out by our data, as acidic substitutions for residue 119 of TRIM5αhu or residue 121 of TRIM5αrh eliminated both the appearance of cytoplasmic bodies and restricting activity. Other studies of TRIM5α-expressing cells in which cytoplasmic bodies have been disrupted or minimized suggest that detectable bodies are not necessary for efficient inhibition of retrovirus infection (23, 27). Acidic substitutions involving arginines 119/121 of the TRIM5α B-box 2 domain, or deletion of this domain, may coincidentally affect cytoplasmic-body formation and another property (e.g., association with self or another factor) required for retroviral restriction.
Disruption of the B-box 2 domain can result in loss of retrovirus-restricting ability for at least two reasons. First, as shown here, some changes in the TRIM5αrh B-box 2 domain eliminate the ability of the protein to restrict HIV-1 infection without significantly affecting expression, oligomerization, or capsid binding. These mutagenic studies suggest that the TRIM5α B-box 2 domain plays a key role in retroviral restriction separate from the requirements for capsid recognition/binding. In this study and in previous studies (24, 31), we observed the accelerated conversion of particulate retroviral capsids to soluble forms of capsid protein in the cytosol of infected cells expressing a functional TRIM5α protein compared with that in control cells not expressing TRIM5α. Notably, this accelerated uncoating of the viral capsid was not observed in cells expressing the B-box 2 TRIM5α mutants defective in the “effector function” of retroviral restriction. Our studies indicate that the majority of the capsid proteins disassembled during TRIM5α-mediated restriction do not undergo ubiquitylation (or other large modification) and are not degraded. Second, some combinations of changes in several B-box 2 residues have been shown to decrease the binding of TRIM5α proteins to the HIV-1 capsid, apparently by affecting the relationship of the B-box 2 and B30.2(SPRY) domains (14b). Understanding these distinct consequences of B-box 2 domain modification will require additional studies.
The B-box 2 arginine residue identified in this work as important for TRIM5αrh and TRIM5αhu effector function appears to be less critical for TRIMCyp-mediated retroviral restriction. Deletion or disruption of the B-box 2 domain of TRIMCyp can lead to a reduction in antiretroviral activity (4). In general, however, it appears that TRIMCyp anti-HIV-1 activity is less dependent on the integrity of the B-box 2 domain than is the virus-restricting activity of TRIM5α proteins. This is consistent with recent observations that moderate levels of antiretroviral activity can be achieved simply by oligomerizing the cyclophilin A domain, a function that does not require the TRIM5 B-box 2 domain (12a, 36).
The arginine 119/121 residue identified here as being important for TRIM5α antiretroviral function, cytoplasmic-body formation, and TRIM5α turnover is predicted to be exposed on the modeled TRIM5α trimer. This raises the possibility that the region surrounding arginines 119/121 may be a docking site for association of TRIM5α with itself or with TRIM5α cofactors that support retroviral restriction or rapid turnover. The panel of mutants generated in this study should assist the investigation of these possibilities.
Acknowledgments
We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation.
This study was supported by the National Institutes of Health (AI063987 and a Center for AIDS Research Award AI60354), the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Borden, K. L. 1998. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76:351-358. [DOI] [PubMed] [Google Scholar]
- 1a.Chatterji, U., M. D. Bobardt, P. Gaskill, D. Sheeter, H. Fox, and P. A. Gallay. 2006. Trim5α accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J. Biol. Chem. 281:37025-37033. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Griffero, F., S. A. Hoschander, and J. Brojatsch. 2002. Endocytosis is a critical step in entry of subgroup B avian leukosis viruses. J. Virol. 76:12866-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz-Griffero, F., X. Li, H. Javanbakht, B. Song, S. Welikala, M. Stremlau, and J. Sodroski. 2006. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349:300-315. [DOI] [PubMed] [Google Scholar]
- 4.Diaz-Griffero, F., N. Vandegraaff, Y. Li, K. McGee-Estrada, M. Stremlau, S. Welikala, Z. Si, A. Engelman, and J. Sodroski. 2006. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology 351:404-419. [DOI] [PubMed] [Google Scholar]
- 5.Fiser, S. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374:461-491. [DOI] [PubMed] [Google Scholar]
- 6.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 7.Ganser-Pornillos, B. K., U. K. von Schwedler, K. M. Stray, C. Aiken, and W. I. Sundquist. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue, K., F. Hayashi, and S. Yokoyama. 2005. Solution structure of the zf-B-box type 2 domain of human tripartite motif protein TRIM29 isoform alpha, pdb 2CSV. RCSB Protein Data Bank. http://www.rcsb.org/pdb.
- 10.Ishikawa, H., H. Tachikawa, Y. Miura, and N. Takahashi. 2006. TRIM11 binds to and destabilizes a key component of the activator-mediated cofactor complex (ARC105) through the ubiquitin-proteasome system. FEBS Lett. 580:4784-4792. [DOI] [PubMed] [Google Scholar]
- 11.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 12.Javanbakht, H., W. Yuan, D. F. Yeung, B. Song, F. Diaz-Griffero, Y. Li, X. Li, M. Stremlau, and J. Sodroski. 2006. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353:234-246. [DOI] [PubMed] [Google Scholar]
- 12a.Javanbakht, H., F. Diaz-Griffero, W. Yuan, D. F. Yeung, X. Li, B. Song, and J. Sodroski. 13 June 2007. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology. doi: 10.1016/j.virol.2007.04.034. [DOI] [PMC free article] [PubMed]
- 13.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, X., B. Gold, C. O'Huigin, F. Diaz-Griffero, B. Song, Z. Si, Y. Li, W. Yuan, M. Stremlau, C. Mische, H. Javanbakht, M. Scally, C. Winkler, M. Dean, and J. Sodroski. 2007. Unique features of TRIM5α among closely related human TRIM family members. Virology 360:419-433. [DOI] [PubMed] [Google Scholar]
- 14a.Li, X., Y. Li, M. Stremlau, W. Yuan, B. Song, M. Perron, and J. Sodroski. 2006. Functional replacement of the RBCC domains of TRIM5α by heterologous TRIM domains. J. Virol. 80:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b.Li, X., B. Song, S.-H. Xiang, and J. Sodroski. 2007. Functional interplay between the B-box 2 and the B30.2(SPRY) domains of TRIM5α. Virology. doi: 10.1016/j.virol.2007.04.022. [DOI] [PMC free article] [PubMed]
- 14c.Li, Y., X. Li, M. Stremlau, M. Lee, and J. Sodroski. 2006. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 80:6738-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meroni, G., and G. Diez-Roux. 2005. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27:1147-1157. [DOI] [PubMed] [Google Scholar]
- 16.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niikura, T., Y. Hashimoto, H. Tajima, M. Ishizaka, Y. Yamagishi, M. Kawasumi, M. Nawa, K. Terashita, S. Aiso, and I. Nishimoto. 2003. A tripartite motif protein TRIM11 binds and destabilizes Humanin, a neuroprotective peptide against Alzheimer's disease-relevant insults. Eur. J. Neurosci. 17:1150-1158. [DOI] [PubMed] [Google Scholar]
- 18.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nisole, S., J. P. Stoye, and A. Saib. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev. Microbiol. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 20.Ohkura, S., M. W. Yap, T. Sheldon, and J. P. Stoye. 2006. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80:8554-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen, J. C. 1998. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 5:1481-1487. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Caballero, D., T. Hatziioannou, F. Zhang, S. Cowan, and P. D. Bieniasz. 2005. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 79:15567-15572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perron, M. J., M. Stremlau, M. Lee, H. Javanbakht, B. Song, and J. Sodroski. 2007. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 81:2138-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saenz, D. T., W. Teo, J. C. Olsen, and E. M. Poeschla. 2005. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5α proteins. J. Virol. 79:15175-15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song, B., F. Diaz-Griffero, D. H. Park, T. Rogers, M. Stremlau, and J. Sodroski. 2005. TRIM5α association with cytoplasmic bodies is not required for antiretroviral activity. Virology 343:201-211. [DOI] [PubMed] [Google Scholar]
- 28.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79:6111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song, B., H. Javanbakht, M. Perron, D. H. Park, M. Stremlau, and J. Sodroski. 2005. Retrovirus restriction by TRIM5α variants from Old World and New World primates. J. Virol. 79:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 31.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toniato, E., X. P. Chen, J. Losman, V. Flati, L. Donahue, and P. Rothman. 2002. TRIM8/GERP RING finger protein interacts with SOCS-1. J. Biol. Chem. 277:37315-37322. [DOI] [PubMed] [Google Scholar]
- 34.Towers, G. J. 2005. Control of viral infectivity by tripartite motif proteins. Hum. Gene Ther. 16:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 36.Yap, M. W., G. B. Mortuza, I. A. Taylor, and J. P. Stoye. 2007. The design of artificial retroviral restriction factors. Virology. doi: 10.1016/j.virol.2007.04.005. [DOI] [PubMed]
- 37.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]