Abstract
Human cytomegalovirus (HCMV) is the leading viral cause of birth defects, affecting primarily the central nervous system (CNS). To further understand this CNS pathology, cells from glioblastoma cell lines T98G and A172, the astrocytic glioblastoma cell line CCF-STTG1 (CCF), and the neuroblastoma cell line SH-SY5Y (SY5Y) were infected with HCMV. CCF and SY5Y cells were fully permissive for infection, while A172 cells were nonpermissive. In T98G cells, the majority of cells showed viral deposition into the nucleus by 6 h postinfection (hpi); however, viral immediate-early gene expression was observed in only ∼30% of cells in the first 72 h. In viral antigen (Ag)-positive cells, although the development of complete viral replication centers was delayed, fully developed centers formed by 96 hpi. Interestingly, even at very late times postinfection, a mixture of multiple small, bipolar, and large foci was always present. The initial trafficking of input pp65 into the nucleus was also delayed. Titer and infectious-center assays showed a small number of T98G cells shedding virus at very low levels. Surprisingly, both Ag-positive and Ag-negative cells continued to divide; because of this continuous division, we adopted a protocol for passaging the T98G cells every third day to prevent overcrowding. Under this protocol, detectable infectious-virus shedding continued until passage 5 and viral gene expression continued through eight passages. This evidence points to T98G cells as a promising model for long-term infections.
Human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus harmless to most of the immunocompetent population. However, HCMV congenital infection is the most common cause of virus-induced birth defects, particularly disorders of the central nervous system (CNS). Each year, ∼1% of all newborns are congenitally infected, and of these infants, 5 to 10% manifest signs of serious neurological defects, including deafness, mental retardation, blindness, microencephaly, hydrocephalus, and cerebral calcification (1, 3, 32). An additional 10% of congenitally infected infants are asymptomatic at birth and subsequently develop brain disorders (9, 24).
It has been suggested previously that the severity of the neuropathological changes and clinical outcomes may be associated with the stage of CNS development at which congenital infection occurs (2); however, the mechanism of HCMV neuropathogenesis in the developing CNS is not well understood. Many studies using mouse and human CMVs with cells ranging from embryonic stem cells to neural precursors to cells of carcinogenic cell lines have been performed in an attempt to more clearly define this pathogenesis. This task is made particularly difficult by the range of permissiveness of the various cell types within the brain.
During the infection of fully permissive cells, the HCMV genome is temporally expressed. HCMV immediate-early (IE) proteins are synthesized quickly and activate the early (E) genes, whose products are essential for viral replication. Expression of the late (L) proteins follows replication, and these products are structural components of the virion. Permissive cells infected with HCMV readily produce large amounts of virus and undergo lytic infection. Cell types not considered to be permissive have been described previously as semi- or nonpermissive. The latter category is fairly easily defined and includes cells either not producing a full complement of proteins or not allowing any viral protein synthesis and, hence, no subsequent production of virions. Cells in the semipermissive category span a much broader range, generally having delayed kinetics of protein expression and very little measurable viral output.
As CMV has strict species specificity, studies using model systems can provide insights into the mechanisms of HCMV-associated effects in the developing brain. Work with the mouse system by researchers in the lab of Tsutsui has indicated likely cell types to investigate. Shinmura et al. (30) found that the injection of murine CMV into the cerebral ventricles of mouse embryos causes a disturbance in neuronal migration and a marked loss of neurons. They proposed that these events may be a cause of microencephaly due to CMV infection. A later paper from Kosugi et al. (15) suggested that neurons in the cortex can be infected, produce low levels of virus, and persist in the infected mouse brain. These neurons appear to escape detection by natural killer cells and macrophages.
Scant research using human fetal tissue has been performed, and the results are somewhat conflicting; despite these differences, some clear messages can be gleaned from these studies. First, neural progenitor cells, from which all of the main CNS cell types are derived, are permissive for infection and likely release limited amounts of virus (6, 20-22). Second, differentiated neural progenitor cells establish three cell types in culture: astrocytes (positive for glial fibrillary acidic protein), microglial cells (CD68+), and neurons (positive for neurofilament). Astrocytes have previously been shown to be fully permissive (analogous to fibroblasts) and release significant quantities of virus (6, 17). The data on both microglial cells (16, 17) and neurons (6, 17, 20) are conflicting; some of the studies claim that they are somewhat permissive (shedding low levels of virus), and others claim that both cell types are nonpermissive.
Due to the inherent difficulty of obtaining and working with primary human neural cultures, much work has been done with established human cell lines. This includes work with several different astrocytoma/glioblastoma and neuroblastoma cell lines (14), oligodendroglioma (HOG) cells (31), and the untransformed neuronal HCN-1A cells (25). From these studies, together with the work on primary human cells, it appears that several neural cell types are permissive (at least to some extent) for HCMV infection. However, the state of differentiation of the cell appears to have a dramatic bearing on permissiveness. Is it possible that in the fetal brain, within which a tremendous amount of differentiation is occurring, the cellular environment favors viral gene expression? A cell's degree of permissiveness for HCMV infection can change as differentiation signaling changes, but whether the virus oscillates between a permissive or semipermissive infection state and a latent infection state remains to be determined.
To begin assessing long-term infection in the brain, we have examined viral gene expression and virion release in the semipermissive cell line T98G. We found that this cell type expresses all classes of viral proteins and produces small amounts of virus for extended periods (up to 1 month). In addition, we found direct evidence of the continued division of antigen (Ag)-positive cells. This, to the best of our knowledge, is the first clear evidence of this phenomenon. Implications for the potential of T98G cells as a model for studying long-term infection are discussed.
MATERIALS AND METHODS
Cell lines and cell culture.
T98G (human glioblastoma cell line; ATCC CRL-1690) cells and human foreskin fibroblasts (HFFs; obtained from the University of California, San Diego, Medical Center) were grown in minimal essential medium (GIBCO BRL); A172 (human glioblastoma cell line; ATCC CRL-1620) and SH-SY5Y (SY5Y; human neuroblastoma cell line; ATCC CRL-2266) cells were grown in Dulbecco's minimal essential medium (GIBCO BRL); and CCF-STTG1 (CCF; human astrocytic glioblastoma cell line; ATCC CRL-1718) cells were grown in RPMI 1640 (GIBCO BRL). All media were supplemented with 10% fetal bovine serum, penicillin (200 U/ml) and streptomycin (200 μg/ml), l-glutamine (2 mM), and amphotericin B (Fungizone; 1.5 μg/ml). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
In order to visualize cells in metaphase, dividing cells were arrested by treatment with ethidium bromide (5 μg/ml) for 20 min, followed by colchicine (0.1 μg/ml) for an additional 30 min prior to the time of harvest (10). These cells were then fixed, permeabilized, and processed as described below. In one set of experiments, cells were pulsed with 10 μM bromodeoxyuridine (BrdU) for 30 min and then washed and allowed to incubate for a further 30 min (chase) in fresh medium prior to treatment with ethidium. This BrdU incubation was performed to allow the localization of actively replicating viral DNA within infected cell nuclei (11).
Virus and virus infection.
HCMV strain Towne (ATCC VR977) was used in these studies. Virus was propagated on a monolayer of HFFs and titrated by plaque assays as described previously (33). All cell types were serum starved (in 0% serum) for 2.5 days to synchronize them in G0. Prior to infection, cells were replated and allowed 2 h to attach. All cell types were infected at a multiplicity of infection (MOI) of 10. The cells were incubated with virus for 12 to 16 h (unless otherwise noted; see below), and subsequently, the inoculum was removed and cells were re-fed. Cells were harvested at the indicated times postinfection (pi). BrdU-labeled virus was prepared as previously described (28). In studies using BrdU-labeled virus, cells were incubated with virus for either 1 or 6 h prior to the removal of the inoculum.
Passaging experiments.
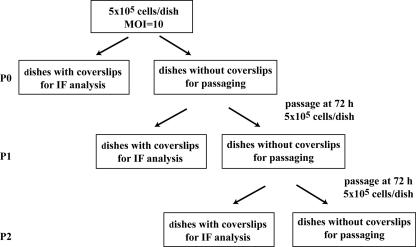
To allow continual investigation of HCMV infection in T98G cells without overcrowding, cultures were serially passaged at 3-day intervals. Following serum starvation to synchronize cells, aliquots of cells (5 × 105) were seeded onto 10-cm dishes, some with poly-l-lysine-coated glass coverslips and others without coverslips, and the cells on each dish were infected at an MOI of 10. The initial infection was designated passage 0 (P0). At 1-day intervals, the coverslips were harvested, cells were fixed with 3% formaldehyde, and the coverslips were stored at 4°C for later analysis. On day 3, the remaining infected cells (from the dishes without coverslips) were harvested, combined, counted, and reseeded in the same manner (5 × 105 cells/dish) as in the first round. This first subculture was designated P1. The subsequent subcultures (P2 through P8) were performed in the same manner at 3-day intervals. A specific time point within a certain passage can be expressed as the number of days postinfection (dpi) by multiplying the passage number (P#) by 3 days and adding the time point in days (tpd) within that passage, as follows: ([P# × 3] + tpd). For example, day 2 of P4 can be expressed as (4 × 3) + 2, or 14, days dpi. A flow chart of the passaging of the infected cells is shown in Fig. 1.
FIG. 1.
Flow chart of the passaging scheme used in this study. See Materials and Methods and the remainder of the text for details.
Preparation of cell lysates and Western blotting.
Cells were rinsed with phosphate-buffered saline (PBS) once, trypsinized, harvested, and washed again with ice-cold PBS. Cells were then resuspended in ice-cold PBS, counted, and spun into a pellet. Cell pellets were snap-frozen in liquid nitrogen and then stored at −80°C until the time course was completed. An additional set of experiments (see Fig. 2C) was conducted by following the same protocol (harvesting cells every 24 h), in which a single culture was allowed to progress for 216 h total. Whole-cell lysates were prepared as described previously (18).
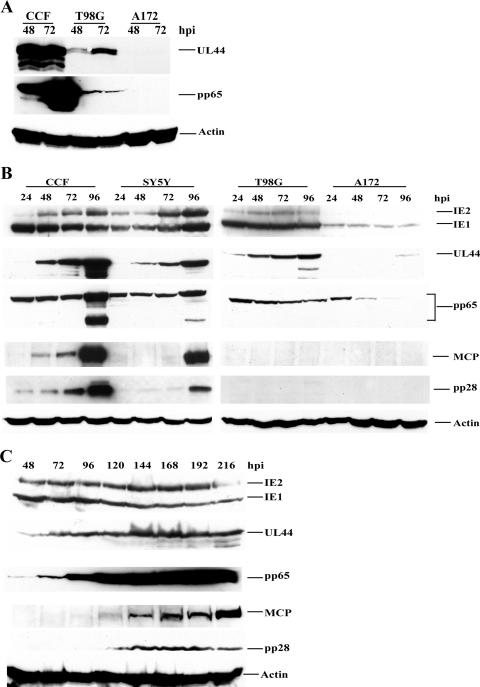
FIG. 2.
Various levels and timing of viral gene expression in four different cell lines. Serum-starved CCF (permissive), SY5Y (permissive), T98G (semipermissive), and A172 (nonpermissive) cells were infected with HCMV Towne at an MOI of 10 and harvested at the indicated times pi. Equal amounts of cell lysates (from 2 × 105 cells) were used for all lanes in all blots. Abs used are described in Materials and Methods. (A) Comparative levels of viral gene expression in CCF, T98G, and A172 cells. All cell lysates were loaded onto the same blot for the direct comparison of expression levels. An actin loading control was included in all three panels to confirm the loading of equal sample amounts. (B) As protein levels differed dramatically among cell types, optimized concentrations for each primary Ab were determined. Lysates from permissive cells (CCF and SY5Y) were loaded onto one blot, and lysates from semi- and nonpermissive cells (T98G and A172) were loaded onto another. To evaluate IE1 and IE2, UL44, and pp65 in CCF and SY5Y cells, primary Abs were diluted 1:6,000. For the parallel T98G and A172 blots, Abs were diluted 1:2,000 (a threefold difference in Ab concentrations). MCP and pp28 primary Abs were diluted 1:2 for all blots (no differences among cell types). Secondary Ab was used at the same dilution of 1:3,000 for all blots. Blots probed for the same viral protein were exposed using enhanced-chemiluminescence reagents at the same sensitivity levels for identical time periods. (C) Prolonged time course for T98G cells. The infection of T98G cells was extended through 216 h (without passaging), and cells were harvested at the indicated times pi. Primary and secondary Ab concentrations were identical to those given for panel B; however, enhanced-chemiluminescence development times varied slightly, and therefore, slight variations in the results for a given cell type and a particular Ag as presented in the different panels of Fig. 2 may be observed.
Immunofluorescence (IF) analysis.
The serum-starved T98G and CCF cells were trypsinized and replated onto 100-mm dishes (5 × 105 cells per dish) containing poly-l-lysine-coated coverslips. After infection, the coverslips were harvested at the indicated times pi and rinsed in PBS. The cells on the coverslips were then fixed with 3% formaldehyde (in PBS) at room temperature for 10 min and permeabilized with 1% Triton X-100 (in PBS) at room temperature for 5 min, and the coverslips were washed three times in PBS. Cells were blocked with 30% fetal bovine serum in a blocking solution (PBS with 1% bovine serum albumin and 0.01% Tween 20) for 15 min, and then the coverslips were incubated with primary antibodies (Abs) diluted in blocking solution for 10 min. After extensive washes in PBS, coverslips were incubated with isotype-specific secondary Abs in blocking solution for 10 min, followed by repeated washes in PBS. Coverslips were mounted in glycerol containing paraphenylene diamine to inhibit photobleaching. Nuclei were counterstained with Hoechst dye. The images were obtained using a Nikon Eclipse E800 fluorescence microscope equipped with a Nikon DXM camera and Metavue software. Cells infected with BrdU-labeled virus were treated with HCl as described previously (28) to reveal incorporated BrdU residues. All percentages given for T98G and CCF cells are the averages obtained from at least two trials (unless otherwise noted). At least 100 cells/coverslip were scored for each time point in each experiment.
Release of infectious virions.
The shedding of infectious virions from T98G cells was investigated by two methods. First, we used a standard plaque-forming assay. Supernatant samples were collected from cell cultures and stored at −80°C after the addition of 1% dimethyl sulfoxide. Supernatant samples were serially diluted twofold (1:2) prior to the infection of a monolayer of HFFs to allow the observation of plaques. Second, we used a focus-forming assay. Cell samples (106 cells) from P0 through P8 were harvested and serially diluted 10-fold (1:10). Immediately following dilution, cells were inoculated onto a monolayer of HFFs. Cells were overlaid with agar, and resulting foci were counted after 7 days. The results from supernatant samples were expressed as PFU per milliliter, and the results from cell samples were expressed as percentages of focus-forming cells (focus-forming units per 100 cells). The assays were performed three times, and the averages of the results ± one standard deviation are presented.
Abs.
Primary Abs used included anti-pp65 (immunoglobulin G1 [IgG1]), anti-IE1 and -IE2 (ch16.0; IgG1), and anti-UL44 (IgG1) mouse monoclonal Abs (Rumbaugh-Goodwin Institute for Cancer Research, Inc.); anti-IE1 (IgG2a), anti-pp28 (IgG1), and anti-major capsid protein (anti-MCP; IgG2a) mouse monoclonal Abs (kind gifts from Bill Britt, University of Alabama, Birmingham); anti-actin (clone ACT05) mouse monoclonal Ab (Neomarkers); and anti-BrdU rat monoclonal Ab (Harlan Sera-Lab). Secondary Abs used were as follows: for immunoblot detection, horseradish peroxidase-linked sheep anti-mouse secondary Ab (Amersham Bioscience), and for IF analysis, tetramethyl rhodamine isothiocyanate-conjugated anti-rat, anti-mouse IgG1 and IgG2a (Jackson ImmunoResearch Laboratories) and Alexa Fluor 488-conjugated goat anti-mouse IgG1 and IgG2a (Molecular Probes).
RESULTS
Variable timing of viral gene expression in different cell lines.
In order to investigate the levels and timing of viral gene expression in the four cell lines (CCF, SY5Y, T98G, and A172), cells synchronized in G0 were plated onto 100-mm dishes (5 × 105 cells/dish), infected at an MOI of 10, and then harvested at 24, 48, 72, and 96 hpi (Fig. 2). To determine the relative levels of viral gene expression in the different cell types, 48- and 72-hpi lysates from 2 × 105 cells were electrophoresed, transferred onto Protran nitrocellulose filters, and probed for the viral Ags UL44 and pp65 (Fig. 2A). As can be clearly observed for both of these Ags, the levels of viral gene expression in fully permissive (CCF), semipermissive (T98G), and nonpermissive (A172) cells varied dramatically.
Based on the results from Fig. 2A, we determined optimized primary Ab concentrations for each cell type and thereafter ran two gels in parallel, one with the fully permissive cell samples and the other with semi- and nonpermissive cell samples (Fig. 2B) (see the legend to Fig. 2 for Ab concentrations). When comparing the panels in Fig. 2B, it is therefore important to remember that three times more primary Ab was used to detect the IE1 and IE2, UL44, and pp65 proteins in T98G and A172 cells than to detect these proteins in CCF and SY5Y cells. However, the separate gels were always probed using identical secondary Ab concentrations, and exposure times for detecting the same Ag were also identical. Viral gene products representative of the three temporal classes, IE (IE1 and IE2 proteins), E (UL44 and pp65 proteins), and L (MCP and pp28 proteins) were examined.
IE1 and IE2 expression in CCF and SY5Y cells was somewhat higher than that in T98G cells and dramatically higher than that in A172 cells. In addition, while IE2 was expressed in CCF, SY5Y, and T98G cells between 24 and 96 hpi, expression levels in the CCF and SY5Y cells increased but those in the T98G cells remained constant over the same time course. No IE2 expression in the A172 cells was observed.
UL44, the processivity factor for the viral polymerase, is expressed at E times after infection. Levels of UL44 expression in CCF and SY5Y cells were similar, with very slight expression at 24 hpi and clear expression by 48 hpi, and expression levels continued to increase as the infection progressed. In T98G cells small amounts of UL44 were also, visible by 24 hpi, with only slight increases in levels observed after 48 hpi. Total protein levels in T98G cells were not as high as those in CCF and SY5Y cells (as also apparent in the direct comparison in Fig. 2A). UL44 was not detectable in A172 cells until 96 hpi, when a faint band was observed.
pp65 (UL83), the major tegument protein, has an E-L expression pattern. Its levels of expression in CCF and SY5Y cells were similar, and bands corresponding to this protein remained visible throughout the time course. By 96 hpi, there were increases in protein levels, and multiple isoforms appeared. There was little if any increase in the expression level and no appearance of cleavage products in T98G cells during the 96-hpi time course. In A172 cells, pp65 levels fell over the course of infection, likely indicating the presence of only input virus at the earlier times pi and no de novo synthesis of pp65 in these cells.
MCP (UL86) and tegument protein pp28 (UL99) are important structural proteins and display L expression kinetics. Note that these two proteins were detected with the same concentration of primary Ab on all blots, allowing direct comparison. MCP and pp28 expression in CCF cells was visible earlier than in SY5Y cells, and expression levels in CCF cells increased significantly at 96 hpi. In SY5Y cells, the expression of both proteins was observed primarily at 96 hpi. There was no detectable MCP in either T98G or A172 cells through 96 hpi (Fig. 2B), indicating either very low-level expression of these proteins or delays in expression beyond 96 hpi. This result raised the question of whether there were any L proteins synthesized in these cell types.
As it seemed clear from our experiments that the A172 cells were nonpermissive, we concentrated further on the T98G cells. In order to answer the above question, infected cultures were maintained for 216 h (9 days) without passaging. Cells were harvested every 24 h, and cell lysates were prepared for viral protein level determination by Western blotting (Fig. 2C). Similar to the expression patterns presented in Fig. 2B, the expression of both IE and E proteins (IE1 and IE2, UL44, and pp65) occurred in these cells. Interestingly, no pp65 cleavage products were observed in the T98G cells over this time frame of infection. The L proteins, MCP and pp28, did finally become visible after 120 hpi, and MCP levels increased with infection time. This result indicated that L protein expression in these cells was delayed by at least 72 h in comparison to that in infected permissive CCF cells.
Although viral genomes were delivered to almost all T98G cells, not all cells displayed de novo viral Ag synthesis.
There are several possible explanations for the reduction in viral Ag expression displayed in T98G and A172 cells (Fig. 2) compared to that in permissive cells. First, perhaps all cells did not receive viral genomes. Second, it is possible that all cells were expressing viral Ags but some on a delayed time scale and at reduced levels compared to others. Third, perhaps only a certain percentage of cells were actually carrying out de novo viral Ag synthesis after infection.
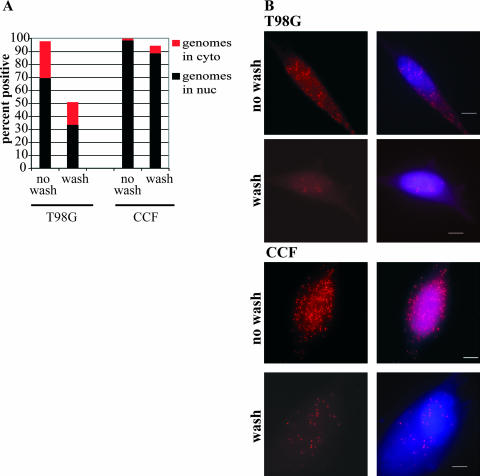
We had previously observed BrdU-labeled-genome deposition (28) into the nuclei of T98G cells infected at a high MOI (18). Luo et al. (18) also found that by 6 hpi, the large majority of T98G cells showed viral genome deposition within the nucleus. In the present study, we repeated these experiments under various infection conditions, with interesting results (Fig. 3). Experiments in which BrdU-labeled virus was allowed to adsorb to the cells for a full 6 h before the cells were harvested (as described in reference 18) displayed results similar to those of our earlier study; virtually all cells were positive for viral genomes, and ∼70% displayed the clear localization of numerous viral genomes within the nucleus at 6 hpi (Fig. 3, no wash). Nearly 100% of permissive CCF cells showed the entry and transit of labeled genomes into the nucleus under these conditions (Fig. 3, no wash). Interestingly, if the virus inoculum was removed and the cells were vigorously washed after a 1-h incubation (as described in reference 28), very different results were obtained. Although 95% of permissive CCF cells still showed the entry of viral genomes and 89% showed the transit of these genomes into the nucleus at 6 hpi under these wash conditions (Fig. 3, wash), far fewer T98G cells (∼50%) were positive for BrdU-labeled genomes at 6 hpi (Fig. 3A). Also, for both cell types, far fewer genomes per cell were present in washed cells than in their unwashed counterparts (Fig. 3B, compare wash to no wash). Only 34% of the washed T98G cells showed clear BrdU labeling in the nucleus at 6 hpi, and the remainder of the BrdU+ cells showed genomes only in the cytoplasm (Fig. 3A). This result indicated a delay in the entry and transit of the virus into the nucleus in T98G cells compared to viral entry and transit in permissive CCF cells under conditions of a short 1-h incubation. Due to these findings, all subsequent experiments performed in this study utilized the extended incubation with the virus inoculum (including those for which results are shown in Fig. 2). Interestingly, even under conditions of extended virus incubation, only ∼35% of A172 cells had genomes in the nucleus at 6 hpi (data not shown). Since such a block may have greatly skewed any further interpretations, we decided to confine our further investigations to T98G cells.
FIG. 3.
The majority of T98G cells showed viral genome deposition into the nucleus by 6 hpi if the cells were subjected to extended incubation with the virus inoculum. (A) Graphic representation of the distribution of BrdU-labeled genomes within the nuclei (black bars) and cytoplasm (red bars) of infected cells at 6 hpi under different infection conditions (see Materials and Methods for details). Percentages are averages from two experiments. (B) T98G and CCF cells were infected at an MOI of 10 with BrdU-labeled virus in order to track input viral genomes. Cells were subjected to either an extended (6-h) incubation with the virus inoculum (no wash) or only a 1-h incubation before extensive washing to remove unadsorbed virus (wash). Cells were harvested at 6 hpi and stained with an Ab to BrdU to monitor the localization of input viral genomes. Nuclei were counterstained with Hoechst. In all panels, the scale bar represents 5 μm.
To investigate the second and third possible reasons for changes in expression levels, we performed IF experiments over the first 72 hpi, first looking for strong expression of one of the IE proteins, IE1. Virtually all infected permissive CCF cells (94.5%) displayed strong IE1 staining in the nucleus by 24 hpi and remained strongly positive throughout the course of infection. We then assessed the percentage of IE1+ cells in the T98G cell population over the first 72 hpi. The level of BrdU-labeled-genome deposition within the nucleus by 6 hpi (at which point ∼70% of the cells were genome positive) had led us to expect a high percentage of IE1 positivity by 24 hpi. This was not the case, and the level of IE1+ cells never rose above 35% in the first 72 hpi (averages of 34.9, 32.5, and 32.4% at 24, 48, and 72 hpi, respectively).
Next, we assessed the presence of the E protein UL44. As the processivity factor for the viral polymerase, UL44 localizes to the viral replication foci within an infected cell nucleus. Although not the first protein to localize to these foci (11), UL44 is a good marker for visualizing them by IF and was therefore used in these studies. Surprisingly, we found an even lower percentage of cells that displayed viral replication foci (as measured by using UL44+ foci) than the percentage that expressed IE1. The average proportion of focus-positive cells within the total population did not rise above 11% for the first 72 hpi (averages of 3.4, 10.5, and 10.7% at 24, 48, and 72 hpi, respectively). Clearly, not all cells were capable of de novo viral Ag synthesis after infection.
Various patterns of viral replication center development in different cell types.
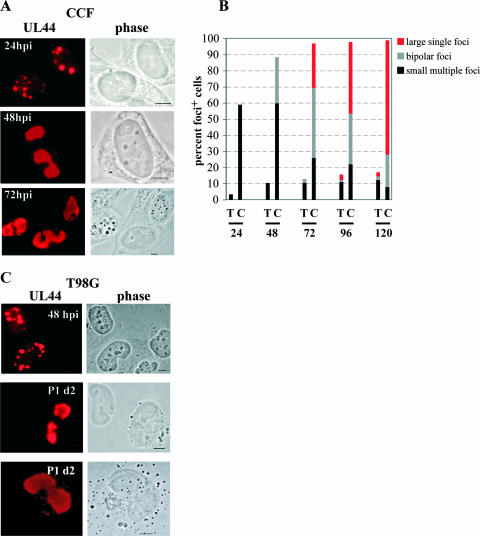
To explore this lack of de novo viral Ag synthesis more fully, we compared the development of UL44 foci in T98G cells and that in fully permissive CCF cells. In CCF cells, multiple small foci were clearly visible by 24 hpi (Fig. 4A). By 48 hpi, these had begun to progress toward the formation of two larger foci (hereafter referred to as bipolar foci) located at opposite poles of the nucleus. Finally, by 72 hpi, the most advanced stage, one very large focus covering the majority of an infected cell's nucleus could be observed in these cells. The distribution of focus-positive CCF and T98G cells into these three categories (those with multiple small foci, bipolar foci, and a single large focus) over the first 120 hpi can be viewed in Fig. 4B.
FIG. 4.
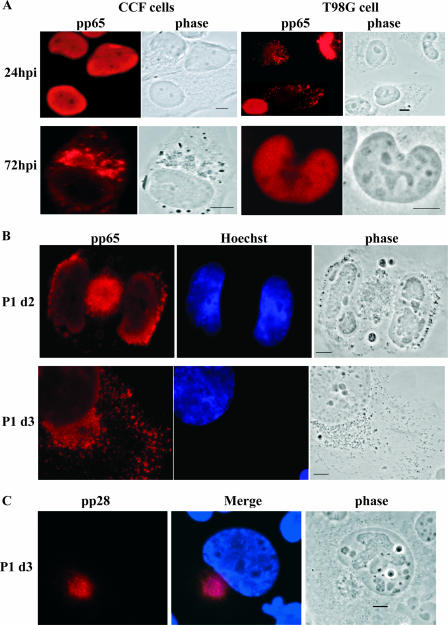
The development of full viral replication centers in T98G cells was delayed compared to that in CCF cells. Serum-starved T98G and CCF cells were seeded onto poly-l-lysine-coated glass coverslips and infected at an MOI of 10. Initially infected T98G cells (P0) were passaged and subcultured every 72 hpi (P1 through P8) as described in Materials and Methods, with one dish from each subculture containing glass coverslips for IF analysis. During the first two passages (P0 and P1), coverslips were harvested every 24 h (at 1-day intervals). Cells were stained for UL44, and cell populations were analyzed for the presence of cells containing UL44 foci as described in the text. (A) Permissive CCF cells showed UL44 foci with the characteristic appearances, from multiple small foci (top panels) to bipolar foci (middle panels) to one large focus (bottom panels), at the expected times pi (24, 48, and 72 hpi, respectively). (B) Graphic representation of the distributions of UL44 focus-positive cells containing multiple small foci (black bars), bipolar foci (gray bars), and a single large focus (red bars). (C) T98G cells do show all three types of foci, but on a protracted time scale. Examples of each type of foci are shown, along with the time point pi at which the image was captured. Scale bars represent 5 μm. P1 d2, day 2 of P1.
As indicated above, we saw very few T98G cells with UL44 foci at either 24 or 48 hpi, and similar to CCF cells at 24 hpi, these cells exhibited only small multiple foci (Fig. 4B). A representative image taken at 48 hpi is shown in Fig. 4C (top panels). At 72 hpi, the large majority of T98G focus-positive cells (∼80%) still had multiple small foci; however, we did observe bipolar foci in the remainder of the focus-positive cells at this time point (Fig. 4B). This finding was essentially the opposite of the result for the CCF cells, among which the majority of focus-positive cells (∼70%) exhibited advanced-stage foci (either bipolar or single large foci) by 72 hpi.
We wanted to monitor T98G cells for a longer period of time to determine if they would develop full replication centers. However, an obstacle that arose during our extended time course (represented in Fig. 2C) had to be addressed. Although essentially all cells contained viral genomes at 6 hpi, many cells did not carry out de novo viral Ag synthesis and, therefore, did not demonstrate arrested cell division as did the permissive cell types (i.e., HFFs and CCF and SY5Y cells) (13). Cellular replication and division continued within the infected T98G cultures, which led to overcrowding on the plates. This situation was not a major obstacle to Western blot analysis, but it seriously impaired the acquisition of accurate information and quality images for IF analysis. To avoid overcrowding, we devised a passaging system in which every 3 days we trypsinized, counted, and reseeded the cultures onto new plates at the same number of cells per plate (5 × 105) as originally plated at time zero (see Materials and Methods for further description). We found a roughly fourfold increase in the number of cells per plate (to 2 × 106) at each 3-day interval. The original 72-h time course was termed P0. After the first passage, we seeded new coverslips and plates (P1) and again split and reseeded the cultures after an additional 3 days to obtain P2. This passaging protocol was continued up through day 3 of P8 (27 dpi). It should be noted that the control CCF cells were not passaged in this way as they did not continue to divide after infection.
This protocol allowed us to monitor the infection in the T98G cells over a longer period. We found that by day 1 of P1 (equivalent to 96 hpi), large single UL44+ foci were present (Fig. 4B). However, allowing the cells to progress for another day (to day 2 of P1, equivalent to 120 hpi) did not substantially increase the proportion of focus-positive cells or affect their phenotypic distribution. At day 2 of P1, the large majority of T98G focus-positive cells (∼70%) still showed multiple small foci. As mentioned earlier, this result was in marked contrast to that for the CCF cells. Figure 4C (middle and bottom panels) shows examples of bipolar and large foci in T98G cells taken during P1.
Our data indicated that in a portion of the T98G population, viral replication centers did fully develop, but full development in HCMV-infected T98G cells was delayed by approximately 24 h compared to that in CCF cells. It should be noted here that even at day 3 of P8 (equivalent to 27 dpi), T98G cells still displayed a mix of phenotypes of UL44 foci, with many cells displaying the less-advanced, multiple-focus phenotype even at the latest times pi.
Transport of input pp65 was delayed in T98G cells, and trafficking of virion-associated protein to the cytoplasm occurred in only a small percentage of cells.
Having observed a delay in full replication center development in the T98G cells, we decided to explore possible delays in tegument protein trafficking as well. The localization of the major tegument protein, pp65 (UL83), was analyzed at the corresponding time points in P0 and P1. As a component of the incoming virion with its own nuclear localization signal, input pp65 transits into the nucleus independently of the genome. In fully permissive cells, this transit occurs quickly, and generally by 2 to 3 hpi (but certainly by 12 hpi), all nuclei are pp65+. For this reason, we have often used this protein as an indicator of virus entry into the infected permissive cell (5, 11, 29). As Fig. 5A (top left panels) shows, the pattern of pp65 localization in CCF cells (which were 98% pp65+) was exclusively nuclear at 24 hpi.
FIG. 5.
The transport of input pp65 in T98G cells was delayed, and the trafficking of virion-associated protein to the cytoplasm occurred in only a small percentage of cells. Cells were seeded and infected as described previously. Passaging was as described in Materials and Methods. (A) Left panels show the patterns of pp65 staining in permissive CCF cells at 24 and 72 hpi. The characteristic trafficking of pp65 to the juxtanuclear space as associated with maturing virions is depicted in the bottom left panels (and observed in 18% of cells). Right panels show the pp65 staining patterns of semipermissive T98G cells at the same time points. The top panels show the delay in the transport of input pp65 to the nucleus (see the text for more details). At 72 hpi, very few cells (<4%) showed trafficking of pp65 protein into the juxtanuclear space. (B) During P1, evidence of proper trafficking of pp65+ particles could be observed in a small percentage (<4%) of T98G cells. P1 d2, day 2 of P1; P1 d3, day 3 of P1. (C) In a small percentage of T98G cells (<6%), proper localization of the tegument protein pp28 could also be observed. An example from day 3 of P1 is shown. Observation during the first four passages revealed the presence of pp28+ cells. Nuclei were counterstained with Hoechst in panels B and C. Scale bars represent 5 μm.
A very different pattern of input-pp65 localization in T98G cells was observed. Again, judging from the BrdU-labeled-genome localization that we observed by 6 hpi, we expected the majority of cells to demonstrate the clear localization of input pp65 into the nucleus by 24 hpi. This was not the case. Instead, as shown in Fig. 5A (top right panels), although virtually all cells (99%) were positive for pp65 by 24 hpi, only one-third (33.3%) exhibited clear nuclear staining, with the rest displaying a mixture of punctate nuclear and cytoplasmic staining or a predominantly punctate cytoplasmic pattern. Since pp65 possesses its own nuclear localization signal, this result may represent a transport defect for this Ag within T98G cells.
pp65 is also synthesized de novo and incorporated into maturing virions; these virions transit from the nucleus and through the cytoplasm via the Golgi apparatus and are released at the plasma membrane. Distinct movement into the juxtanuclear space (or viral assembly complex) (4, 5) is observed for pp65 and several other tegument proteins during the course of infection of a permissive cell. A proportion (18%) of the total population of permissive CCF cells showed this distinct cytoplasmic localization by 72 hpi, and by 96 and 120 hpi, much larger fractions of cells (57 and 76%, respectively) showed this pattern (the lower left panels in Fig. 5A provide an example). Localization in T98G cells never progressed to the point at which most cells showed cytoplasmic pp65 localization. Instead, at 72 hpi, only a very small percentage (3.5%) of T98G cells displayed cytoplasmic localization similar to that in CCF cells, with the very large majority of cells still showing nuclear staining only (lower right panels in Fig. 5A).
IF results from UL44 staining in T98G cells revealed that although full UL44 focus formation was slower in these cells than in permissive cells, the viral replication centers did fully develop. This finding indicated the potential for infectious virus particles to be made and released. To investigate this potential, we assessed the distribution and relocalization of pp65 via IF analysis at later times pi after passaging. As shown in Fig. 5B, as time progressed, further relocalization of pp65 was observed, both into the juxtanuclear space (4, 5) and into the dendrites of infected cells. However, the percentage of cells within the total population with this pattern of pp65 cytoplasmic staining never increased above 4% throughout the passaging scheme (3.45, 2.7, 2.5, 0.45, and 0% as measured for P0 through P4, respectively). We also checked for the proper localization of another structural protein (pp28) into the juxtanuclear space outside the nucleus. As our Western analysis had indicated no synthesis of this protein until approximately 120 hpi, we examined P1 cells and observed a small percentage (<6%; see below) with the proper localization of this protein beginning at day 3 of P1 (Fig. 4C).
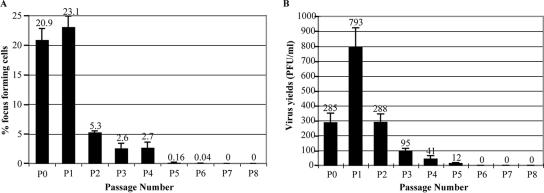
The trafficking of pp65+ particles toward the cell surface, as well as the production and proper localization of pp28, indicated the potential for virus release from these Ag-positive cells. We therefore performed focus-forming assays for virus-producing cells at day 3 of each passage (Fig. 6A) as described in Materials and Methods. Infected T98G cells were harvested at day 3 of the passages indicated in Fig. 6, diluted serially 1:10, and inoculated directly onto HFF monolayers without freezing. In the first 6 dpi (P0 and P1), roughly 20% of the cells were determined to be actively shedding virus (i.e., there were 20 focus-forming units/100 cells) (Fig. 6A). By 9 dpi (day 3 of P2), this percentage dropped to approximately 5% and remained between 2 and 5% through 15 dpi (day 3 of P4). A very small number of cells (<1%) continued to shed virus at detectable levels until 21 dpi (day 3 of P6). By 24 dpi (day 3 of P7), no virus-shedding cells could be detected using these methods.
FIG. 6.
Low levels of infectious virus particles were released from HCMV-infected T98G cells. (A) The percentage of virus-shedding cells was determined by a focus-forming assay as described in Materials and Methods, and results are expressed as percentages of focus-forming cells. (B) Virus yield in the supernatant was determined by a standard plaque-forming assay as described in Materials and Methods, and results are expressed as PFU per milliliter. In both panels, error bars represent one standard deviation based on results from three experiments.
Although the number of virus-shedding cells was relatively low, these cells, in theory, could still shed substantial amounts of virus. To assess this potential, we assayed the viability of the virus released into the supernatant overlaying the infected cultures. Supernatant samples were collected at 3 dpi and at 3 days postreseeding for all subsequent passages. The presence and quantity of virus in the supernatant were determined by plaque-forming assays performed by using 1:2 serial dilutions. Our data indicated that the level of infectious virions released into the supernatant by T98G cells (Fig. 6B) was very low (<1,000 PFU/ml). Small amounts of virus were shed for several passages (through day 3 of P5), but the amount of virus shedding was below our level of detection by day 3 of P6.
HCMV infection does not arrest T98G cells.
In a normal infection in permissive cells, cells actively expressing virus no longer divide and eventually die. However, in our T98G populations, cells continued to divide, as demonstrated by repeated cell crowding at 3-day intervals. Cells were seeded at 5 × 105/plate, and after 3 days, the cell number had increased to ∼2 × 106/plate (a fourfold increase). Although BrdU-labeled viral particles were visible within most nuclei by 6 hpi under conditions of extended incubation, by 20 hpi all nuclei were entirely stained with unincorporated BrdU from the supernatant (data not shown). This result indicated that cellular DNA synthesis was ongoing, even in the presence of the viral genome.
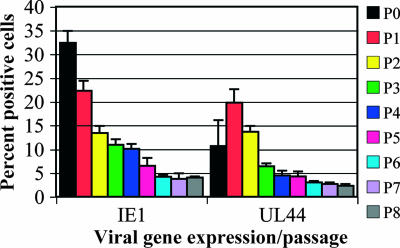
Were these cells producing such small amounts of virus that they were able to survive while actively shedding? We knew that the quantity of virus shed by the T98G cells was very small, and thus, there were two possible explanations for the continued presence of Ag-positive cells. First, Ag-positive cells did not die but also did not divide. In this case, the proportion of Ag-positive cells would decline by roughly a factor of 4 at each passage, from 32% (P0) to 8% (P1) … to 0.03% (P5) to 0.01% (P6), etc. This drop did not occur. Figure 7 shows results for IE1+ cells and cells with UL44 focus-positive cells. In addition, although the measurement was performed only once, we did not observe a decline in pp28 cytoplasmic staining (5.8, 5, 2.5, and 2.7% as assayed at day 3 of P1, P2, P3, and P4, respectively) that would fit this scenario either. Therefore, since percentages of Ag-positive cells did not decline at the fourfold rate, other factors must have contributed to the long-term expression of Ags.
FIG. 7.
Expression of viral Ags continued throughout 27 dpi. Cells were passaged as described in Materials and Methods. Coverslips were harvested for analysis at 3-day intervals, beginning at 72 hpi (day 3 of P0). Error bars represent standard deviations based on results from at least two experiments. Coverslips were stained and scored for the presence of IE1 and UL44+ foci.
Qualitative analysis of these Ag-positive cells during passaging displayed a varied spectrum of sizes of UL44+ foci (through P8) and variation in the intensities of pp65 within the nuclei of cells (particularly through P3). These findings indicated that there was a small increase in Ag-positive cells due to new or reinfection from shed virus. However, due to the very low levels of shed virus, it seemed unlikely that this newly synthesized virus alone could account for the slower-than-expected decline in percentages of Ag-positive cells. A measurement of multiple-micronucleus formation (and of associated apoptosis via a tunnel assay) showed that only a very small fraction of the entire population was dying at any given time (1.7, 1.7, 0.4, 0.9, and 0.9% at day 3 of P0, P1, P2, P3, and P4, respectively [measurements were performed only once]). This finding established that the gradual decline in the number of Ag-positive cells was not due to their death.
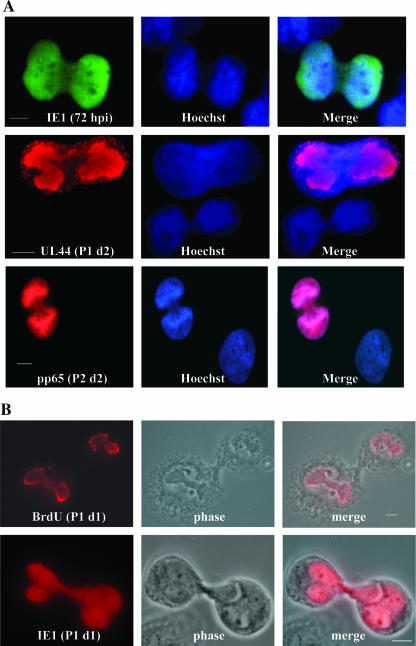
Were the viral-Ag-positive cells dividing? To accurately assess this possibility, we examined the Ag-positive population for dividing cells via a colchicine block (used to arrest cells for metaphase analysis) (10). It became clear that Ag-positive cells were indeed dividing (at a rate quantitated once using IE1+ cells as 19.5, 14, 12, and 16%, respectively, for the first four passages), as can be seen in Fig. 8A (showing examples of cells positive for IE1, UL44-containing foci, and pp65 at various times during infection). Although the actual cytokinetic event was difficult to capture, we were able to visualize it in some cases (Fig. 8B). Importantly, when we pulse-labeled cells with BrdU just prior to metaphase arrest (to label actively replicating viral DNA within the replication centers), we observed equal distributions of viral genomes between the two daughter cells as well (pictured in Fig. 8B, top panels). Therefore, we concluded that the prolonged presence of viral-Ag-positive cells within the T98G population was due to low levels of virus shedding (establishing either newly infected, reinfected, or activated cells), the persistence of Ag-positive cells, and their continued division from the earliest stages of infection.
FIG. 8.
T98G cells that were positive for viral Ag continued to divide. (A) In order to investigate the possibility of Ag-positive-cell division, cells were arrested in metaphase by using a colchicine block. Cells were then stained for the indicated Ags. IE1+ cells were analyzed through P4, and an average of 15% of these IE1+ cells were found to be dividing during each passage. P2 d2, day 2 of P2. (B) Two examples of dividing cells, including phase images, are shown to emphasize the cytokinesis between daughter cells. In the top panels, cells were pulse-labeled with BrdU just prior to treatment with colchicine to visualize actively replicating viral DNA within replication centers. Equal division of viral genomes can be seen. P1 d1, day 1 of P1. Scale bars represent 5 μm.
DISCUSSION
T98G cells are undifferentiated glioblastoma cells that have previously been defined as being semipermissive for HCMV infection (14, 26). This study confirms and extends this assessment. Previously, only the expression of the IE and E proteins had been observed in these cells. Earlier work with T98G cells from our laboratory showed that the HCMV genome was deposited into the nuclei of the large majority of these cells (∼85%) by 6 hpi (18) and that ∼30% of these cells expressed IE proteins within the first 72 hpi (Fig. 3 and 7). Figure 2 clearly demonstrates that the levels of expression of two of the more abundant viral proteins, UL44 and pp65, in fully permissive (CCF), semipermissive (T98G), and nonpermissive (A172) cells varied significantly. These dramatic differences in expression levels made it difficult to visualize or measure the relative protein levels in all different cell types simultaneously; therefore, we developed optimized Ab incubation conditions for each cell type for Western analysis. This may, in part, explain why previous studies did not detect L protein expression in T98G cells. We found that in infected T98G cell cultures allowed to progress beyond the usual permissive cell time course (216 h without passaging), the L proteins (MCP and pp28) appeared at very low levels by 120 hpi (Fig. 2C), as also confirmed by the IF analysis using the passaging protocol. This delay in expression may explain why Jault et al. (13) did not detect L protein expression in T98G cells.
Full development of replication centers into large UL44-positive foci and the initial localization of input pp65 to the nucleus appeared to be delayed in T98G cells (Fig. 4 and 5). Although multiple small foci were observed by 24 hpi, bipolar foci were not seen until 72 hpi (∼24 h later than those in infected permissive CCF cells), with large single foci appearing after an additional 24 h (at day 1 of P1). Two particular details need to be pointed out: the percentage of cells containing UL44+ foci in the first 72 hpi was very small (11% or less of the total population), and even at later times during the course of infection, the focus-positive cell population always included a mix of cells with multiple small and advanced-stage foci.
The proportion of focus-positive cells among the total population was very low (∼10%) during P0 but increased to ∼20% during P1. Thereafter, the level of UL44 focus-positive cells (detectable by IF analysis) paralleled the percentage of IE+ cells (Fig. 7). It is interesting to speculate that there was a delay in E gene expression in cells that were IE+, causing a further delay in the development of replication centers. These delays were very similar to what we previously observed in p53−/− cells (5). Interestingly, the T98G cells possess a mutation in p53 (which affects its DNA binding domain, essentially rendering it nonfunctional [12, 19, 34]), which may have contributed to the delay in kinetics we observed in these cells.
pp65 localization in T98G cells was also different from that in the other cell types. First, the initial transit of input pp65 into the nucleus was delayed in the large majority of cells, despite the deposition of the viral genome into ∼70% of the unwashed cells by 6 hpi. Under these conditions, only ∼30% of the total population of cells showed clear pp65 nuclear staining at 24 hpi. Was this the same 30% of cells that went on to express IE proteins during P0? This group may be linked to the initial entry and transit of the genome into the cellular nucleus. In experiments with BrdU-labeled virus, when cells were washed after 1 h of incubation with virus, only ∼30% of cells contained BrdU-labeled virions within their nuclei at 6 hpi. Also, under these wash conditions, the number of labeled virions per cell was significantly lower. If there was a block of initial binding and entry into the cell, perhaps only in those cells in which the virus very quickly penetrated the plasma membrane and transited into the nucleus did the infection proceed to the point of gene expression. After initial infection and entry, if rapid transit did not occur, perhaps the cell was able to mount an antiviral response not allowing gene expression. Future experiments will be aimed at addressing these questions.
Plaque- and focus-forming assays determined that infectious virus was produced at very low levels in the HCMV-infected T98G cells and lasted through several passages (Fig. 6). Even though Jault et al. stated that viral gene expression was blocked at the E stage (13), they did report low levels of virus release, similar to the levels we observed. Poland et al. reported no release of infectious virus from T98G cells (26). These observed differences may stem from several sources, including differences in the MOI, the number of cells originally seeded, the culture medium volume, or the time pi of sample collection for titer determination. Additionally, the duration of these authors' studies was too brief to observe the L protein synthesis seen in our experiments, which did not occur before 120 hpi. Titers varied by passage, with a high of 793 PFU/ml at day 3 of P1. Titers dropped over the next several passages until they were undetectable after P5 (Fig. 6). Correspondingly, the percentage of virus-shedding cells in the total population, as measured by focus-forming assays, was approximately 20% in P0 and P1 but decreased thereafter until no virus-shedding cells were detectable after P6. As we continued to observe Ag-positive cells through day 3 of P8, it is possible that virus levels below our limits of detection were present even beyond our observational period of 27 dpi. Notwithstanding this conjecture, detectable low-level virus shedding lasted 18 to 21 days, which is markedly longer than previously reported for infections of permissive cells.
HCMV infection of primary fibroblasts and CCF cells caused the arrest of the cells, and eventually these cells died via necrosis. These cells could not be passaged generationally. HCMV infection of T98G cells did not cause arrest, and cell division continued even after viral protein expression began (Fig. 8). The ability of HCMV to persist within dividing T98G cells may promote the sustained shedding of virus.
Of particular interest in the long-term infections of T98G cells was that no single phenotype for UL44 focus size was ever observed, even (or maybe especially) at later passages. Our data indicate that several events may have happened concurrently in our cultures. (i) During most passages, low-level virus release occurred. Therefore, it was possible that Ag-negative cells were infected, reinfected, or activated and thus converted into Ag-positive cells. (ii) Ag-positive cells were dividing (an observation we believe is novel), with both daughter cells expressing viral proteins. (iii) Cells that were initially infected (as judged by the IF-visualized localization of input pp65 and viral genomes within their nuclei), but not IE+, later began de novo viral protein synthesis. A combination of these factors likely contributed to the relatively long-term infection we observed in our experiments.
Our study is not the first report of relatively long-term infections of brain-derived cell types. There have been three previous reports of the establishment of persistent HCMV infections in brain cells (7, 8, 23). Ogura and colleagues established a persistent HCMV infection in cells of the permissive 118MGC (glioma) cell line. They infected at an MOI of 10, but unlike in our experiments, within 6 days 95% of the cells were dead and only a very small subset of cells survived. These cells were continuously passaged for over a year, with low levels of virus shed from <15% of cells. It was shown that a mutation in the viral genome was responsible for the slow growth kinetics of the virus that enabled this persistent infection to proceed.
In 1996 and again in 1998, Cinatl and colleagues established persistent HCMV infections in cells of the neuroblastoma cell lines UKF-NB2 and NB4. Low levels of virus were shed from <20% of the cells for over 2 years. It is not clear from the descriptions in the two papers whether, as in the case of the Ogura study, a very small subpopulation of initially infected cells survived cell lysis. However, judging from the report that cultures were grown for 22 days before initial subculturing began, this is a reasonable assumption. Our cultures were inherently different in this respect from the cultures in these studies. From the outset, T98G cells displayed a level of infection characteristic of semipermissive cells, in which all cells in the population receive viral genomes but individually expressed viral Ags at very different rates.
We realize that IF analysis is not the most sensitive assay and that we may be missing a small percentage of Ag-positive cells within the population by using this form of analysis. However, it is clear from our studies that the number of detectable Ag-positive cells did decline over the time course. We speculate that these cultures could express viral Ags for a longer period and at higher percentages if they did not require continuous subculturing. Virion-producing cells would continuously shed into localized pockets, increasing the likelihood of neighboring cells' becoming infected, reinfected, or activated and producing regions of Ag-positive cells. In our passaging protocol, Ag-positive cells were dispersed every 3 days, inhibiting their opportunity to create islands of Ag-positive cells. Initial experiments addressing this possibility (starting with a smaller number of cells), although preliminary, do indicate the development of large clusters of Ag-positive cells that appear to maintain a higher percentage of Ag positivity than the passaged cells over the same time frame (data not shown).
In vivo, latent or persistent CMV infection is found principally in white blood cells, the genital tract, salivary glands, etc. (as reviewed in reference 27). Latently or persistently CMV-infected cells may serve as a reservoir for infecting neighboring tissues or organs. In the isolated compartment of the brain, where most cell types are susceptible to CMV (at least to some degree), such a virus reservoir may be very harmful, especially during fetal development. As previously described for the mouse model of congenital infection, perhaps reservoirs for the virus that remain Ag positive exist, shedding very low levels of virus and thus going undetected and undisturbed by the immune system (15).
We feel that T98G cells may provide a new model system to study latent viral infection within the brain. This system has the characteristics of long-term, low-level gene expression and viral shedding important for such a system. We observed Ag-positive cells through 27 dpi (day 3 of P8) by using IF analysis as our assay system; however, low-level expression that was below our limit of detection may persist for longer periods. Future studies will utilize in situ hybridization to look for latent viral genomes within the late-passage populations. In addition, we will try to reactivate viral gene expression using various techniques. Utilizing a virus that expresses an IE2-green fluorescent protein fusion, we also plan to sort IE+ from IE− cell populations at early times pi. In this way, we will be better able to monitor the distinct populations and assess the potential for reactivation in the initially IE− but genome-positive cells. We will also be able to assess the rates of cell division in both populations and monitor the IE+ population in real time.
In summary, our findings demonstrate that the course of HCMV infection in T98G cells is delayed compared to that in fully permissive cells but that all temporal viral protein classes are expressed. They also show that full viral replication center development occurs and that infectious virus particles are shed for several passages (to P6). Perhaps most importantly, our studies show that viral Ag-positive and virus-shedding cells continue to divide and that their daughter cells remain Ag positive.
Acknowledgments
This work was supported by NIH grants RO1-AI51463 and P20 RR015587 (COBRE program) to E.A.F.
We thank John O'Dowd for critical reading of the manuscript and Melinda Ouwerkerk for help with sample processing.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Bale, J. F., Jr. 1984. Human cytomegalovirus infection and disorders of the nervous system. Arch. Neurol. 41:310-320. [DOI] [PubMed] [Google Scholar]
- 2.Barkovich, A. J., and C. E. Lindan. 1994. Congenital cytomegalovirus infection of the brain: imaging analysis and embryologic considerations. AJNR Am. J. Neuroradiol. 15:703-715. [PMC free article] [PubMed] [Google Scholar]
- 3.Becroft, D. M. 1981. Prenatal cytomegalovirus infection: epidemiology, pathology and pathogenesis. Perspect. Pediatr. Pathol. 6:203-241. [PubMed] [Google Scholar]
- 4.Britt, W. J., and L. Vugler. 1987. Structural and immunological characterization of the intracellular forms of an abundant 68,000 Mr human cytomegalovirus protein. J. Gen. Virol. 68:1897-1907. [DOI] [PubMed] [Google Scholar]
- 5.Casavant, N. C., M. H. Luo, K. Rosenke, T. Winegardner, A. Zurawska, and E. A. Fortunato. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 80:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheeran, M. C., S. Hu, H. T. Ni, W. Sheng, J. M. Palmquist, P. K. Peterson, and J. R. Lokensgard. 2005. Neural precursor cell susceptibility to human cytomegalovirus diverges along glial or neuronal differentiation pathways. J. Neurosci. Res. 82:839-850. [DOI] [PubMed] [Google Scholar]
- 7.Cinatl, J., Jr., J. Cinatl, J. U. Vogel, R. Kotchetkov, P. H. Driever, H. Kabickova, B. Kornhuber, D. Schwabe, and H. W. Doerr. 1998. Persistent human cytomegalovirus infection induces drug resistance and alteration of programmed cell death in human neuroblastoma cells. Cancer Res. 58:367-372. [PubMed] [Google Scholar]
- 8.Cinatl, J., Jr., J. U. Vogel, J. Cinatl, B. Weber, H. Rabenau, M. Novak, B. Kornhuber, and H. W. Doerr. 1996. Long-term productive human cytomegalovirus infection of a human neuroblastoma cell line. Int. J. Cancer 65:90-96. [DOI] [PubMed] [Google Scholar]
- 9.Conboy, T. J., R. F. Pass, S. Stagno, W. J. Britt, C. A. Alford, C. E. McFarland, and T. J. Boll. 1986. Intellectual development in school-aged children with asymptomatic congenital cytomegalovirus infection. Pediatrics 77:801-806. [PubMed] [Google Scholar]
- 10.Fortunato, E. A., M. L. Dell'Aquila, and D. H. Spector. 2000. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 97:853-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukami, T., S. Nakasu, K. Baba, M. Nakajima, and M. Matsuda. 2004. Hyperthermia induces translocation of apoptosis-inducing factor (AIF) and apoptosis in human glioma cell lines. J. Neuro-oncol. 70:319-331. [DOI] [PubMed] [Google Scholar]
- 13.Jault, F. M., J. M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jault, F. M., S. A. Spector, and D. H. Spector. 1994. The effects of cytomegalovirus on human immunodeficiency virus replication in brain-derived cells correlate with permissiveness of the cells for each virus. J. Virol. 68:959-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosugi, I., H. Kawasaki, Y. Arai, and Y. Tsutsui. 2002. Innate immune responses to cytomegalovirus infection in the developing mouse brain and their evasion by virus-infected neurons. Am. J. Pathol. 161:919-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecointe, D., C. Hery, N. Janabi, E. Dussaix, and M. Tardieu. 1999. Differences in kinetics of human cytomegalovirus cell-free viral release after in vitro infection of human microglial cells, astrocytes and monocyte-derived macrophages. J. Neurovirol. 5:308-313. [DOI] [PubMed] [Google Scholar]
- 17.Lokensgard, J. R., M. C. Cheeran, G. Gekker, S. Hu, C. C. Chao, and P. K. Peterson. 1999. Human cytomegalovirus replication and modulation of apoptosis in astrocytes. J. Hum. Virol. 2:91-101. [PubMed] [Google Scholar]
- 18.Luo, M. H., K. Rosenke, K. Czornak, and E. A. Fortunato. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto, H., M. Shimura, T. Omatsu, K. Okaichi, H. Majima, and T. Ohnishi. 1994. p53 proteins accumulated by heat stress associate with heat shock proteins HSP72/HSC73 in human glioblastoma cell lines. Cancer Lett. 87:39-46. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy, M., D. Auger, and S. R. Whittemore. 2000. Human cytomegalovirus causes productive infection and neuronal injury in differentiating fetal human central nervous system neuroepithelial precursor cells. J. Hum. Virol. 3:215-228. [PubMed] [Google Scholar]
- 21.Odeberg, J., N. Wolmer, S. Falci, M. Westgren, A. Seiger, and C. Soderberg-Naucler. 2006. Human cytomegalovirus inhibits neuronal differentiation and induces apoptosis in human neural precursor cells. J. Virol. 80:8929-8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odeberg, J., N. Wolmer, S. Falci, M. Westgren, E. Sundtrom, A. Seiger, and C. Soderberg-Naucler. 2007. Late human cytomegalovirus (HCMV) proteins inhibit differentiation of human neural precursor cells into astrocytes. J. Neurosci. Res. 85:583-593. [DOI] [PubMed] [Google Scholar]
- 23.Ogura, T., J. Tanaka, S. Kamiya, H. Sato, H. Ogura, and M. Hatano. 1986. Human cytomegalovirus persistent infection in a human central nervous system cell line: production of a variant virus with different growth characteristics. J. Gen. Virol. 67:2605-2616. [DOI] [PubMed] [Google Scholar]
- 24.Pass, R. F., S. Stagno, G. J. Myers, and C. A. Alford. 1980. Outcome of symptomatic congenital cytomegalovirus infection: results of long-term longitudinal follow-up. Pediatrics 66:758-762. [PubMed] [Google Scholar]
- 25.Poland, S. D., L. L. Bambrick, G. A. Dekaban, and G. P. Rice. 1994. The extent of human cytomegalovirus replication in primary neurons is dependent on host cell differentiation. J. Infect. Dis. 170:1267-1271. [DOI] [PubMed] [Google Scholar]
- 26.Poland, S. D., P. Costello, G. A. Dekaban, and G. P. Rice. 1990. Cytomegalovirus in the brain: in vitro infection of human brain-derived cells. J. Infect. Dis. 162:1252-1262. [DOI] [PubMed] [Google Scholar]
- 27.Reddehase, M. J., J. Podlech, and N. K. Grzimek. 2002. Mouse models of cytomegalovirus latency: overview. J. Clin. Virol. 25(Suppl. 2):S23-S36. [DOI] [PubMed] [Google Scholar]
- 28.Rosenke, K., and E. A. Fortunato. 2004. Bromodeoxyuridine-labeled viral particles as a tool for visualization of the immediate-early events of human cytomegalovirus infection. J. Virol. 78:7818-7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenke, K., M. A. Samuel, E. T. McDowell, M. A. Toerne, and E. A. Fortunato. 2006. An intact sequence-specific DNA-binding domain is required for human cytomegalovirus-mediated sequestration of p53 and may promote in vivo binding to the viral genome during infection. Virology 348:19-34. [DOI] [PubMed] [Google Scholar]
- 30.Shinmura, Y., I. Kosugi, S. Aiba-Masago, S. Baba, L. R. Yong, and Y. Tsutsui. 1997. Disordered migration and loss of virus-infected neuronal cells in developing mouse brains infected with murine cytomegalovirus. Acta Neuropathol. (Berlin) 93:551-557. [DOI] [PubMed] [Google Scholar]
- 31.Spiller, O. B., L. K. Borysiewicz, and B. P. Morgan. 1997. Development of a model for cytomegalovirus infection of oligodendrocytes. J. Gen. Virol. 78:3349-3356. [DOI] [PubMed] [Google Scholar]
- 32.Stagno, S., R. F. Pass, G. Cloud, W. J. Britt, R. E. Henderson, P. D. Walton, D. A. Veren, F. Page, and C. A. Alford. 1986. Primary cytomegalovirus infection in pregnancy. Incidence, transmission to fetus, and clinical outcome. JAMA 256:1904-1908. [PubMed] [Google Scholar]
- 33.Tamashiro, J. C., L. J. Hock, and D. H. Spector. 1982. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J. Virol. 42:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ullrich, S. J., W. E. Mercer, and E. Appella. 1992. Human wild-type p53 adopts a unique conformational and phosphorylation state in vivo during growth arrest of glioblastoma cells. Oncogene 7:1635-1643. [PubMed] [Google Scholar]