Abstract
Early region 3 genes of human adenoviruses contribute to the virus life cycle by altering the trafficking of cellular proteins involved in adaptive immunity and inflammatory responses. The ability of early region 3 genes to target specific molecules suggests that they could be used to curtail pathological processes associated with these molecules and treat human disease. However, this approach requires genetic dissection of the multiple functions attributed to early region 3 genes. The purpose of this study was to determine the role of targeting on the ability of the early region 3-encoded protein RIDα to downregulate the EGF receptor. A fusion protein between the RIDα cytoplasmic tail and glutathione S-transferase was used to isolate clathrin-associated adaptor 1 and adaptor 2 protein complexes from mammalian cells. Deletion and site-directed mutagenesis studies showed that residues 71-AYLRH of RIDα are necessary for in vitro binding to both adaptor complexes and that Tyr72 has an important role in these interactions. In addition, RIDα containing a Y72A point mutation accumulates in the trans-Golgi network and fails to downregulate the EGF receptor when it is introduced into mammalian cells as a transgene. Altogether, our data suggest a model where RIDα is trafficked directly from the trans-Golgi network to an endosomal compartment, where it intercepts EGF receptors undergoing constitutive recycling to the plasma membrane and redirects them to lysosomes.
Adenoviruses (Ads) are nonenveloped DNA viruses that replicate and assemble in the host cell nucleus (13). Ads are responsible for approximately 5% of acute upper respiratory tract (3) and 15% of lower respiratory tract (1) infections in infants and children. Similar to other DNA viruses, Ads are also capable of establishing persistent infections, because they have evolved numerous strategies to evade host antiviral surveillance mechanisms. Some of these same mechanisms also facilitate survival of the virus during an acute infection. Thus, a thorough understanding of viral genes that control host immune and inflammatory responses is fundamental to our ability to prevent and treat virus-induced disease at multiple levels. These mechanisms have also influenced strategies for designing Ad-based vectors for gene therapy (14).
The early region 3 (E3) transcription genes of human Ads encode several proteins that exploit the intracellular trafficking machinery to modify host immune or inflammatory responses (11, 29). Thus, E3 proteins have served as novel probes of membrane transport mechanisms in addition to providing insights to Ad pathophysiology. The most abundant E3 protein, E3/19K, suppresses the host adaptive immunity response by retaining class I MHC molecules in the endoplasmic reticulum. Other E3 proteins affect the functions of molecules involved in proliferation and apoptosis, intracellular cell signaling events linked to NF-κB, and secretion of proinflammatory chemokines. Two E3 proteins, RIDα (also called E3-10.4 and E3-13.7) and RIDβ (also called E3-14.5), that act to remove several receptors from the surface of host cells, including death receptors FAS and TRAIL, tumor necrosis factor receptor 1 (TNFR1), and EGF receptor (EGFR), have been identified. These two E3 proteins have independent functions and are also capable of forming a molecular complex that has been named the receptor internalization and degradation (RID) complex. However, mutations that affect the ability of the RID complex to downregulate one receptor molecule have no effect on expression with a second class of receptors, suggesting that regulation of different targets by the RID complex involves multiple mechanisms (9).
RIDα is a type II membrane protein expressed by all human Ad serotypes (27). Its basic structural organization consists of a short N-terminal cytosolic domain (∼4 residues), two transmembrane domains connected by an exocytic loop domain (∼17 residues), and a C-terminal cytosolic tail domain (∼28 residues). We have shown previously that RIDα is sufficient to downregulate the EGFR (25) and that it localizes to the endocytic pathway, where it diverts EGFRs in a constitutive recycling loop to lysosomes (10). However, other laboratories have reported that RIDα functions only in the context of the RID complex that localizes to the plasma membrane, where it regulates EGFR internalization (43). Although the precise role of EGFR downregulation during an Ad infection is not completely understood, the EGFR has been linked to TNF-α-dependent NF-κB activity (22) and to the release of proinflammatory chemokines (38) in other biological settings.
The RIDα cytosolic tail has several consensus sorting signals that likely engage the host cell machinery necessary for its localization and function. The goal of this study was to identify these sorting signals and to disable them by site-directed mutagenesis. The study focused on potential interactions with clathrin adaptor protein (AP) complexes involved in membrane protein sorting at the level of the trans-Golgi network (TGN) (AP class 1 [AP-1]) and the plasma membrane (AP-2) (42). Our results indicate that RIDα residues 71-AYLRH bind to both classes of APs. In addition, we show that a Y72A point mutation traps RIDα in the TGN but not the plasma membrane, suggesting that the Ad protein encounters AP-1 first in the biosynthetic pathway. Our data also support a model where AP-2 serves as a quality control function to capture RIDα molecules that leak to the plasma membrane and redirect them back to endosomes, similar to many other membrane cargoes with overlapping AP binding sites (2). These studies were carried out with RIDα transgenes, confirming the original Ad mapping studies that demonstrated the ability of this protein to modulate EGFR trafficking independent of other E3 proteins (7).
MATERIALS AND METHODS
Cell lines, antibodies, Ad stocks, and transfections.
Human hepatocellular carcinoma-derived N-PLC-PRF/5 cells (6) and fetal lung-derived WI-38 cells (20) were maintained in minimal essential medium (MEM). Human epithelial-derived A431 carcinoma cells (12) and human embryonic kidney 293 (HEK-293) cells (16) were maintained in Dulbecco's modified MEM. Chinese hamster ovary (CHO) cells (40) were maintained in MEM alpha modification. Human lung carcinoma-derived A549 cells (12) were maintained in Ham's F12 medium. All media were supplemented with 10% fetal bovine serum and 2 mM glutamine. Cells were grown at 37°C with 5% CO2 and were subcultured with 0.25% trypsin-0.1% EDTA in phosphate-buffered saline (PBS).
The following antibodies were used: α-adaptin, γ-adaptin, FLAG M2, and FLAG-BioM2 monoclonal antibodies from Sigma (St. Louis, MO); transferrin receptor (TfR) monoclonal antibody from Zymed (San Francisco, CA); the human-specific EGFR EGF-R1 monoclonal antibody (46); RIDα rabbit polyclonal antibody produced with a synthetic peptide corresponding to residues 76 to 91 (27); furin convertase rabbit polyclonal antibody from ABR (Golden, CO); glutathione S-transferase (GST) goat polyclonal antibody from Amersham-Pharmacia (Piscataway, NJ); and Golgi gp125 antibody obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, The University of Iowa, Iowa City, IA. Secondary antibodies were purchased from Amersham-Pharmacia (horseradish peroxidase [HRP]) or Jackson ImmunoResearch (fluorescent and HRP; West Grove, PA).
Ad stocks were grown in HEK-293 cells, and titers were determined by plaque assay, using standard techniques. The Ad2-Ad5 recombinant Ad mutant that overproduces the RIDα protein has been described elsewhere (7). Cells were acutely infected with approximately 200 PFU per cell, according to established protocols (26).
CHO cells were transiently transfected using TransIt-CHO (Mirus, Madison, WI) as directed by the manufacturer, and HEK-293 cells were transfected using FuGENE 6 (Roche, Basel, Switzerland) following the manufacturer's instructions.
Cloning and mutagenesis.
Wild-type and modified RIDα carboxyl-terminal sequences were subcloned into pGEX-3X (Amersham-Pharmacia) as follows. RIDα residues Asp64 to Leu91 (see Fig. 1A) were subcloned in the M13mp18 single-stranded DNA phage vector (New England BioLabs, Ipswich, MA). Y72A and Y79A point mutations were introduced using the M13mp18 RIDα template, a Sculptor in vitro mutagenesis system (Amersham-Pharmacia), and the following mutagenic primers to introduce tyrosine-to-alanine substitutions (in boldface type): 5′-TGTGCGCATTGCGGCCCTCAGGCACCAT-3′ for Y72A, and 5′-GCACCATCCGCAAGCCAGAGACAGGACT-3′ for Y79A. Wild-type and mutated sequences were amplified by PCR using a forward primer (5′-CAUCAUCAUCAUGATATCGACTGGGTTTGTGTGCGCATTGC-3′) designed to anneal to the codon for Asp64 (in boldface type) and a reverse primer (5′-CUACUACUACUAGATATCTTTCGGAACCACCATCAAAACAGGATTTTCG-3′) to anneal to M13mp18 vector sequences 3′ to the polylinker region. Both primers incorporate EcoRV restriction sites (underlined) and also have noncomplementary dU repeats on the 5′ ends (in italics). The dU repeats in the PCR products were depurinated with uracyl DNA glycosidase to expose single-stranded 3′ ends and enable base-pairing to complementary ends in the linear shuttle vector pAMP (Invitrogen, Gaithersburg, MD). EcoRV fragments were subcloned in frame at the SmaI site in pGEX-3X, which reconstituted the RIDα Ile63 codon. The GST fusion plasmid encoding RIDα carboxyl-terminal sequences truncated to residues Tyr70 or Tyr79 were prepared using PCR and full-length RIDα cloned in the pcDNA1/Amp eukaryotic expression vector (Clontech, Mountain View, CA) as a template. Sequences were amplified with a forward primer 5′-CTAACTAGAGAACCCACTGC-3′ designed to anneal to vector sequences 5′ to the RIDα coding region, and reverse mutagenic primer 5′-AGTGGGATCCTAGTATTGCGGGTGGTGCCTGA-3′ to incorporate an A71Stop codon or 5′-CCTAGGATCCTAAATGCGCACACAAACCCA-3′ to incorporate an R80Stop codon, respectively (both stop codons are in boldface type). The reverse mutagenic primers also incorporated BamHI sites (underlined) compatible with the pcDNAI/Amp polylinker. The cytoplasmic tail sequences were amplified using forward (5′-GTTTCCCGGGATTGACTGGGTTTGTG-3′) and reverse (5′-GAGTCCCGGGTTGAGAGTCAGCAGTAGC-3′) primers complementary to Ile63 and sequences in pcDNAI/Amp 3′ to the RIDα coding region, respectively. Both primers incorporate SmaI sites (underlined) to facilitate in-frame cloning at the SmaI site in the pGEX-3X polylinker region. The GST fusion plasmid encoding RIDα carboxyl-terminal sequences truncated to residue His75 was created using an overlap PCR extension technique, the RIDα/pcDNA1/Amp template, and the following four primers. Primers 1 and 4 are forward and reverse flanking primers that anneal to unique restriction enzymes in the polylinker region at the 5′ and 3′ ends of the RIDα coding region that were already described, and primers 2 and 3 are forward (5′-ACCTCAGGCATAACCGCAATACA-3′) and reverse (5′-CTGTATTGCGGTTAGTGCCTGAG-3′) mutagenic primers that replace His76 with a stop codon (in boldface type). The first round of PCRs were carried out with primers 1 and 3 or primers 2 and 4, creating two overlapping PCR products. Both fragments were gel purified and used as templates for a second PCR with the flanking primers to generate a 627-bp product with the H76Stop mutation. This fragment was inserted into pcDNA1/Amp using restriction enzymes incorporated in the ends of the product compatible with restriction sites in the polylinker region. Carboxyl-terminal sequences encoding the H76Stop were amplified and subcloned in pGEX-3X exactly as described for the A71Stop and R80Stop constructs.
FIG. 1.
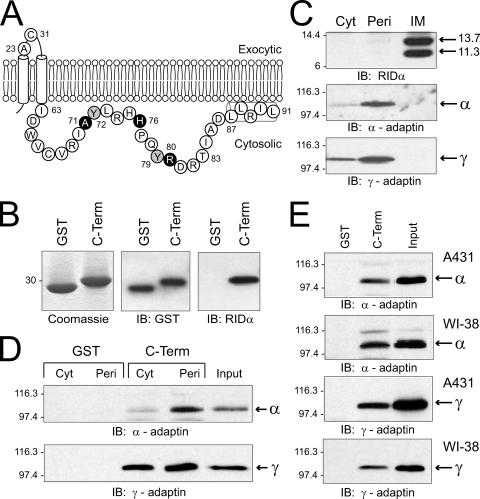
Binding of RIDα cytosolic tail to AP complexes. (A) Schematic showing membrane topology of RIDα and the complete amino acid sequence (in single-letter code) of the carboxyl terminus (Ile63-Leu91) that was linked to GST for expression in Escherichia coli. Tyr72 and Tyr79 or residues that were changed to stop codons (Fig. 2) are highlighted in gray or black, respectively. Nuclear magnetic resonance studies have shown that residues 87-LLRIL comprise an amphipathic helix stabilized by interactions with membrane mimetic micelles (45). Two additional residues are presented in the exocytic loop connecting the two membrane spanning helices: Ala23, comprising the amino terminus of the 11.3-kDa cleaved form of the protein produced cotranslationally by signal peptidase, and Cys31, which forms intermolecular disulfide bonds (27). (B) GST or the C-terminal (C-Term) fusion protein was purified from E. coli by glutathione affinity chromatography and resolved by SDS-PAGE for detection by Coomassie staining or by immunoblotting (IB) with GST or RIDα-specific antibodies. (C) Ad-infected cells were fractionated into crude cytosol (Cyt) and peripheral (Peri) and integral membrane (IM) proteins, as described in Materials and Methods. Equal aliquots of total cell equivalents were resolved by SDS-PAGE and immunoblotted with antibodies to RIDα, α-adaptin, or γ-adaptin. (D) GST or C-terminal fusion proteins were incubated with crude cytosol or peripheral membrane proteins isolated from N-PLC-PRF/5 cells, and bound proteins were immunoblotted with adaptin-specific antibodies. (E) Fusion proteins were incubated with peripheral membrane proteins from A431 or WI-38 cells and analyzed as described for panel D. Molecular weight standards: β-galactosidase, 116,300; phosphorylase B, 97,400; carbonic anhydrase, 30,000; lysozyme, 14,400; aprotinin, 6,000.
The cDNAs encoding full-length wild-type RIDα and RIDα with a single Y72A or Y79A amino acid substitution for expression in eukaryotic cells were constructed as follows. Wild-type RIDα cloned in pcDNAI/Amp was used as template for a PCR using forward (5′-ATCGTAAAGATCTTGATTCCTCGAGTTCTTATATTATTG-3′) and reverse (5′-CTAAGATCTCCTTAAAGAATTCTGAGAAGATCAGCTATAGTCCTG-3′) primers to amplify the RIDα open reading frame and incorporate flanking BglII restriction sites (underlined). PCR products were digested with BglII and ligated to an amino-terminal FLAG epitope in the polylinker region of the pExchange2 plasmid (Stratagene, La Jolla, CA) digested with the same restriction enzyme. The Y72A and Y79A point mutations were incorporated into pExchange2/RIDα using a QuikChange XL site-directed mutagenesis kit (Stratagene), and PCR primers 5′-GTGTGCGCATTGCGGCCCTCAGGCACCATC-3′ (forward) and 5′-GATGGTGCCTGAGGGCCGCAATGCGCACAC-3′ (reverse) (Y72A substitution in boldface type), or 5′-AGGCACCATCCGCAAGCCAGAGACAGGACTATAG-3′ (forward) and 5′-CTATAGTCCTGTCTCTGGCTTGCGGATGGTGCCT-3′ (reverse) (Y79A substitution in boldface type), according to the manufacturer's instructions.
PCR primers were designed using the DNASTAR software package (DNASTAR, Inc., Madison, WI). PCR amplifications were carried out using a RoboCycler 40 temperature cycler (Stratagene, La Jolla, CA). All PCR products and religated recombinant products were sequenced by automated DNA sequencing (Cleveland Clinic Foundation Genomics Core, Cleveland, OH).
Fusion proteins.
GST fusion proteins were purified from BL21 cells that were freshly transformed with pGEX-3X plasmids. Bacteria were cultured at 37°C to an optical density at 600 nm of approximately 0.6, induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h at room temperature, and collected by low-speed centrifugation. Cells were subjected to one freeze-thaw cycle, resuspended in a solution of 50 mM Tris (pH 7.7), 0.1 M NaCl, 0.2 mM EDTA, and protease inhibitors (0.2 mM phenylmethylsulfonyl fluoride and 1 μM leupeptin), and then digested with lysozyme (0.1 mg/ml) for 1 h at room temperature. MgSO4 was added to a final concentration of 3 mM, and bacterial lysates were digested for an additional 1 h at room temperature with 0.02 mg (each) of DNase and RNase. Lysates were adjusted to pH 7.4 and incubated with 1.5% l-lauryl sarcosine to solubilize inclusion bodies for 15 min on ice, followed by centrifugation at 12,000 × g for 10 min at 4°C. Supernatants were adjusted to 3% Triton X-100 and incubated with glutathione-Sepharose beads (Amersham-Pharmacia) for 20 min at 4°C with rotation. Beads with attached fusion proteins were washed three times with a solution of 50 mM Tris (pH 7.4), 10 mM MgCl2, and 1% Triton X-100 supplemented with 0.4 M NaCl (high-salt wash) and twice with the same solution supplemented with 0.15 M NaCl (low-salt wash). Beads with attached fusion proteins were incubated with crude subcellular fractions enriched for clathrin adaptors, by using the method described in the next paragraph, for 20 min at 4°C, and the beads were washed three times with the high-salt solution and then twice with the low-salt solution. Fusion protein complexes were solubilized with Laemmli buffer and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for immunoblotting using standard methods.
Cell fractionation methods.
Crude subcellular fractionation was carried out essentially as described in reference 41. Briefly, cells were rinsed twice and then scraped in PBS supplemented with 2 mM EDTA, 5 mM EGTA, and protease inhibitors. Cells were resuspended in 0.1 M morpholineethansulfonic acid (MES; pH 6.5), 0.2 M EDTA, 0.5 mM MgCl2, 0.02% NaN3, 10 mg/ml bovine serum albumin, and protease inhibitors and then lysed with 1% NP-40 for 5 min at room temperature. Postnuclear supernatants were centrifuged at 60,000 × g for 30 min at 4°C in an Optima TL Ultracentrifuge (Beckman Instruments, Inc., Palo Alto, CA) to collect the supernatant corresponding to crude cytosol. The resulting membrane pellet was resuspended in the NP-40 lysis buffer and incubated with 0.5 M Na2CO3 for 5 min on ice to release peripheral membrane proteins. The mixture was then centrifuged at 50,000 × g for 20 min at 4°C, and the peripheral membrane protein-enriched supernatant was incubated with 0.5 M KH2PO4 for 1 h on ice. Integral membrane proteins were solubilized by incubating membrane pellets with radioimmunoprecipitation assay (RIPA) detergent (1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) in 50 mM Tris, pH 8.0, supplemented with 150 mM NaCl, 2 mM EDTA, 5 mM EGTA, and protease inhibitors, for 30 min on ice.
Cell homogenates were also fractionated on Percoll (Amersham-Pharmacia) gradients essentially as described in reference 18. Briefly, cells were rinsed twice with PBS supplemented with 2 mM EDTA and 5 mM EGTA and then scraped in ice-cold homogenization buffer (HB) consisting of 10 mM HEPES (pH 7.5), 0.25 M sucrose, 1 mM EDTA, and protease inhibitors. Cells were collected by centrifugation, resuspended in HB, and homogenized with 22 strokes of a Dounce homogenizer. The homogenate was diluted with an equal volume of fresh HB and centrifuged at 400 × g for 10 min at 4°C to pellet unbroken cells and nuclei. Postnuclear supernatants were adjusted to a final concentration of 27% Percoll in 0.25 M sucrose using a 90% Percoll stock solution and then layered over a 1-ml sucrose cushion consisting of 10× HB. Gradients were centrifuged for 90 min at 25,000 × g in an SS34 fixed-angle rotor (Sorvall Instruments, Newtown, CT) without braking. A total of nine 1.2-ml fractions were collected manually, starting from the top of the gradient. Fractions were analyzed by comparing equal aliquots of total cellular protein determined by Bradford assay (Bio-Rad Laboratories, Hercules, CA) or by immunoprecipitation after membranes were solubilized with the RIPA lysis buffer and centrifuged at 100,000 × g for 30 min at 4°C in a TL 100.3 fixed-angle rotor (Beckman Instruments) to precipitate Percoll. Alternatively, fractions were assayed for β-hexosaminidase activity by incubating 20 μg of protein in a solution of 0.1 M MES (pH 6.5), 1 mM p-nitrophenyl-β-d-glucosaminide, and 0.2% Triton X-100 for 90 min at 37°C. The reaction was quenched with 0.5 M glycine, pH 10, and absorbance was read at 405 nm using a model 3550 automatic microplate reader (Bio-Rad). Gradients were also collected from cells that had been incubated with 125I-EGF (50 ng/ml, 150 μCi/μg; PerkinElmer-NEN) or 125I-transferrin (200 ng/ml, 0.5 μCi/μg; PerkinElmer-NEN) to label plasma membrane receptors. Fractions from these gradients were analyzed by gamma counting (Cobra Auto-Gamma counter; Packard Instruments, Meriden, CT).
Immunoprecipitations and immunoblotting.
Immunoprecipitations were carried out using antibodies adsorbed to protein A-Sepharose CL-4B beads (Sigma), or streptavidin agarose (Pierce, Rockford, IL). Immune complexes were centrifuged at 14,000 × g for 15 min at 4°C, washed five times with lysis buffer, solubilized with Laemmli buffer, and resolved by SDS-PAGE using standard methods. Immunodepleted supernatants were centrifuged a second time to ensure removal of immune complexes before they were added to GST fusion protein pull-down assays. Gels with radioactive proteins were treated with En3Hance (PerkinElmer-NEN, Wilmington, DE) for fluorography. Immunoblotting was carried out using proteins transferred to nitrocellulose membranes by standard methods. Nitrocellulose filters were incubated with primary antibodies and the appropriate HRP-conjugated secondary antibodies (Amersham Life Sciences, Inc., Arlington, IL; Jackson ImmunoResearch Laboratories, Inc.) for detection by enhanced chemiluminescence (Amersham Life Sciences). For quantification, immunoblots were incubated with 125I-labeled goat anti-mouse secondary antibody (1 × 106 cpm/ml, 8.07 μCi/μg; PerkinElmer-NEN) for 1 h at room temperature. Blots were air-dried after extensive washing, and radiolabeled proteins were quantified by phosphor storage autoradiography. Digitized images were analyzed using the ImageQuant software package (Molecular Dynamics, Sunnyvale, CA), which averages five measurements of light emission for each pixel location, to give a pixel value that is proportional to the amount of stored radiation.
Confocal microscopy.
HEK-293 cells or CHO cells transiently transfected with RIDα expression plasmids were prepared for staining essentially as described in reference 8. Briefly, cells were permeabilized with 0.5% β-escin in a solution of 80 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, supplemented with 5 mM EGTA and 1 mM MgCl2 for 5 min and then fixed with 3% paraformaldehyde-PBS for 15 min. Cells were stained with primary antibody or secondary antibodies and 4′,6′-diamidino-2-phenylindole (DAPI) for 45 min at room temperature. Antibodies were diluted in a solution containing 0.5% β- escin and 3% radioimmunoassay-grade bovine serum albumin and were blocked with a solution containing 1% normal serum from the host animal used to generate the secondary antibody. Cells were examined with a Leica TCS SP2 with AOBS confocal microscope (Leica, Mannheim, Germany) using the 405-nm wavelength line of a UV laser and the 488/568-nm lines of an argon-krypton laser. Image resolution using a Leica 100×, 1.4-numerical-aperture oil immersion lens and Leica LCS software was 512 by 512 pixels. Some cells were pretreated with nocodazole (100 μM; Sigma) or vehicle (dimethyl sulfoxide) for 30 min prior to staining. Phase contrast images were collected, and the outline of the cells was drawn using Metamorph (Molecular Devices, Sunnyvale, CA) and overlaid onto the respective confocal image.
Surface reduction of extracellular disulfide bonds.
Cells were pulse-labeled for 1 h with l-[35S]cysteine (50 mCi/ml, 1,075 Ci/mmol; PerkinElmer-NEN) in cysteine-free MEM supplemented with 10% dialyzed fetal bovine serum and 0.2% bovine serum albumin and then incubated in chase medium (complete Dulbecco's MEM supplemented with 500 μM nonradioactive cysteine) for 3 h to allow proteins to achieve steady-state localization. Radiolabeled cells were incubated twice (25 min/incubation) with an ice-cold solution of 80 mM l-cysteine, 75 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 0.5 N NaOH, and 1% fetal bovine serum to reduce external disulfide bonds of surface proteins (36). Cells were rinsed twice with PBS containing iodoacetamide (1 mg/ml) and then incubated with the RIPA lysis buffer supplemented with iodoacetamide (1 mg/ml) and protease inhibitors for analysis by immunoprecipitation. Immunoprecipitates were solubilized in Laemmli buffer supplemented with 1 mg/ml iodoacetamide and separated by SDS-PAGE under reducing (0.1 M dithiothreitol [DTT]) or nonreducing (no DTT) conditions.
EGFR protein stability assay.
CHO cells, which lack endogenous EGFRs, were cotransfected with plasmids encoding human EGFR (23) and FLAG-tagged wild-type or Y72A or Y79A RIDα plasmids or empty vector. Cells were radiolabeled with [35S]cysteine for 30 min starting at 48 h posttransfection as described previously. Cells were harvested either immediately or following a 5-h chase in medium supplemented with a 10-fold excess of nonradioactive cysteine. Cells were lysed with RIPA buffer and analyzed by immunoprecipitation, SDS-PAGE, and fluorography.
RESULTS
The C-terminal cytoplasmic tail of RIDα interacts with AP complexes.
We have shown previously that RIDα localizes to the endocytic pathway, where it redirects constitutively recycling EGFRs to lysosomes in Ad-infected cells (10, 26). The RIDα cytoplasmic tail contains a number of putative sorting motifs that may regulate these sorting properties (2), including a dileucine motif at residues 87-LL and two possible tyrosine-based sorting motifs containing Tyr72 and Tyr79, respectively (Fig. 1A). To determine whether the RIDα cytoplasmic tail interacts with clathrin AP complexes involved in membrane protein sorting, we utilized a fusion protein consisting of the 30-amino-acid cytoplasmic domain of RIDα fused to GST (characterized in Fig. 1B) to pull down complexes from mammalian cell extracts. The mammalian cell fractionation scheme involved purification of a peripheral membrane protein fraction to enrich for functional AP complexes according to the method described in reference 41. Figure 1C validates this purification scheme, showing that α-adaptin and γ-adaptin (subunits of AP-2 and AP-1, respectively) are both enriched in the peripheral membrane fraction, but not in integral membrane fractions that include both of the known molecular weight forms (e.g., 13.7-kDa and 11.3-kDa) of the RIDα protein. A small amount of each of these adaptin molecules was also present in crude cytosol, as expected for AP complexes that cycle on and off membranes. Thus, the crude cytosol and peripheral membrane protein fractions were mixed with GST or the C-terminal fusion protein, precipitated with glutathione agarose, washed, and subjected to SDS-PAGE and immunoblot analysis with monoclonal antibodies to α-adaptin and γ-adaptin. As shown in Fig. 1D, α-adaptin and γ-adaptin from a human hepatocellular cell line were both coprecipitated with the C-terminal fusion protein in contrast to GST alone. Similar results were obtained using peripheral membrane protein fractions from a number of other cell lines, including epithelial carcinoma-derived A431 cells and normal human diploid WI-38 fibroblasts (Fig. 1E) and human lung carcinoma-derived A549 cells and HEK cells (data not shown).
Analysis of RIDα cytoplasmic tail mutants.
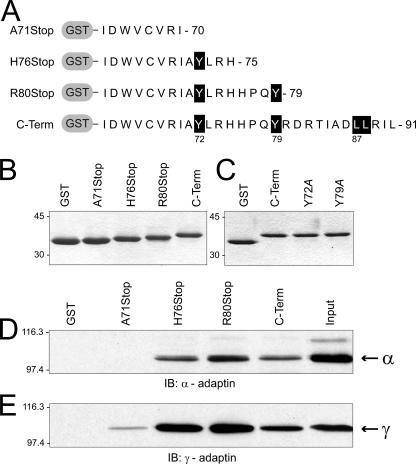
Our next goal was to identify the sequence(s) within the cytoplasmic tail of RIDα that interacts with each of the AP complexes identified in the first set of experiments. As already noted, RIDα contains putative dileucine and tyrosine motifs that could be involved in adaptor complex formation. Thus, we constructed a series of fusion proteins with cytoplasmic tail truncations that are shown in Fig. 2A and B to assess a role for each of these putative signals. The fusion protein with an R80Stop substitution has the carboxyl-terminal dileucine motif deleted, the H76Stop has the distal tyrosine residue Tyr79 deleted, and the A71Stop has all putative sorting motifs deleted. In vitro pull-down assays with these fusion proteins revealed that RIDα residues 71-AYLRH are responsible primarily for binding both types of AP complexes (Fig. 2D and E). These data also show that the carboxyl-terminal dileucine motif does not bind APs directly, contradicting published reports from another laboratory (21). Consistent with the results shown here, we have also observed that the conserved 679-LLRIL sequence localized to the EGFR juxtamembrane region that regulates ligand-induced postendocytic sorting to lysosomes (32, 33) does not bind APs in vitro (data not shown).
FIG. 2.
Mapping the sites of interaction of RIDα with AP complexes using truncation mutants. (A) Schematic showing sequences of cytoplasmically truncated GST fusion proteins used in the mapping studies. Critical residues in putative sorting signals are highlighted by a black background. (B and C) Proteins were purified from E. coli by glutathione affinity chromatography and resolved by SDS-PAGE for detection by Coomassie staining. Proteins in panel B were used in panels D to E of this figure, and those in panel C were used to obtain the data in Table 1. (D and E) Fusion proteins shown in panel B were incubated with peripheral membrane proteins isolated from N-PLC-PRF/5 cells, and bound proteins were immunoblotted (IB) with adaptin-specific antibodies. C-Term, C terminal.
The AP binding site identified in the in vitro mapping studies has one tyrosine at residue Tyr72. To assess its role in AP binding, fusion proteins were constructed with Y72A as well as Y79A point mutations incorporated into the full-length cytoplasmic tail (Fig. 2C). These fusion proteins were incubated with peripheral membrane protein fractions, and adaptin binding was quantified by immunoblotting with 125I-labeled secondary antibodies and phosphor storage autoradiography after the bound proteins were resolved by SDS-PAGE and transferred to nitrocellulose filters. Results from one set of experiments are summarized in Table 1, and similar results were obtained in three separate determinations. We found that the Y72A substitution was associated with reduced binding to both AP-1 and AP-2, although its effect on AP-2 binding was consistently greater than that on AP-1 binding. Interestingly, the Y79A substitution adjacent to the primary AP binding site identified in Fig. 2 was associated with increased binding to AP-2 but reduced binding to AP-1, suggesting that residues outside of the binding motif may be important for AP interactions.
TABLE 1.
Quantification of adaptin binding to fusion proteins
| Fusion proteina | α-Adaptinb
|
γ-Adaptin
|
||
|---|---|---|---|---|
| Amt boundc | % Differenced | Amt bound | % Difference | |
| C terminal | 657 ± 33 | NA | 618 ± 54 | NA |
| Y72A | 140 ± 8 | −79 | 222 ± 19 | −64 |
| Y79A | 1,009 ± 97 | +53 | 373 ± 23 | −40 |
Peripheral membrane proteins were incubated with 100 μg of fusion protein attached to glutathione beads. The fusion proteins tested had either wild-type RIDα Ile63-Leu91 sequences or Ile63-Leu91 sequences with Y72A or Y79A substitutions.
Bound proteins were immunoblotted with adaptin-specific antibodies followed by a 125I-labeled secondary antibody, and blots were exposed to a phosphor screen.
Volume integration of pixel values in a defined area with background subtracted. Pixel values are arbitrary units that are proportional to the amount of radiation stored and re-emitted from the phosphor screen. Values in the table are the means from three independent determinations for each band ± standard errors of the means and have been multiplied by 10−3. Similar results were obtained in two independent determinations.
Percent difference relative to the amount of protein bound to C-terminal fusion protein, which is set to 100%.
AP-1 and AP-2 bind to the RIDα cytoplasmic tail with distinct properties.
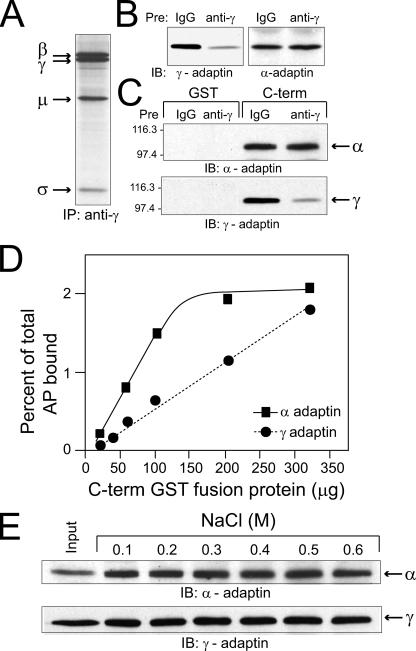
Results presented to this point suggest that AP-1 and AP-2 bind overlapping but distinct regions in the RIDα cytoplasmic tail. To further test this hypothesis, studies were carried out to determine whether there is competition between AP-1 and AP-2 for binding to the C-terminal fusion protein. These studies were possible because the γ-adaptin antibody immunoprecipitates intact AP-1 complexes comprised of all four of the protein subunits (Fig. 3A). Thus, peripheral membrane protein fractions were immunodepleted for AP-1 prior to incubation with GST or the C-terminal fusion protein. AP-2 binding was essentially identical when the C-terminal fusion protein was incubated with cell fractions that were precleared with an isotype-matched nonspecific immunoglobulin G versus the γ-adaptin specific antibody; in comparison, AP-1 binding was substantially reduced in the immunodepleted samples in the same experiment (Fig. 3B). Although the reverse experiment was not possible since immunoprecipitations with the α-adaptin antibody must be carried out under denaturing conditions, these results strongly suggest that AP-1 and AP-2 binding to the RIDα cytoplasmic tail is noncompetitive.
FIG. 3.
Lack of competition between AP-1 and AP-2 for RIDα C-terminal binding. (A) Peripheral membrane proteins from cells that were metabolically labeled with l-[35S]cysteine for 16 h were incubated with the γ-adaptin monoclonal antibody, which immunoprecipitates (IP) intact AP-1 complexes comprised of β, μ, and σ subunits in addition to γ-adaptin. (B and C) Peripheral membrane fractions were preincubated with nonspecific mouse immunoglobulin G (IgG) or with the γ-adaptin monoclonal antibody, and the immunodepleted samples were then incubated with GST or C-terminal (C-term) fusion proteins. Aliquots of the immunodepleted samples (B) and proteins bound to glutathione beads (C) were analyzed by immunoblotting (IB) with adaptin-specific antibodies. (D) Equal aliquots of peripheral membrane proteins were added to increasing concentrations of the C-terminal fusion protein indicated on the x axis. Bound proteins were transferred to nitrocellulose filters, which were incubated with 125I-labeled secondary antibodies after an initial incubation with primary antibodies to α-adaptin or γ-adaptin for quantification by phosphor storage autoradiography. The amount of bound protein was calculated as a percentage of total adaptin proteins in the peripheral membrane fractions, based on immunoblots of total protein quantified using the same method. The experiment was carried out twice with similar results. (E) C-terminal/adaptin complexes were washed five times with wash buffers supplemented with various concentrations of NaCl, as indicated in the figure.
Figure 3D provides further evidence that AP-1 and AP-2 bind to RIDα by distinct mechanisms. This experiment was carried out by incubating cell fractions with increasing amounts of the C-terminal fusion protein, followed by quantitative immunoblotting for α-adaptin and γ-adaptin binding exactly as described for Table 1. Data are represented as the percent of total adaptin protein detected in combined crude cytosol and peripheral membrane fractions that were also quantified by immunoblotting with 125I-labeled secondary antibodies and phosphor storage autoradiography. This experiment demonstrates that α-adaptin binding is saturated rapidly compared to γ-adaptin. To evaluate the relative importance of ionic interactions in the binding of AP complexes to the cytosolic tail of RIDα, the in vitro binding assay was carried out in the presence of increasing concentrations of salt or by changing the pH of the binding solution. Ionic interactions are highly sensitive to changes in salt or pH, while hydrophobic interactions are not. Binding of α-adaptin and γ-adaptin to RIDα cytosolic tail sequences was not appreciably affected by changes in salt concentration (Fig. 3E) or in pH (data not shown), suggesting that these protein-protein interactions are enabled primarily by hydrophobic amino acids with nonpolar side-chains. Thus, Ala71 and/or Leu73 in the 71-AYLRH motif may be important for AP binding; in addition, hydrophobic residues in the surrounding tail sequences may not be necessary but nevertheless contribute to the overall strength of AP binding.
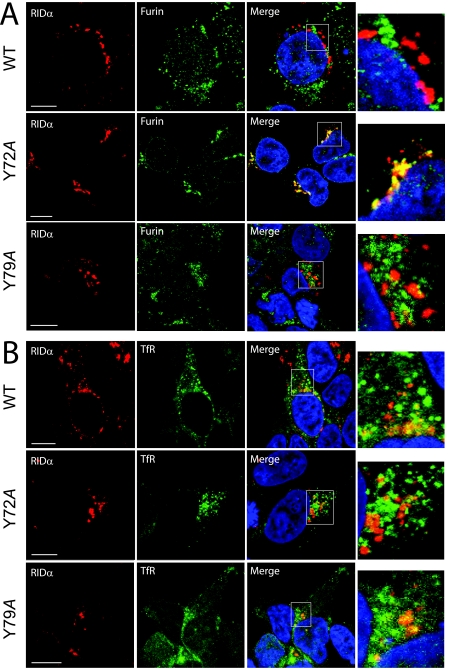
The Y72A point mutation alters subcellular localization of RIDα in mammalian cells.
The Y72A and Y79A substitutions were engineered into the full-length viral protein to study their expression and localization in mammalian cells. Wild-type and mutant proteins were transiently expressed in HEK-293 cells, and subcellular localization was determined by collecting confocal images of cells stained for the viral protein and then costained for furin or the TfR, which are well-characterized markers of the biosynthetic and endocytic compartments, respectively. The wild-type protein was largely excluded from the biosynthetic compartment defined by furin that is enriched in the TGN (Fig. 4A) and was found to colocalize with the TfR (Fig. 4B), a marker for endocytic vesicles as previously shown (10). The Y72A mutant, however, colocalized almost exclusively with the TGN marker furin (Fig. 4A) and did not colocalize with the TfR (Fig. 4B), consistent with the idea that the Y72A mutation blocks exit from the biosynthetic compartment. Similar to the wild type, the Y79A mutant did not colocalize with furin (Fig. 4A), but it partially colocalized with the TfR (Fig. 4B). This result indicates that while the Y79A mutation does affect AP-1 binding in vitro (Table 1), the protein is able to efficiently exit the biosynthetic compartment.
FIG. 4.
Colocalization of wild-type (WT), Y72A, and Y79A RIDα with membrane-compartment markers. HEK-293 cells transiently expressing wild-type, Y72A, or Y79A mutant proteins were costained with antibodies to the FLAG-tagged viral protein (red channel) and to the TGN marker furin (A) or the endocytic marker TfR (B) (green channel) and DAPI (blue channel) for analysis by confocal microscopy. Red and green channels were merged, and “yellow” indicates the overlap of red and green fluorescence. Higher-magnification images of outlined areas are shown to the right of each set of panels. Size markers, 10 μm.
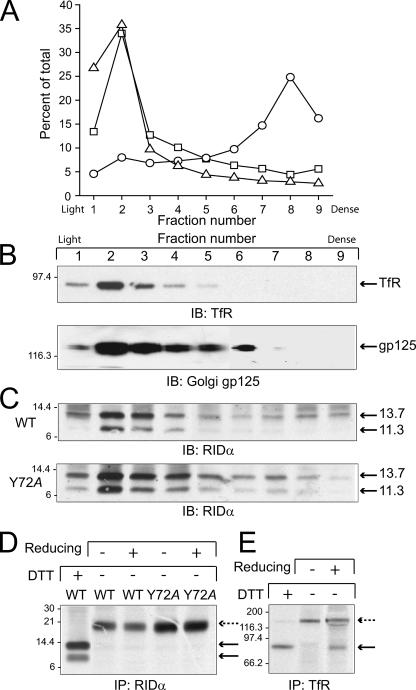
The subcellular distribution of wild-type and mutant proteins was confirmed by cell fractionation of membrane compartments on Percoll gradients. Fractions were initially characterized by quantifying radioactivity in fractions from cells that were incubated with 125I-EGF or 125I-transferrin to label plasma membrane receptors, by assaying fractions for β-hexosaminidase activity enriched in lysosomes (Fig. 5A), and by immunoblotting with antibodies to well-characterized markers of biosynthetic (gp125) and endocytic (TfR) compartments (Fig. 5B). Based on these analyses, we concluded that biosynthetic compartments are concentrated in fractions 2 through 5, the plasma membrane is enriched in fractions 1 and 2, and TfR-positive endosomes are concentrated in fractions 2 and 3. Both molecular weight forms of the wild-type protein exhibited a subcellular distribution that is most similar to TfR in light fractions 2 and 3, and distribution of the mutant protein was most similar to the Golgi glycoprotein gp125 (Fig. 5C). Thus, these data are consistent with the images obtained by confocal microscopy showing that the wild-type protein localizes to an endocytic compartment, compared to the mutant protein, which localizes to the biosynthetic pathway.
FIG. 5.
Subcellular distribution of wild-type and Y72A RIDα proteins. (A) CHO cells were fractionated on Percoll gradients, and individual fractions were assayed for β-hexosaminidase activity (open circles). Cells were also fractionated after they had been incubated with 125I-transferrin for 30 min on ice (open squares) or after they were transfected with a human EGFR expression plasmid and incubated with 125I-EGF for 30 min on ice 48 h later (open triangles). Enzyme activity or radioactivity was plotted as a percentage of total activity or radioactivity detected across the entire gradient. (B) Equal aliquots of total cellular protein from individual cell fractions were immunoblotted (IB) with antibodies to well-characterized markers of the Golgi body (gp125) and of the plasma membrane and early endosomes (TfR). (C) CHO cells were transfected with wild-type (WT) or Y72A RIDα plasmids, and immune complexes from individual Percoll gradient fractions were analyzed by immunoblotting with a RIDα-specific antibody. (D and E) CHO cells transfected as described for panel C and nontransfected CHO cells were pulse-labeled and then switched to chase medium to allow proteins to reach their steady-state localization. Proteins were immunoprecipitated and analyzed by SDS-PAGE under standard reducing conditions (+DTT) or were immunoprecipitated from cells subjected to surface reducing (+) or sham (−) conditions followed by SDS-PAGE under nonreducing conditions (−DTT).
In contrast to results shown here and published elsewhere (10, 24, 26, 28), other laboratories have reported that RIDα is localized to the plasma membrane (21, 43). Thus, the observation that the wild-type protein is enriched in fraction 2 could also be interpreted as plasma membrane localization, based on the distribution of radioactivity in gradients obtained from cells labeled with 125I-labeled ligands (Fig. 5A). In addition, disabling a critical AP-2 binding site could result in plasma membrane accumulation of the Y72A mutant protein. Additional experiments were therefore carried out to exclude the possibility that RIDα is targeted to the plasma membrane. RIDα exists in two molecular weight forms: one corresponding to the full-length molecule, which has two membrane-spanning α-helices and cytosolic amino and carboxy termini, and a lower-molecular-weight species lacking the amino-terminal α-helix, which is cleaved by signal peptidase in the endoplasmic reticulum (Fig. 1A) (27). Sequences connecting the α-helices should be exposed at the cell exterior in either form of the molecule located at the plasma membrane. However, this region cannot be labeled by conventional surface biotinylation, since it lacks residues with free amino groups. We therefore took advantage of the fact that both RIDα molecular weight species form disulfide-linked dimers at Cys31 (highlighted in Fig. 1A) (27). Hence, disulfide bonds on proteins located at the cell surface should be reduced if cells are exposed to an external reducing agent, such as cysteine, while intracellular proteins are protected (4, 36). Transfected cells were subjected to pulse-chase metabolic labeling and then incubated with cysteine-containing medium and lysed in the presence of iodoacetamide to prevent de novo disulfide bond formation. When RIDα immune complexes were resolved under nonreducing conditions (i.e., in the absence of DTT), radiolabeled proteins migrated as high-molecular-weight dimers, regardless of whether or not cells were exposed to extracellular cysteine (Fig. 5D). Introduction of the Y72A mutation had no effect on the resistance of high-molecular-weight dimers to surface reduction. This result was in contrast to that for disulfide-linked TfRs, which were partially reduced by external cysteine (Fig. 5E), consistent with approximately 20% to 45% of this molecule exhibiting plasma membrane localization, with the remainder in endocytic compartments (34, 47). These data confirm that the majority of wild-type and mutant proteins are localized to intracellular membrane compartments at steady state.
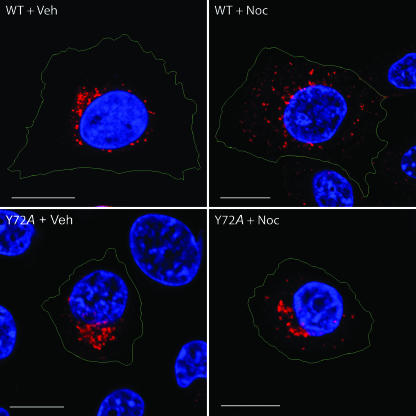
Endosomal compartments are dispersed throughout the cytoplasm following a brief exposure to the microtubule-depolymerizing agent nocodazole (30). We have shown previously that wild-type RIDα displays the same behavior, consistent with the viral protein's localization to endocytic compartments (A. H. Shah, N. L. Cianciola and C. Carlin, submitted for publication). We therefore reasoned that a similar treatment should have little or no effect on localization of the RIDα protein with the Y72A substitution if the mutant protein accumulates in a biosynthetic compartment, as is indicated by the results shown in Fig. 4 and 5. Results in Fig. 6 indicate that nocodazole has a very modest effect on the distribution of the Y72A mutant protein compared to vesicles with wild-type RIDα that were redistributed from the perinuclear region to the peripheral cytosol. Altogether, these data suggest that the mutant protein is retained in the biosynthetic pathway and mostly excluded from a nocodazole-sensitive endocytic pathway.
FIG. 6.
Effect of nocodazole on RIDα protein subcellular localization. CHO cells transiently expressing wild-type or Y72A mutant proteins were pretreated with dimethyl sulfoxide vehicle (Veh) or nocodazole (Noc; 100 μM) for 30 min prior to staining with antibody to the FLAG-tagged viral protein (red channel) and with DAPI (blue channel). Cell outlines (in green) were made with the MetaMorph program using phase contrast images as a guide. Size markers, 10 μm.
Y72A and Y79A point mutations prevent EGFR downregulation by RIDα.
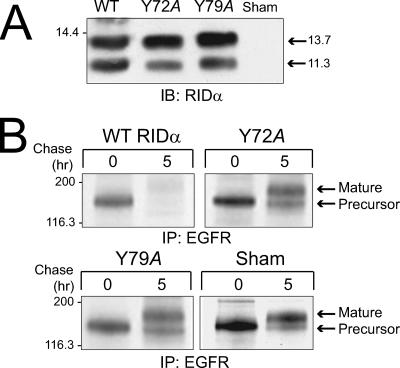
Although many functions have now been associated with this Ad protein, RIDα was first identified because of its ability to specifically reduce EGFR plasma membrane expression (7). Thus, studies were carried out to determine the effect of the Y72A and Y79A mutations in RIDα on EGFR metabolic stability in CHO cells expressing both proteins. Cells were cotransfected with viral protein and human EGFR plasmids and harvested for total cellular protein or pulse-labeled with [35S]cysteine for 30 min to measure protein stability. When viral protein immune complexes were examined by immunoblotting, we observed that wild-type and mutant viral proteins were expressed at similar levels at 48 h posttransfection (Fig. 7A). The EGFR acquires seven to nine N-linked high-mannose oligosaccharides cotranslationally that are processed to complex carbohydrates during Golgi body maturation, resulting in retarded migration on SDS-polyacrylamide gels (5). A molecular weight species corresponding to the high-mannose oligosaccharide EGFR precursor was detected in cells cotransfected with an empty vector control or plasmids encoding wild-type or mutant viral protein in cells that were harvested immediately after a 30-min pulse-label (Fig. 7B), showing that the EGFR is synthesized to similar extents in all four populations of transfected cells. The molecular weight species corresponding to the mature EGFR protein with complex carbohydrates was detected in cells transfected with the empty vector control or with plasmids encoding the Y72A or Y79A mutant proteins after a 5-h chase with nonradioactive amino acid precursors. This was in contrast to cells expressing wild-type RIDα, where mature EGFR protein was not detected, consistent with the known ability of RIDα to target EGFR to lysosomes for degradation in the absence of other Ad proteins (25). Thus, the Y72A mutation appears to block the ability of the viral protein to facilitate EGFR degradation. RIDα with the Y79A mutation also does not downregulate the receptor, even though these mutants appear to localize to endosomes (Fig. 4B). The loss-of-function phenotype associated with the Y79A mutant can likely be attributed to the fact that this region mediates protein-protein interactions with ORP1L (Shah et al., submitted), a Rab7 effector that links late endocytic vesicles to minus end-directed microtubule motor complexes (31).
FIG. 7.
Effect of Y72A and Y79A point mutations on RIDα expression and function in mammalian cells. CHO cells were cotransfected with plasmid expressing the human EGFR along with a plasmid expressing wild-type RIDα (WT), plasmids expressing RIDα with a Y72A or Y79A substitution, or an empty vector (Sham). (A) Equal aliquots of total cell protein were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with a RIDα-specific antibody. (B) The cells were radiolabeled with [35S]cysteine for 30 min, starting at 48 h posttransfection, and harvested either immediately or following a 5-h chase in medium with an excess of nonradioactive cysteine. EGFR immune complexes were resolved by SDS-PAGE and detected by autoradiography. IP, immunoprecipitation.
DISCUSSION
The E3 region is not required for viral replication; nevertheless, it plays a critical role in Ad pathogenesis (15). The importance of this region is underscored by the fact that the first generation of Ad gene therapy vectors which contained large E3 deletions were ultimately deemed unsafe (14). E3 genes encode integral membrane proteins that regulate a variety of host cell functions involved in innate immunity and inflammatory responses. The ability of these proteins to modify host cell function is due in part to cytosolic tail sequences that interact with sorting machinery and target membrane proteins to specific intracellular compartments. The E3 protein RIDα was originally identified because of its ability to downregulate the EGFR (7) and other related receptor tyrosine kinases (35). In this study, we have demonstrated that RIDα residues 71-AYLRH comprise a binding site for AP complexes and that Tyr72 is required for RIDα localization to endosomes and its ability to downregulate the EGFR. These findings support previous studies concluding that RIDα acts by targeting EGFRs undergoing constitutive recycling to the plasma membrane (10, 26, 28). The fact that 71-AYLRH is precisely conserved in all Ad serotypes that have been sequenced except for Ad12 suggests its fundamental importance in a majority of Ad-induced diseases (7).
Although the mutation of Tyr72 to an alanine residue leads to a clear reduction in AP binding in vitro (Table 1), 71-AYLRH does not conform to classical tyrosine-dependent YXXØ motifs that have a preference for hydrophobic residues with bulky side chains at the Ø position (39). Instead, it has a histidine residue that is mildly basic and hydrophilic. However, many other factors contribute to sorting signal recognition, including the position of the signal relative to the membrane and to the carboxyl terminus and the presence of amino acid residues in areas adjacent to the signal. We have reported previously that 71-AYLRH exists in an amphipathic helix that is stabilized by interactions with a membrane-mimetic phospholipid micelle surface based on data obtained using nuclear magnetic resonance spectroscopic methods (45). This close degree of membrane association could have an important role in regulating the availability of the signal for interaction with APs. Thus, 71-AYLRH may be masked when the cytoplasmic tail is intimately associated with membrane, while cellular events that result in its translocation into the cytosol could make it available for binding APs. Some examples that could bring about such a change include modulation by another membrane protein, changes in the endosomal pH or ionic environment as RIDα traverses different cellular compartments, posttranslational modification, or a dramatic shift in the tilt or membrane placement of the adjacent transmembrane helix. For example, we have recently demonstrated that RIDα function is highly dependent on reversible palmitoylation at a residue in the cytosolic tail (N. L. Cianciola and C. Carlin, unpublished data), suggesting that the association of the RIDα cytosolic tail with membranes is tightly regulated in cells (17). The possibility that 71-AYLRH availability is regulated by a transmembrane mechanism is particularly intriguing since the RIDα loop domain connecting its two membrane spanning domains resides in compartmental lumens. Thus, the transmembrane domain could act as a conduit to fine-tune 71-AYLRH recognition at specific subcellular organelles.
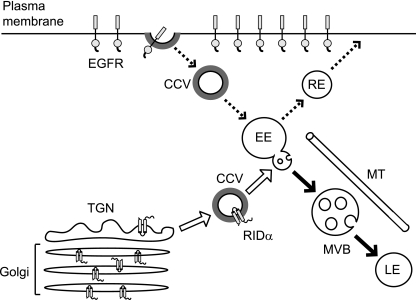
Even though 71-AYLRH recognizes two different classes of APs, we have demonstrated that AP-1 and AP-2 do not compete for binding to RIDα in vitro (Fig. 3C) and that mutation of adjacent residue Tyr79 leads to increased binding of AP-2 but diminished binding to AP-1 (Table 1). Thus, it seems likely that AP-1 and AP-2 recognize distinct but overlapping sets of tyrosine-based sorting signals in RIDα. The Y72A mutation traps RIDα in the TGN (Fig. 4B) but not the plasma membrane (Fig. 5D) suggesting the Ad protein encounters AP-1 first in the biosynthetic pathway. AP-2 is known to interact with a majority of tyrosine-based signals identified for other molecules, in agreement with studies showing that most naturally occurring signals mediate internalization (37). The broad specificity of AP-2 recognition implies that it serves a quality control function to retarget membrane proteins to their correct intracellular location that escape to the plasma membrane (2). This would suggest that the plasma membrane may not be an obligatory membrane transport destination for RIDα and that a majority of RIDα is delivered directly to endosomes (see summary model in Fig. 8). Accordingly, even though RIDα can be detected on the plasma membrane, this localization correlates with high levels of protein expression and constitutes a relatively minor fraction of the total protein in the cell (10). This is entirely consistent with the quality control role for AP-2 that has been proposed for membrane proteins that leak to the plasma membrane.
FIG. 8.
Summary model. Data presented in this study suggest that newly synthesized RIDα is delivered directly from the TGN to endosomes (open arrows) where it encounters EGFRs undergoing constitutive recycling to the plasma membrane (dashed arrows). Since RIDα proteins with a defective AP binding site accumulate in the Golgi body and TGN, we conclude that newly synthesized RIDα proteins encounter AP-1 necessary for incorporation into clathrin-coated vesicles (CCV), which are necessary for exiting the biosynthetic compartment. Data also show that RIDα is delivered directly to endosomes and not the plasma membrane, suggesting that the ability of the viral protein to bind AP-2 serves a quality control function in which proteins that “leak” to the plasma membrane are quickly endocytosed and retargeted to their appropriate intracellular locations. Although the EGFR and RIDα are both transported to late endosomes (LE) via multivesicular body (MVB) transport intermediates (solid arrows), only EGFR is degraded. EE and RE, early and recycling endosomes, respectively; MT, microtubule.
In addition to the Tyr72-based sorting motif, RIDα also has a potential dileucine-based motif located at residues 87-LL (Fig. 1A). Although another laboratory has published a study saying that these residues constitute an AP-2 binding site (21), those results were not substantiated in the present study. We did observe that AP binding was remarkably insensitive to pH or salt (Fig. 3D), supporting a role for hydrophobic interactions either within the signal itself or in adjacent regions. It is possible that the dialanine substitution at 87-LL analyzed in reference 21 lowers the overall strength of AP binding. Interestingly, 87-LL is part of a larger motif that is precisely conserved in the EGFR (26), suggesting its involvement in cargo selection and/or targeting EGFRs to lysosomes. This conjecture is supported by evidence that this sequence in the EGFR is necessary for ligand-induced trafficking to lysosomes (32, 33) and also RIDα-mediated diversion of recycling EGFRs to lysosomes (44). Thus, although 87-LL may not be directly involved in AP recognition, it undoubtedly has an important role in RIDα function at least as it relates to EGFR downregulation. The 87-LL motif shared with the EGFR is found in group C Ad2 and Ad5; however, this region is not precisely conserved in other serotypes (7). Thus, different Ads may vary in their ability to specifically target the EGFR.
RIDα has been associated with other activities besides EGFR downregulation. For example, the RID complex (comprised of RIDα and RIDβ) downregulates death receptors, including TNFR1 and FAS (9). However, mutagenesis studies support a model where the RID complex acts on TNFR1 at the plasma membrane, in contrast to FAS, where the functional interaction occurs intracellularly. These seemingly paradoxical results are best understood by considering the many steps involved in receptor downregulation. These include cargo selection, sorting to specific endocytic compartments involved in transport to lysosomes, and coupling to microtubules necessary for transporting MVB intermediates to the perinuclear region (19). We have already discussed the idea that the molecular basis for RIDα-mediated EGFR cargo selection likely involves the dileucine motif that is conserved in EGFR and RIDα encoded by Ad2 and Ad5. In addition, we have recently discovered that RIDα interacts with Rab7 effectors, including RILP and ORP1L, which are necessary for microtubule-dependent transport (Shah et al., submitted). Thus, it is likely that RIDα regulates EGFR downregulation at multiple levels. Other cargoes could require accessory molecules to deliver specific receptors to endosomes once the maturation process is under way. Thus, RIDβ may promote TNFR1 uptake to endosomes, whereupon they are then sorted to lysosomes by a mechanism involving RIDα-dependent endosomal maturation. This conjecture is consistent with the observation that RIDβ binds AP-2 and that AP-2 is required for RID-mediated downregulation of TNFR1 but not FAS (9). The ability to “mix and match” different aspects of RID function may have evolved to allow Ads to fine-tune RID activity in different cell types or during acute versus persistent infections.
Acknowledgments
These studies were done with the support of the Pediatric Imaging Center of University Hospitals of Cleveland, with the help of Sarah Richer Dawson. We also thank Kim Preston for technical assistance and Song J. Kil for critically reviewing the manuscript.
This work was supported by Public Health Service grant RO1 GM64243. N.L.C. was supported in part by NIH HL-007653, and A.H.S. was supported in part by a Cell and Molecular Biology Training Grant awarded through the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 18 July 2007.
REFERENCES
- 1.Avila, M. M., G. Carballal, H. Rovaletti, B. Ebekian, M. Cusminsky, and M. Weissenbacher. 1989. Viral etiology in acute lower respiratory infections in children from a closed community. Am. Rev. Respir. Dis. 140:634-637. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino, J., and L. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, C. D., H. W. Kim, A. J. Vargosko, B. C. Jeffries, J. O. Arrobio, B. Rindge, R. H. Parrott, and R. M. Chanock. 1969. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. Am. J. Epidemiol. 90:484-500. [DOI] [PubMed] [Google Scholar]
- 4.Bretscher, A. 1989. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes in A431 cells induced by EGF. J. Cell Biol. 108:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlin, C. R., and B. B. Knowles. 1986. Biosynthesis and glycosylation of the epidermal growth factor receptor in human tumor-derived cell lines A431 and Hep 3B. Mol. Cell. Biol. 6:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlin, C. R., D. Simon, J. Mattison, and B. B. Knowles. 1988. Expression and biosynthetic variation of the epidermal growth factor receptor in human hepatoma-derived cell lines. Mol. Cell. Biol. 8:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlin, C. R., A. E. Tollefson, H. A. Brady, B. L. Hoffman, and W. Wold. 1989. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell 57:135-144. [DOI] [PubMed] [Google Scholar]
- 8.Chavrier, P., R. G. Parton, H. P. Hauri, K. Simons, and M. Zerial. 1990. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62:317-329. [DOI] [PubMed] [Google Scholar]
- 9.Chin, Y. R., and M. S. Horwitz. 2005. Mechanism for removal of tumor necrosis factor receptor 1 from the cell surface by the adenovirus RIDα/β complex. J. Virol. 79:13606-13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crooks, D., S. J. Kil, J. M. McCaffery, and C. Carlin. 2000. E3-13.7 integral membrane proteins encoded by human adenoviruses alter epidermal growth factor receptor trafficking by interacting directly with receptors in early endosomes. Mol. Biol. Cell 11:3559-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fessler, S., F. Delgado-Lopez, and M. Horwitz. 2004. Mechanisms of E3 modulation of immune and inflammatory responses. Curr. Top. Microbiol. Immunol. 273:113-135. [DOI] [PubMed] [Google Scholar]
- 12.Giard, D. J., S. A. Aaronson, G. J. Todaro, P. Arnstein, J. H. Kersey, H. Dosik, and W. P. Parks. 1973. In vitro cultivation of human tumors. J. Natl. Cancer Inst. 51:1417-1423. [DOI] [PubMed] [Google Scholar]
- 13.Ginsberg, H. 1999. The life and times of adenoviruses. Adv. Virus Res. 54:1-13. [DOI] [PubMed] [Google Scholar]
- 14.Ginsberg, H. 1996. The ups and downs of adenovirus vectors. Bull. N.Y. Acad. Med. 73:53-58. [PMC free article] [PubMed] [Google Scholar]
- 15.Ginsberg, H. S., and G. A. Prince. 1994. The molecular basis of adenovirus pathogenesis. Infect. Agents Dis. 3:1-8. [PubMed] [Google Scholar]
- 16.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 17.Greaves, J., and L. H. Chamberlain. 2007. Palmitoylation-dependent protein sorting. J. Cell Biol. 176:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green, S. A., K. P. Zimmer, G. Griffiths, and I. Mellman. 1987. Kinetics of intracellular transport and sorting of lysosomal membrane and plasma membrane proteins. J. Cell Biol. 105:1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu, F., and J. Gruenberg. 1999. Biogenesis of transport intermediates in the endocytic pathway. FEBS Lett. 452:61-66. [DOI] [PubMed] [Google Scholar]
- 20.Hayflick, L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 21.Hilgendorf, A., J. Lindberg, Z. Ruzsics, S. Honing, A. Elsing, M. Lofqvist, H. Engelmann, and H.-G. Burgert. 2003. Two distinct transport motifs in the adenovirus E3 proteins act in concert to down-modulate apoptosis receptors and the epidermal growth factor receptor. J. Biol. Chem. 278:51872-51884. [DOI] [PubMed] [Google Scholar]
- 22.Hirota, K., M. Murata, T. Itoh, J. Yodoi, and K. Fukuda. 2001. Redox-sensitive transactivation of epidermal growth factor receptor by tumor necrosis factor confers the NF-kappa B activation. J. Biol. Chem. 276:25953-25958. [DOI] [PubMed] [Google Scholar]
- 23.Hobert, M. E., S. Kil, and C. R. Carlin. 1997. The cytoplasmic juxtamembrane domain of the epidermal growth factor receptor contains a novel autonomous basolateral sorting signal. J. Biol. Chem. 272:32901-32909. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman, B. L., K. Takishima, M. R. Rosner, and C. Carlin. 1993. Adenovirus and protein kinase C have distinct molecular requirements for regulating epidermal growth factor receptor trafficking. J. Cell. Physiol. 157: 535-543. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman, B. L., A. Ullrich, W. Wold, and C. Carlin. 1990. Retrovirus-mediated transfer of an adenovirus gene encoding an integral membrane protein is sufficient to down regulate the receptor for epidermal growth factor. Mol. Cell. Biol. 10:5521-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman, P., and C. Carlin. 1994. Adenovirus E3 protein causes constitutively internalized EGF receptors to accumulate in a prelysosomal compartment, resulting in enhanced degradation. Mol. Cell. Biol. 14:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman, P., M. B. Yaffe, B. L. Hoffman, S. Yei, W. S. M. Wold, and C. Carlin. 1992. Characterization of the adenovirus E3 protein that down-regulates the epidermal growth factor receptor. J. Biol. Chem. 267:13480-13487. [PubMed] [Google Scholar]
- 28.Hoffman, P. H., P. Rajakumar, B. Hoffman, R. Heuertz, W. S. M. Wold, and C. R. Carlin. 1992. Evidence for intracellular down-regulation of the epidermal growth factor receptor during adenovirus infection by an EGF-independent mechanism. J. Virol. 66:197-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horwitz, M. S. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med. 6(Suppl. 1):S172-S183. [DOI] [PubMed] [Google Scholar]
- 30.Husain, M., and B. Moss. 2003. Intracellular trafficking of a palmitoylated membrane-associated protein component of enveloped vaccinia virus. J. Virol. 77:9008-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson, M., N. Rocha, W. Zwart, I. Jordens, L. Janssen, C. Kuiji, V. M. Olkkonen, and J. Neefjes. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor βlll spectrin. J. Cell Biol. 176:459-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kil, S., and C. Carlin. 2000. EGF receptor residues Leu679, Leu680 mediate selective sorting of ligand-receptor complexes in early endocytic compartments. J. Cell. Physiol. 185:47-60. [DOI] [PubMed] [Google Scholar]
- 33.Kil, S. J., M. E. Hobert, and C. Carlin. 1999. A leucine-based determinant in the EGF receptor juxtamembrane domain is required for the efficient transport of ligand-receptor complexes to lysosomes. J. Biol. Chem. 274:3141-3150. [DOI] [PubMed] [Google Scholar]
- 34.Klausner, R. D., J. Van Renswoude, G. Ashwell, C. Kempf, A. N. Schecter, A. Dean, and K. R. Bridges. 1983. Receptor-mediated endocytosis of transferrin in K562 cells. J. Biol. Chem. 258:4715-4724. [PubMed] [Google Scholar]
- 35.Kuivinen, E., B. L. Hoffman, P. A. Hoffman, and C. R. Carlin. 1993. Structurally related class I and class II receptor protein tyrosine kinases are down-regulated by the same E3 protein coded by human group C adenoviruses. J. Cell Biol. 120:1271-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Low, S. H., B. L. Tang, S. H. Wong, and W. Hong. 1992. Selective inhibition of protein targeting to the apical domain of MDCK cells by brefeldin A. J. Cell Biol. 118:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marks, M. S., H. Ohno, T. Kirchhausen, and J. S. Bonifacino. 1997. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 7:124-128. [DOI] [PubMed] [Google Scholar]
- 38.Monick, M. M., K. Cameron, J. Staber, L. S. Powers, T. O. Yarovinsky, J. G. Koland, and G. W. Hunninghake. 2005. Activation of the epidermal growth factor receptor by respiratory syncytial virus results in increased inflammation and delayed apoptosis. J. Biol. Chem. 280:2147-2158. [DOI] [PubMed] [Google Scholar]
- 39.Ohno, H., R. C. Aguilar, D. Yeh, D. Taura, T. Saito, and J. S. Bonifacino. 1998. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 273:25915-25921. [DOI] [PubMed] [Google Scholar]
- 40.Puck, T. T., S. J. Cieciura, and A. Robinson. 1958. Genetics of somatic mammalian cells. III. Long-term cultivation of euploid cells from human and animal subjects. J. Exp. Med. 108:945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson, M. 1993. Assembly and targeting of adaptin chimeras in transfected cells. J. Cell Biol. 123:67-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson, M. S., and J. S. Bonifacino. 2001. Adaptor-related proteins. Curr. Opin. Cell Biol. 13:444-453. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, A., A. Tollefson, P. Krajcsi, S. Yei, and W. Wold. 1995. The adenovirus E3 10.4K and 14.5K proteins, which function to prevent cytolysis by tumor necrosis factor and to down-regulate the epidermal growth factor receptor, are localized in the plasma membrane. J. Virol. 69:172-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsacoumangos, A., S. J. Kil, L. Ma, F. D. Sonnichsen, and C. Carlin. 2005. A novel dileucine lysosomal-sorting-signal mediates intracellular EGF-receptor retention independently of protein ubiquitylation. J. Cell Sci. 118:3959-3971. [DOI] [PubMed] [Google Scholar]
- 45.Vinogradova, O., C. R. Carlin, F. D. Sonnichsen, and C. R. Sanders. 1998. A membrane setting for the sorting motifs present in the adenovirus E3-13.7 protein which down-regulates the epidermal growth factor receptor. J. Biol. Chem. 273:17343-17350. [DOI] [PubMed] [Google Scholar]
- 46.Waterfield, M. D., E. L. V. Mayes, P. Stroobant, P. L. P. Bennet, S. Young, P. N. Goodfellow, G. S. Banting, and B. Ozanne. 1982. A monoclonal antibody to the human epidermal growth factor receptor. J. Cell. Biochem. 20:149-161. [DOI] [PubMed] [Google Scholar]
- 47.Weiel, J. E., and T. A. Hamilton. 1984. Quiescent lymphocytes express intracellular transferrin receptors. Biochem. Biophys. Res. Commun. 119:598-603. [DOI] [PubMed] [Google Scholar]