Abstract
Disease-associated PrP fragments produced upon in vitro or in vivo proteolysis can provide significant insight into the causal strain of prion disease. Here we describe a novel molecular strain typing assay that used thermolysin digestion of caudal medulla samples to produce PrPres signatures on Western blots that readily distinguished experimental sheep bovine spongiform encephalopathy (BSE) from classical scrapie. Furthermore, the accumulation of such PrPres species within the cerebellum also appeared to be dependent upon the transmissible spongiform encephalopathy (TSE) strain, allowing discrimination between two experimental strains of scrapie and grouping of natural scrapie isolates into two profiles. The occurrence of endogenously produced PrP fragments, namely, glycosylated and unglycosylated C2, within different central nervous system (CNS) regions is also described; this is the first detailed description of such scrapie-associated fragments within a natural host. The advent of C2 fragments within defined CNS regions, compared between BSE and scrapie cases and also between two experimental scrapie strains, appeared to be largely dependent upon the TSE strain. The combined analyses of C2 fragments and thermolysin-resistant PrP species within caudal medulla, cerebellum, and spinal cord samples allowed natural scrapie isolates to be separated into four distinct molecular profiles: most isolates produced C2 and PrPres in all CNS regions, a second group lacked detectable cerebellar C2 fragments, one isolate lacked both cerebellar PrPres and C2, and a further isolate lacked detectable C2 within all three CNS regions and also lacked cerebellar PrPres. This CNS region-specific deposition of disease-associated PrP species may reflect the natural heterogeneity of scrapie strains in the sheep population in the United Kingdom.
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are fatal, neurodegenerative diseases. TSEs include scrapie in goats and sheep, bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease in deer and elk, and Creutzfeldt-Jakob disease (CJD) in humans. The “protein-only” hypothesis dictates that the central event in TSE disease is the conversion of cellular prion protein (PrPC) into a pathological isoform, PrPSc (33). TSE agents exist with diverse phenotypic characteristics, and strong scientific evidence suggests that PrPSc carries strain-specific properties within the conformational structure of the PrP molecule within the context of the peptide backbone and/or within its glycoform composition (7, 12, 27, 34). For many decades, it has been understood that classical scrapie is a number of distinct strains and more than 15 have been characterized by mouse bioassay (9). However, the possibility that strains may be modified upon transmission across the species barrier, where the host PrP sequence is distinct, makes it difficult to know whether such a degree of diversity of strains actually exists in the sheep population. Thus, little is known about the prevalence, geographical distribution, and possible relevance to human health of sheep scrapie strains. Importantly, while there is the possibility that BSE may be present in the sheep population, it is imperative to understand the biological and molecular basis of sheep scrapie strains in the natural host.
There are a growing number of diagnostic tests that distinguish classical ovine scrapie and ovine experimental BSE. The gold standard remains bioassay in inbred mice, where differences in the incubation period and patterns of the pathological lesions in the central nervous system (CNS) are characteristic of a given strain. Studies of pathological lesion profiling in sheep to identify TSE strains have been equivocal (4, 30, 42). Immunohistochemical (IHC) methods (so-called “peptide mapping” or “PrPd profiling”) detail PrPSc accumulation to distinct regions and cell types as well as the morphological characteristics of PrPSc deposition (19, 20, 24). Such methods have allowed the distinction of experimental ovine BSE from various scrapie isolates and experimental scrapie strains. However, the methodology is laborious and requires the availability of good quality tissue from large areas of the brain. The advent of more rapid biochemical tests that can distinguish ovine TSEs would facilitate the assessment of whether BSE is present within sheep and also assist in the epidemiological study of scrapie strains within the natural host.
To date, rapid biochemical strain-typing tests have focused on the proteinase K (PK)-resistant species of PrPSc, designated PrPres. Such species have been reported to differ among certain TSE strains in terms of their glycoform ratios and the sites of PK cleavage (2, 12, 22, 23, 36, 37). However, the efficacy of glycoform ratio analysis to distinguish classical scrapie from experimental BSE remains debatable (3, 22, 36). Furthermore, differences in the apparent molecular mass of PK-resistant PrPres can be as little as 0.4 kDa between ovine BSE and scrapie (21, 22, 36, 37) and such subtle differences are not easily detected on Western blots (23, 29, 41). An alternative method determines such differences in PK cleavage sites by staining Western blots with the monoclonal antibody P4 which shows a quantitative difference in the staining of scrapie and BSE samples due to a reduction in the presence of the P4 epitopes in BSE PrPres (36, 37). However, this strain typing method also shows considerable variation between laboratories (1, 36).
The application of these rapid tests has provided conflicting results in terms of the natural heterogeneity of scrapie isolates. A number of studies found uniformity in a range of isolates (36, 37, 38), while others have shown considerable variation in the PrPres type (22, 23). Also of interest is the description of aberrant full-length and N-terminally truncated PrP species within scrapie-affected mice and CJD-affected humans (11, 32). The detailed study of such TSE-associated PrP species within sheep has not been reported and may provide further tools to study the heterogeneity of ovine TSE strains.
Here we apply a novel strain typing method that uses the thermostable protease thermolysin to produce PrPres fragments that readily distinguish experimental BSE from natural scrapie isolates. With those samples tested to date, the assay is unaffected by PrP genotype, and upon analysis of multiple regions of the CNS, it categorizes natural scrapie isolates into two groups, where disparate neuroanatomical depositions of PrPres are observed. The analysis of endogenous disease-associated, N-terminally truncated PrP fragments is also described. Such analysis further highlights the distinct deposition of disease-associated PrP conformers in different CNS regions between experimental TSE strains and between natural scrapie isolates.
MATERIALS AND METHODS
Materials.
CNS material from healthy and TSE-affected sheep was obtained from the Veterinary Laboratories Agency TSE-Archive (VLA; Addlestone, Surrey, United Kingdom). Experimental BSE- and scrapie-infected animals were dosed orally as described elsewhere (5). BSE-infected animals were a primary passage of bovine BSE into sheep (5). CH1641 and SSBP1 were kindly supplied by N. Hunter (Institute for Animal Health, Neuropathogenesis Unit, Edinburgh, United Kingdom). The CH1641 scrapie isolate originated from a sheep in the Institute for Animal Health, Neuropathogenesis Unit flock in 1970 and subsequently was passaged three times in Cheviot sheep (17). Following a fourth passage (24), CH1641 was inoculated into AHQ/AHQ Cheviot sheep at the VLA (CH1641-VLA1). Consistency of strain characteristics was determined by Western blot analysis. SSBP1 as originally described (14) was passaged at least 24 times through Cheviot and Cheviot crosses, including a final passage in VRQ/VRQ Cheviot sheep at the VLA (SSBP1-VLA1). All TSE-affected animals displayed clinical disease and were confirmed as TSE positive by IHC, histology, and Western blot analysis (36). The exception was animal 0210, which was confirmed as TSE positive by IHC and histology. Spinal cord material was from segments C1 to C2.
All TSE-affected animals were euthanized, and samples taken postmortem were snap-frozen for storage at −80°C either within 60 min or within a 12-h autopsy (Table 1 presents individual sample details). Material from healthy animals was frozen within a 12-h autopsy procedure.
TABLE 1.
Molecular profiling of natural and experimental ovine TSE strainsa
| Animal | Genotypeb | Breed (age in yr) | PrPres type within CM | PrPres withinc:
|
C2 withinc,d:
|
|||
|---|---|---|---|---|---|---|---|---|
| SC | Cer | CM | SC | Cer | ||||
| 0455e | VRQ/VRQ | Swaledale (6) | Scrapie | + | + | + | + | − |
| 0226e | ARQ/VRQ | Swaledale (5) | Scrapie | + | + | + | + | − |
| 0456e | ARQ/VRQ | Swaledale (3) | Scrapie | + | − | + | + | − |
| 0284f | AHQ/AHQ | Finn Dorset (unknown) | Scrapie | + | − | − | − | − |
| 0615e | ARQ/VRQ | Swaledale (5) | Scrapie | + | + | + | + | + |
| 0923g | VRQ/VRQ | Welsh Hill Speckled (2) | Scrapie | + | + | + | + | + |
| 0925g | VRQ/VRQ | Welsh Hill Speckled (5) | Scrapie | + | + | + | + | + |
| 0836e | ARQ/VRQ | Mule (2) | Scrapie | + | + | + | + | + |
| 1276g | VRQ/VRQ | Welsh Mountain (2) | Scrapie | + | + | + | + | + |
| 1563g | VRQ/VRQ | Bleu De Maine (3) | Scrapie | + | + | + | + | + |
| 1275g | VRQ/VRQ | Welsh Mountain (2) | Scrapie | + | + | + | + | + |
| 0210e | ARQ/ARQ | Warborough (2) | Scrapie | ND | + | ND | ND | ND |
| 0635e | ARQ/ARQ | Charollais Cross (3) | Scrapie | ND | + | ND | ND | ND |
| 0678e | ARQ/ARQ | Suffolk Cross (3) | Scrapie | ND | + | ND | ND | ND |
| 0575e | ARQ/ARQ | Cambridge (3) | Scrapie | ND | ND | ND | ND | ND |
| 0392g,h | ARQ/ARQ | Romney (3) | BSE | ND | + | − | ND | − |
| 1693g,h | ARQ/ARQ | Romney (2) | BSE | ND | + | − | ND | − |
| 0654g,h | ARQ/ARQ | Romney (3) | BSE | ND | + | − | ND | − |
| 1842g,h | ARQ/ARQ | Romney (unknown) | BSE | ND | ND | ND | ND | ND |
| CH1641g,h | AHQ/AHQ | Cheviot (1) | ND | ND | − | ND | ND | − |
| SSBP1g,h | VRQ/VRQ | Cheviot (2) | ND | ND | + | ND | ND | + |
Shown are thermolysin PrPres profiles and results for the detection of truncated PrP C2 species. +, PrPres or C2 fragment was detected; −, PrPres or C2 fragment was not detected; ND, not determined.
Amino acid residues at positions 136, 154, and 171.
Thermolysin-resistant PrP or C2 when present within spinal cord (SC) and cerebellum (Cer) samples gave BSE- or scrapie-specific molecular weight profiles as indicated for the caudal medulla (CM) samples.
A total of 3.3 μl of 2% (wt/vol) brain homogenate was analyzed. All samples were analyzed at least in triplicate.
Tissues were frozen within 1 h of death.
For animal 0284, how long after death tissues were snap-frozen is not known; however, no autolysis was noted for the tissue.
Tissues were frozen within 12 h of death.
Experimentally infected animals (all other animals are natural infections).
CNS homogenate preparation.
Homogenates were prepared by cutting up 100 to 300 mg of CNS tissue and adding lysis buffer (0.5% [vol/vol] Nonidet P-40 and 0.5% [wt/vol] sodium deoxycholate in 15 mM KH2PO4, 81 mM Na2HPO4, 137 mM NaCl, and 3 mM KCl) to dilute the sample to 10% (wt/vol). The resulting suspension was passed through needles of decreasing diameters (19-, 21-, and then 23-gauge needles), and the samples were centrifuged for 5 min at 400 × g to remove cellular debris. All homogenates had pH values between 7.40 and 7.50.
Sucrose gradient centrifugation.
Brain homogenates (10% [wt/vol], 200 μl, and diluted to a final volume of 720 μl 1× phosphate-buffered saline [PBS]) were loaded onto 10 to 60% sucrose step gradients comprised of 0.72 ml each of 10, 15, 20, 25, 30, and 60% sucrose TNE (10 mM Tris, pH 7.4, 20 mM NaCl, 1 mM EDTA). Samples were spun at 200,000 × g for 1 h at 4°C in a Beckman L8-70 M ultracentrifuge using Beckman Ultra-Clear centrifuge tubes (13 by 15 mm) in a Beckman SW50.1 rotor. Fractions (12 by 0.42 ml) were collected from the top of the tube, and 18 μl of each fraction was analyzed on Western blots.
Protease digestion of CNS homogenates.
Aliquots (25 to 50 μl) of 10% (wt/vol) brain homogenates were digested. Thermolysin (Sigma, Poole, United Kingdom) digestions were carried out with 150 μg/ml/h protease for a total of 8 h. After each hour, 150 μg/ml protease was added and the reaction mixture was stirred and incubated at 70°C. PK digestions were carried out with 50 μg/ml protease for 1 h at 37°C. Two volumes of NuPAGE 2× LDS sample buffer containing 5% (vol/vol) β-mercaptoethanol (Invitrogen, Paisley, United Kingdom) was added to the digested homogenates, and the samples were heated to 100°C for 10 min. Typically, 3.3 μl of digested 10% (wt/vol) brain homogenate was analyzed by Western blotting. For the analysis of C2 fragments, samples were analyzed directly on Western blots without protease treatment; routinely 3.3 μl of 2% (wt/vol) brain homogenate was analyzed.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
Discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis was carried out through a running gel containing 12% (wt/vol) total acrylamide using NuPAGE precast Bis-Tris gels (Invitrogen, Paisley, United Kingdom). Separated proteins were transferred to a polyvinylidene difluoride membrane (Roche, Lewes, United Kingdom) by using a NuPAGE blot module (Invitrogen, Paisley, United Kingdom) at 30 V for 1 h. Blots were probed with monoclonal antibody, anti-mouse horseradish peroxidase conjugate (DakoCytomation, Glostrup, Denmark), and BM chemiluminescence blotting substrate (Roche, Lewes, United Kingdom). Primary antibody titers were determined for each antibody: for SAF32, 1 in 40,000 (gift from J. Grassi, CEA Saclay, Gif/Yvette, France), and for P4, 1 in 1,000 (R-Biopharm Rhone Ltd.). Gels were visualized by using an ICCD225 camera (Photek Ltd., East Sussex, United Kingdom), and densitometric analysis was carried out by using IFS32 image software. SAF32 binds to a repeated epitope within the octapeptide repeat region of PrP with the most C-terminal site of this sequence at amino acid residues 86 to 91, P4 binds within the region 89 to 104, and L42 binds within the region 145 to 163. The numbering of PrP amino acid residues corresponds to that of ovine PrP throughout the study and is that used by Goldmann and coworkers (18).
RESULTS
Distinct PrPres fragments for ovine TSE strains produced with thermolysin digestion.
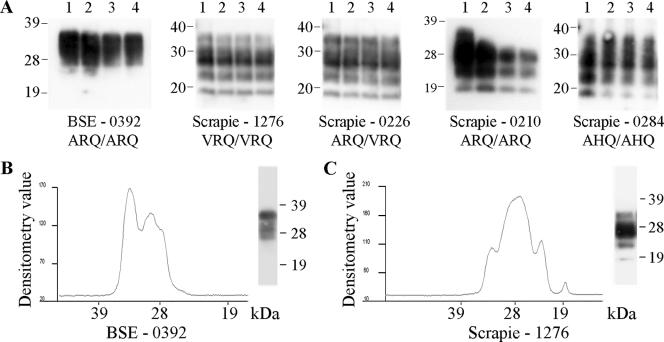
Caudal medulla homogenates from TSE-affected sheep were subjected to a novel TSE strain typing assay: repeat additions of high concentrations of the protease thermolysin. Western blot analysis using the monoclonal antibody P4 revealed distinct PrPres profiles for natural scrapie compared to those for experimental BSE (Fig. 1A). At the 8-h time point, scrapie samples produced bands with apparent molecular masses of approximately 36, 28, 23, and 19 kDa, while BSE samples produced poorly resolved bands with apparent molecular masses of approximately 36 and 28 kDa. Such Western blot banding patterns were obtained with four experimental BSE animals of PrP genotype ARQ/ARQ as well as 15 scrapie field isolates with a range of PrP genotypes: ARQ/ARQ, VRQ/VRQ, ARQ/VRQ, and AHQ/AHQ (Table 1). For all of these animals, both PrP genotype and sheep breed appeared to have no effect on molecular strain typing. It was noted that the detection in scrapie samples of the band with the highest molecular mass varied between homogenate preparations. Individual samples were subjected to multiple analyses (up to two separate homogenate preparations analyzed up to eight times) and gave equivalent results between analyses. Similar digestions of healthy tissue resulted in the complete removal of any PrPC bands between 19 and 36 kDa after 1 h of digestion.
FIG. 1.
Ovine TSE strain discrimination by thermolysin digestion. Caudal medulla samples from TSE-affected sheep of various PrP genotypes were digested by the addition of 150 μg/ml thermolysin every hour for a total of 8 h. (A) Resulting PrP fragments after 1, 4, 6, and 8 h of digestion are shown in lanes 1 to 4, respectively. Densitometric analysis of the Western blots is also shown for the 8-h time points for BSE (B) and scrapie (C) samples. TSE type, animal references, and PrP genotype are indicated. Digested brain homogenate was loaded at 3.3 μl of 10% (wt/vol) per lane, and PrP was detected on Western blots with monoclonal antibody P4. Molecular mass markers are indicated in kilodaltons.
The differences between BSE and scrapie samples were further highlighted upon densitometric analysis of the Western blots, where scrapie samples produced the 23- and 19-kDa bands and the most prevalent band was that at 28 kDa (Fig. 1C). Conversely, BSE did not produce 23- and 19-kDa bands and the band at 36 kDa was at least equivalent in intensity to that at 28 kDa (Fig. 1B). These criteria were applied to routinely distinguish ovine BSE and scrapie.
CNS region deposition of thermolysin-resistant PrP in TSE-affected sheep.
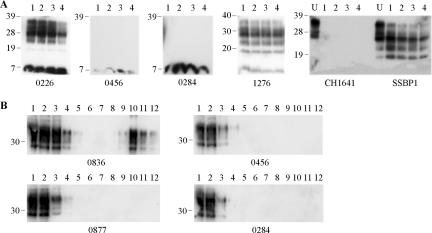
In order to determine the CNS distribution of thermolysin-resistant PrP in TSE-affected sheep, the thermolysin strain typing assay was applied to caudal medulla, spinal cord, and/or cerebellum samples from the same animals (Table 1). Spinal cords from 11 scrapie cases were assayed and, in all instances, gave the same scrapie-specific PrPres molecular profile as that of the caudal medulla samples. Similarly, for BSE samples, the caudal medulla and cerebellum gave identical BSE-specific PrPres profiles. However, the analysis of cerebellum samples from 14 scrapie-affected animals revealed two distinct outcomes; 12 of the samples produced scrapie-specific molecular signatures for PrPres, but two samples, from animals 0456 and 0284, did not yield any detectable P4-reactive, thermolysin-resistant PrP (Fig. 2A; Table 1). Western blots of the cerebellar digests from animals 0456 and 0284 were also probed with monoclonal antibody L42, confirming the absence of thermolysin-resistant PrP within these samples under the assay conditions used (data not shown). It should be noted that a 7-kDa P4-reactive band was present in all samples, including those from TSE-affected and healthy animals (data not shown), and represents an N-terminal fragment of PrP that does not contain favored thermolysin cleavage sites (31). To further investigate the presence of disease-associated PrP species within the cerebellum of animals 0284 and 0456, homogenates were analyzed on sucrose gradients (Fig. 2B). Cerebellum homogenates from these two animals gave profiles indistinguishable from those of similar samples from healthy animals (animal 0877 [Fig. 2B]) and did not contain disease-associated PrP aggregates that were readily identified in the cerebellum of other scrapie-affected animals (animal 0836 [Fig. 2B, lanes 9 to 12]).
FIG. 2.
Analysis of ovine cerebellum samples from clinical scrapie-affected animals. (A) Samples were digested by the addition of 150 μg/ml thermolysin every hour for a total of 8 h. Resulting digests after 1, 4, 6, and 8 h of digestion are shown in lanes 1 to 4, respectively. For experimental scrapie strains SSBP1 and CH1641, undigested controls (U) are also shown. Digested brain homogenate was loaded at 3.3 μl of 10% (wt/vol) per lane, and undigested samples were loaded at 3.3 μl of 2% (wt/vol) per lane. PrP was detected on Western blots with monoclonal antibody P4. (B) Samples (200 μl of 10% [wt/vol] cerebellum homogenate) were resolved through a 10 to 60% sucrose step gradient and 12 0.42-ml fractions were collected. Each fraction (18 μl) was analyzed by Western blotting (lanes 1 to 12 for fractions taken from the top to bottom of the sucrose gradients), and PrP was detected with monoclonal antibody SAF32. For all blots, animal reference numbers and molecular mass markers (kilodaltons) are indicated. All samples in panel A are from clinical scrapie-affected animals; in panel B, animals 0836, 0456, and 0284 are clinical scrapie cases, and animal 0877 is a healthy control.
Cerebellum was also analyzed from clinical animals experimentally infected with scrapie strains CH1641 and SSBP1. Material from these animals produced very different depositions of PrPres within the cerebellum: under the assay conditions used, SSBP1 gave scrapie-associated banding patterns of 36, 28, 23, and 19 kDa, whereas the CH1641 sample produced no detectable PrPres bands after only 1 h of digestion with thermolysin (Fig. 2A).
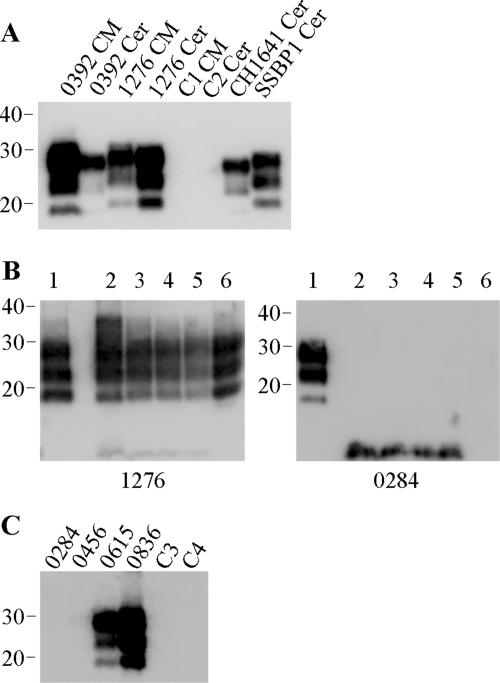
The presence of PK-resistant PrP within all samples was also assessed and correlated strongly with the presence of thermolysin-resistant PrP species (Fig. 4). Such analyses confirmed a lack of PrPres within cerebellum material from animals 0284 and 0456 (Fig. 4B and C). Interestingly, the analysis of cerebellum samples from the CH1641-infected animal revealed a discrepancy in the correlation of PrPres after PK and thermolysin digestions; thermolysin-resistant PrP was not detected (Fig. 2A), yet PK-resistant PrP was readily identified (Fig. 4A). PK-resistant PrP from the CH1641-infected animal displayed molecular profiles associated with this strain of scrapie (36): apparent molecular masses which were indistinguishable from BSE samples but lower than those for PK-resistant PrP from other scrapie-affected animals.
FIG. 4.
PK digestion of ovine TSE samples. (A) Caudal medulla (CM) or cerebellum (Cer) samples from BSE (0392)-affected or scrapie (1276, CH1641, and SSBP1)-affected sheep, along with healthy controls (C1 and C2), were digested by the addition of 50 μg/ml PK for 1 h. (B) Caudal medulla (lane 1) or cerebellum (lanes 2 to 6) samples from scrapie-affected sheep (animals 1276 and 0284) were digested by the addition of 50 μg/ml PK for 1 h (lanes 1 and 6) or by the addition of 150 μg/ml thermolysin every hour for a total of 8 h. For thermolysin digestions, resulting PrP fragments after 1, 4, 6, and 8 h of digestion are shown in lanes 2 to 5, respectively. (C) Cerebellum samples from scrapie-affected sheep (animals 0284, 0456, 0615, and 0836), along with healthy controls (C3 and C4), were digested by the addition of 50 μg/ml PK for 1 h. For all lanes, digested brain homogenate was loaded at 3.3 μl of 10% (wt/vol) per lane, and PrP was detected on Western blots with monoclonal antibody P4 (B and C) or L42 (A). Molecular mass markers are indicated in kilodaltons.
Endogenous PrP cleavage within the CNSs of TSE-affected sheep.
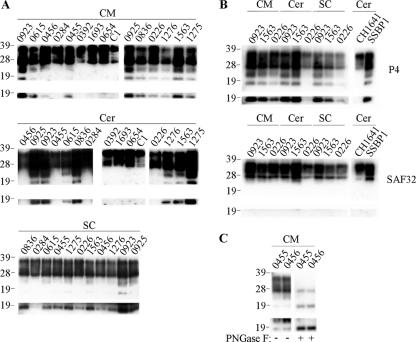
The occurrence of N-terminally truncated, disease-associated PrP species has been described for scrapie-infected mice (32) and CJD-affected humans (11). These PrP species are reported to be the glycosylated and nonglycosylated versions of a single PrP fragment, designated C2. Here, we investigated whether such PrP species were also present within a natural scrapie host. Caudal medulla, spinal cord, and cerebellum samples from 11 scrapie-affected sheep of various PrP genotypes were analyzed for the presence of C2 PrP fragments (Fig. 3A and Table 1). In seven animals, truncated PrP fragments with apparent molecular masses of 23 and 19 kDa were readily detected within all CNS samples. A further three animals (0456, 0455, and 0226) produced such fragments in samples from the spinal cord and caudal medulla but not the cerebellum. A further scrapie isolate (0284) did not produce any detectable truncated PrP species in any of the CNS regions tested. Interestingly, the two animals that did not contain PrPres in their cerebella, 0284 and 0456, also lacked detectable, endogenously produced, truncated PrP species within this brain region. In addition, we examined BSE-affected sheep and found that C2 PrP species were not detected in either the cerebellum or caudal medulla brain region (animals 0392, 1693, and 0654) (Fig. 3A). Similarly, healthy animals with various PrP genotypes did not produce any detectable truncated PrP species (e.g., animal C1 in Fig. 3A) in the spinal cord (n = 11), cerebellum (n = 3), or caudal medulla (n = 14). Cerebellum samples from experimentally infected scrapie animals were also analyzed. The SSBP1-infected animal produced truncated PrP species within the cerebellum, whereas the CH1641-infected animal did not produce any detectable C2 fragments (Fig. 3B). These results highlight the apparent strain-specific nature of the endogenous cleavage of PrP within TSE-affected sheep.
FIG. 3.
C2 PrP species within the CNS of TSE-affected sheep. Samples from clinical scrapie- or BSE-affected sheep of various PrP genotypes were analyzed. Brain homogenates (3.3 μl of 2% [wt/vol]) were analyzed directly (panels A and B) or were deglycosylated before analysis (panel C as indicated). PrP was detected on Western blots with P4 (all panels) or SAF32 (panel B as indicated). Included were caudal medulla (CM), spinal cord (SC), and cerebellum (Cer) samples. Material from field isolates of scrapie (from animals 0923, 0615, 0456, 0284, 0455, 0925, 0836, 0226, 1276, 1563, and 1275) as well as experimental scrapie (from strains CH1641 and SSBP1), experimental BSE (from animals 0392, 1693, and 0654), and samples from a healthy control animal (C1) are shown. Prolonged exposures of the blots are shown in the bottom panels, highlighting the presence of unglycosylated C2 fragment. For all blots, animal reference numbers and molecular mass markers (kilodaltons) are indicated. −, absence of PNGase F treatment; +, presence of PNGase F treatment.
In order to determine the location of the cleavage of scrapie-associated PrP that yields bands of 23 and 19 kDa, samples were probed with either P4 or SAF32 monoclonal antibodies (Fig. 3B). Truncated PrP species were detected with only P4, indicating that both the 23- and 19-kDa bands contain similar N-terminal cleavage sites and that these sites are located between the SAF32 and P4 epitopes. The minimum binding sequence for SAF32 is QPHGGG (31), the most C-terminal site of this repeated sequence within PrP is at residues 86 to 91, and P4 binds within the region of residues 89 to 104. Therefore, endogenous cleavage of scrapie-associated ovine PrP appears to be within the region spanned by residues 86 and 104. This is in agreement with these ovine PrP fragments being C2, which in other species are reported to be cleaved at residue Trp93 (11, 26, 32, 43).
The apparent molecular masses for C2 in the present study appear to be inconsistent with published data describing human and murine PrP, where C2 has an apparent molecular mass of ∼21 kDa and a glycosylated form of ∼26 kDa (11, 26, 32, 43). These reports also note that the C2 fragment corresponds in apparent molecular mass and antibody recognition to the unglycosylated band produced upon PK digestion of PrPSc. Here, we confirmed that the 19-kDa band possessed these traits and therefore corresponds to fragment C2 (data not shown). Also, upon treatment of samples with PNGase F, the 23- and 19-kDa bands produced a single band of approximately 19 kDa (Fig. 3C), indicating that the 23-kDa band is likely to correspond to the 26-kDa glycosylated C2 fragment previously described (32). These inconsistencies in apparent molecular mass are most likely due to methodological differences and highlight the difficulty of comparing the apparent molecular masses of PrP fragments between studies.
DISCUSSION
Here we describe a novel strain typing test that uses the protease thermolysin to produce PrPres Western blot profiles. In total, 15 scrapie isolates were compared to isolates of four BSE animals, and in all cases, caudal medulla samples yielded scrapie or BSE-specific PrPres fragment patterns. With all samples tested to date, PrP genotype and sheep breed appeared to have no effect on the molecular profiles of the scrapie samples. This test has several advantages over previously reported rapid strain typing tests (21, 36, 37): it does not rely on subtle differences in the molecular masses of PrPres bands resulting from PK digestion of BSE and scrapie samples; it does not require the multiple analyses of samples as does molecular mass and glycoform ratio determination; and similarly, it does not rely on the analysis of samples on multiple gels to allow the assessment of quantitative differences in the binding of anti-PrP antibodies.
It was noted that the analysis of caudal medulla samples from multiple scrapie field isolates produced indistinguishable PrPres profiles. To further investigate the deposition and molecular profiles of PrPres in scrapie-affected sheep, we applied the thermolysin strain typing test to caudal medulla, cerebellum, and spinal cord samples of 11 scrapie isolates and to caudal medulla and cerebellum samples from a further 3 isolates. All spinal cord and caudal medulla samples produced typical scrapie PrPres profiles. However, the analysis of cerebellum samples produced two distinct PrPres deposition patterns: 12 isolates produced a scrapie-type PrPres profile with significant PrPres signals present after eight sequential 1-h digestions with thermolysin, and in contrast, two isolates lacked any detectable cerebellar PrPres after just 1 h of digestion with the protease. This lack of PrPres within these samples was confirmed by Western blot analysis of PK digests. Furthermore, the cerebellar samples of these animals lacked any detectable disease-associated PrP aggregates upon sucrose gradient analysis, indicating a lack of any disease-associated PrP in this brain region.
It has been suggested that different PrPSc strains target different brain regions (13), and it seems plausible that the distinct scrapie isolates described in this study are indeed different naturally occurring scrapie strains. Such cases would represent variants of “classical” scrapie, as PrPSc, where detected, possessed resistance to relatively high concentrations of protease and produced “typical” PrPres banding patterns (Fig. 1 and 4). This is in contrast to “atypical” scrapie cases which display disparate PrPres distribution within the CNS compared to that of classical scrapie but have PrPres with relatively low protease resistance and often produce PrPres fragments that are distinct from those of typical scrapie (6, 15, 28). The current study highlights that caution is necessary for the selection of brain material to determine the TSE status of small ruminants, as disease-associated PrP may well be absent from certain CNS regions in TSE-infected animals.
Cerebellum samples were also analyzed from clinical sheep infected experimentally with two characterized scrapie strains, SSBP1 and CH1641. These samples also produced very different depositions of PrPres within the cerebellum: SSBP1 gave scrapie-associated banding patterns even after eight repeat digestions with thermolysin, whereas the CH1641 sample did not produce any PrPres bands after just 1 h of digestion with protease. Interestingly, while this sample lacked any detectable thermolysin-resistant PrP, PK-resistant PrP species were readily detected. This result is in contrast to those with all other samples analyzed which contained PrP with similar qualitative resistance to both proteases under the assay conditions used. Such discrepancy in protease susceptibility appears to be confined to CH1641 and may provide the basis for the rapid molecular differentiation of this scrapie strain from ovine BSE. These data appear to support the hypothesis that the CNS region-specific deposition of disease-associated PrP, along with its biochemical properties, is linked to TSE strain.
It has been suggested for CJD that the exact profile of PrPSc fragments, and not merely the characteristics of protease-resistant cores, correlates with neuropathological phenotypes (26). To date, two major PrP fragments have been described within both human and murine tissues. First is the C1 fragment of PrP, which is the major in vivo-generated fragment of PrPC, resulting from disintegrin-mediated cleavage at residue His114 or Val115 (40). Second is the C2 fragment, which is present in very small amounts in healthy individuals but accumulates during TSE disease and is due to calpain-dependent cleavage of PrP at residue Trp93 (11, 26, 32, 43). The C2 fragment is known to copurify with PrPSc aggregates, and unlike the C1 fragment, it contains the so-called amyloidogenic region of PrP purported to play a critical role in the conversion of PrPC to PrPSc (16, 39). As such, it has been speculated that the accumulation of the C2 fragment may represent an important pathological event in prion diseases (11, 26, 32, 43). To investigate the occurrence and CNS deposition of TSE-associated PrP fragments within sheep, we analyzed caudal medulla, spinal cord, and cerebellar homogenates for the presence of P4-reactive-truncated PrP. Antibody P4 is predicted to bind towards the extreme N terminus of C2 and not recognize C1 fragments. Samples from a range of healthy sheep did not contain any detectable C2 fragment. Similarly, C2 fragment was not detected in cerebellum and caudal medulla samples from BSE-affected sheep. For natural scrapie isolates, seven animals produced C2 PrP in all CNS regions analyzed. A further three scrapie-affected animals produced C2 fragments in samples from the spinal cord and caudal medulla but not from the cerebellum, and one further animal did not produce readily detectable C2 fragments in any of the CNS regions. As far as we are aware, this is the first detailed description of the advent of C2 fragments within a natural host of scrapie and builds upon the identification of scrapie-associated C2 observed with murine scrapie and the preliminary observation of such a PrP fragment in a single ovine scrapie sample (8). Furthermore, IHC analysis results by “PrP peptide mapping” (24, 25) have described the endogenous cleavage of intracellular PrPSc in scrapie-affected animals, which results in truncated PrPSc that contains the P4 epitope. This intracellular truncated PrPSc represents one potential source of the C2 PrP described in the present study.
The present study also indicates that the extent of C2 PrP accumulation within defined CNS regions may be TSE strain specific. The C2 fragment was present within SSBP1 cerebellum but was not detectable in the equivalent sample from a CH1641-affected sheep. Furthermore, C2 PrP fragment was detected within the caudal medulla of 10 out of 11 scrapie field isolates; this result is in contrast to isolates from the three BSE-affected sheep that were analyzed, which lacked detectable C2 in this brain region. These results appear to support those of a previous study that demonstrated quantitative differences in the deposition of the C2 fragment within whole murine brains from animals affected by different scrapie strains (32). In addition, human Gerstmann-Sträussler-Scheinker and CJD samples have been shown to be distinguishable through endogenously generated disease-associated PrP fragments (26). Previous IHC studies show that intracellular BSE PrPSc is truncated C terminal of the P4 epitope and can be cleaved at different sites, depending upon the cell type (20, 24, 25). The present study further demonstrates that endogenously truncated, P4-reactive PrPSc is not readily detectable in BSE-infected CNS material. Furthermore, the CNS region-specific deposition of the C2 fragment within scrapie-affected animals is in agreement with the reported distinct PrPSc processing within different cell types and brain regions (19, 20, 24, 25, 35).
The present study offers evidence that links the neuroanatomical deposition of C2 and PrPres to TSE strain. However, for naturally infected animals, certain caveats remain. It is unknown what effect factors such as route of infection, infectivity dose, or age of exposure may have on the presentation of disease-associated PrP within the host. In addition, for the analysis of C2 fragments, the occurrence of postmortem autolysis may result in the production of PrP fragments. However, several murine and human studies have examined biopsy samples and, along with the use of cell-culture systems, have demonstrated that the production of the C2 fragment is an in vivo event (10, 11, 26, 43). In the present study, no healthy or BSE samples produced any detectable C2 fragments and samples from these animals were harvested and snap-frozen during a 12-h postmortem procedure. For scrapie-affected animals, samples were taken by using a procedure identical to the 12-h postmortem procedure or during a 1-h autopsy. There was no correlation between the autopsy procedure and the occurrence and levels of detectable C2 fragments. In addition, homogenates were prepared in the presence or absence of protease inhibitors and gave identical C2 profiles (data not shown). Together, these data indicate that C2 fragments are likely to be produced in vivo and are not due to differences in postmortem sample processing.
In the present study, we developed a novel strain typing assay that can readily differentiate ovine BSE from all scrapie samples; moreover, the presence of thermolysin-resistant PrPres and PrP C2 fragment has been used to characterize the molecular profiles of disease-associated PrP species within sheep affected by scrapie or BSE. The distinct deposition of scrapie-associated PrP species within three neuroanatomical regions allowed the grouping of classical scrapie field cases into four groups: the majority of isolates produced C2 and PrPres in all CNS regions, a second group lacked detectable cerebellar C2 fragment, a single isolate is described that lacked both PrPres and detectable C2 within the cerebellum, and a single isolate lacked detectable C2 within all CNS regions analyzed and also lacked cerebellar PrPres. These groups may represent strains of classical scrapie. Further study of these groups by experimental passage should elucidate whether they produce distinct pathology under experimental conditions and whether their molecular traits are transmissible. Such analysis would validate the use of PrPres and C2 profiles as markers of classical scrapie strains within the natural host.
Acknowledgments
The work was funded by the Department for Environment, Food and Rural Affairs (United Kingdom) (project SE1792).
We thank J. Grassi (CEA Saclay, Gif/Yvette, France) for the generous gift of SAF32 monoclonal antibody and the TSE Archive at the VLA (Addlestone, Surrey, United Kingdom) for the provision of tissue samples. We also thank N. Hunter (IAH Neuropathogenesis Unit, Edinburgh, United Kingdom) for her kind permission to use the CH1641 sample.
Footnotes
Published ahead of print on 25 July 2007.
REFERENCES
- 1.Acutis, P. L., F. Martucci, M. Mazza, S. Nodari, C. Maurella, G. Ru, C. Casalone, and M. Caramelli. 2006. Molecular typing of transmissible spongiform encephalopathy from Italian disease outbreaks in small ruminants. Vet. Rec. 159:746-747. [DOI] [PubMed] [Google Scholar]
- 2.Baron, T., J.-Y. Madec, and D. Cavalos. 1999. Similar signature of the prion protein in natural sheep scrapie and bovine spongiform encephalopathy-linked diseases. J. Clin. Microbiol. 37:3701-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, T., J.-Y. Madec, D. Calavas, Y. Richard, and F. Barillet. 2000. Comparison of French natural scrapie isolates with bovine spongiform encephalopathy and experimental scrapie infected sheep. Neurosci. Lett. 284:175-178. [DOI] [PubMed] [Google Scholar]
- 4.Begara-McGorum, I., L. González, M. Simmons, N. Hunter, F. Houston, and M. Jeffrey. 2002. Vacuolar lesion profile in sheep scrapie: factors influencing its variation and relationship to disease-specific PrP accumulation. J. Comp. Pathol. 127:59-68. [DOI] [PubMed] [Google Scholar]
- 5.Bellworthy, S. J., S. A. Hawkins, R. B. Green, I. Blamire, G. Dexter, I. Dexter, R. Lockey, M. Jeffrey, S. Ryder, C. Berthelin-Baker, and M. M. Simmons. 2005. Tissue distribution of bovine spongiform encephalopathy infectivity in Romney sheep up to the onset of clinical disease after oral challenge. Vet. Rec. 156:197-202. [DOI] [PubMed] [Google Scholar]
- 6.Benestad, S. L., P. Sarradin, B. Thu, J. Schonheit, M. A. Tranulis, and B. Bratberg. 2003. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet. Rec. 153:202-208. [DOI] [PubMed] [Google Scholar]
- 7.Bessen, R. A., and R. F. Marsh. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68:7859-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossers, A., P. B. G. M. Belt, G. J. Raymond, B. Caughey, R. De Vries, and M. A. Smits. 1997. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl. Acad. Sci. USA 94:4931-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruce, M. E., I. McConnell, H. Fraser, and A. G. Dickinson. 1991. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 72:595-603. [DOI] [PubMed] [Google Scholar]
- 10.Caughey, B., G. J. Raymond, D. Ernst, and R. E. Race. 1991. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol. 65:6597-6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, S. G., D. B. Teplow, P. Parchi, J. K. Teller, P. Gambetti, and L. Autilio-Gambetti. 1995. Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 270:19173-19180. [DOI] [PubMed] [Google Scholar]
- 12.Collinge, J., K. C. L. Sidle, J. Meads, J. Ironside, and A. F. Hill. 1996. Molecular analysis of prion strain variation and the aetiology of “new variant” CJD. Nature 383:685-690. [DOI] [PubMed] [Google Scholar]
- 13.DeArmond, S. J., Y. Qiu, H. Sanchez, P. R. Spilman, A. Ninchak-Casey, D. Alonso, and V. Daggett. 1999. PrPc glycoform heterogeneity as a function of brain region: implications for selective targeting of neurons by prion strains. J. Neuropathol. Exp. Neurol. 58:1000-1009. [DOI] [PubMed] [Google Scholar]
- 14.Dickinson, A. G. 1976. Scrapie in sheep and goats, p. 209-239. In R. H. Kimberlin (ed.), Slow virus diseases of animals and man. North-Holland Publishing Company, Amsterdam, The Netherlands.
- 15.Everest, S. J., L. Thorne, D. A. Barnicle, J. C. Edwards, H. Elliott, R. Jackman, and J. Hope. 2006. Atypical prion protein in sheep brain collected during the British scrapie-surveillance programme. J. Gen. Virol. 87:471-477. [DOI] [PubMed] [Google Scholar]
- 16.Forloni, G., N. Angeretti, R. Chiesa, E. Monzani, M. Salmona, O. Bugiani, and F. Tagliavini. 1993. Neurotoxicity of a prion protein fragment. Nature 362:543-546. [DOI] [PubMed] [Google Scholar]
- 17.Foster, J. D., and A. G. Dickinson. 1988. The unusual properties of CH1641, a sheep-passaged isolate of scrapie. Vet. Rec. 123:5-8. [DOI] [PubMed] [Google Scholar]
- 18.Goldmann, W., N. Hunter, J. D. Foster, J. M. Salbum, K. Beyreuther, and J. Hope. 1990. Two alleles of neural protein gene linked to scrapie in sheep. Proc. Natl. Acad. Sci. USA 87:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González, L., S. Martin, I. Begara-McGorum, N. Hunter, F. Houston, M. Simmons, and M. Jeffrey. 2002. Effects of agent strain and host genotype on PrP accumulation in the brain of sheep naturally and experimentally affected with scrapie. J. Comp. Pathol. 126:17-29. [DOI] [PubMed] [Google Scholar]
- 20.González, L., S. Martin, and M. Jeffrey. 2003. Distinct profiles of PrPd immunoreactivity in the brain of scrapie- and BSE-infected sheep: implications for differential cell targeting and PrP processing. J. Gen. Virol. 84:1339-1350. [DOI] [PubMed] [Google Scholar]
- 21.Gretzschel, A., A. Buschmann, M. Eiden, U. Ziegler, G. Lühken, G. Erhardt, and M. H. Groschup. 2005. Strain typing of German transmissible spongiform encephalopathies field cases in small ruminants by biochemical methods. J. Vet. Med. Ser. B 52:55-63. [DOI] [PubMed] [Google Scholar]
- 22.Hill, A. F., K. C. Sidle, S. Joiner, P. Keyes, T. C. Martin, M. Dawson, and J. Collinge. 1998. Molecular screening of sheep for bovine spongiform encephalopathy. Neurosci. Lett. 255:159-162. [DOI] [PubMed] [Google Scholar]
- 23.Hope, J., S. C. Wood, C. R. Birkett, A. Chong, M. E. Bruce, D. Cairns, W. Goldmann, N. Hunter, and C. J. Bostock. 1999. Molecular analysis of ovine prion protein identifies similarities between BSE and an experimental isolate of natural scrapie, CH1641. J. Gen. Virol. 80:1-4. [DOI] [PubMed] [Google Scholar]
- 24.Jeffrey, M., L. Gonzalez, A. Chong, J. Foster, W. Goldmann, N. Hunter, and S. Martin. 2006. Ovine infection with the agents of scrapie (CH1641 isolate) and bovine spongiform encephalopathy: immunochemical similarities can be resolved by immunohistochemistry. J. Comp. Pathol. 134:17-29. [DOI] [PubMed] [Google Scholar]
- 25.Jeffrey, M., S. Martin, and L. Gonzalez. 2003. Cell-associated variants of disease-specific protein immunolabelling are found in different sources of sheep transmissible spongiform encephalopathy. J. Gen. Virol. 84:1033-1046. [DOI] [PubMed] [Google Scholar]
- 26.Jiménez-Huete, A., P. M. Lievens, R. Vidal, P. Piccardo, B. Ghetti, F. Tagliavini, B. Frangione, and F. Prelli. 1998. Endogenous proteolytic cleavage of normal and disease-associated isoforms of the human prion protein in neural and non-neural tissues. Am. J. Pathol. 153:1561-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalili-Shirazi, A., L. Summers, J. Linehan, G. Mallinson, D. Anstee, S. Hawke, G. S. Jackson, and J. Collinge. 2005. PrP glycoforms are associated in a strain-specific ratio in native PrPSc. J. Gen. Virol. 86:2635-2644. [DOI] [PubMed] [Google Scholar]
- 28.Le Dur, A., V. Béringue, O. Andréoletti, F. Reine, L. L. Thanh, T. Baron, B. Bratberg, J.-L. Vilotte, P. Sarradin, S. L. Benestad, and H. Laude. 2005. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc. Natl. Acad. Sci. USA 102:16031-16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lezmi, S., S. Martin, S. Simon, E. Comoy, A. Bencsik, J. P. Deslys, J. Grassi, M. Jeffrey, and T. Baron. 2004. Comparative molecular analysis of the abnormal prion protein in field scrapie cases and experimental bovine spongiform encephalopathy in sheep by use of Western blotting and immunohistochemical methods. J. Virol. 78:3654-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ligios, C., M. Jeffrey, S. J. Ryder, S. J. Bellworthy, and M. M. Simmons. 2002. Distinction of scrapie phenotypes in sheep by lesion profiling. J. Comp. Pathol. 127:45-57. [DOI] [PubMed] [Google Scholar]
- 31.Owen, J. P., B. C. Maddison, G. C. Whitelam, and K. C. Gough. 2007. Use of thermolysin in the diagnosis of prion diseases. Mol. Biotechnol. 35:161-170. [DOI] [PubMed] [Google Scholar]
- 32.Pan, T., P. Wong, B. Chang, C. Li, R. Li, S. C. Kang, T. Wisniewski, and M. S. Sy. 2005. Biochemical fingerprints of prion infection: accumulations of aberrant full-length and N-terminally truncated PrP species are common features in mouse prion disease. J. Virol. 79:934-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA 95:13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safar, J., H. Wille, V. Itri, D. Groth, H. Serban, M. Torchia, F. E. Cohen, and S. B. Prusiner. 1998. Eight prion strains have PrPSc molecules with different conformations. Nat. Med. 4:1157-1165. [DOI] [PubMed] [Google Scholar]
- 35.Somerville, R. A. 1999. Host and transmissible spongiform encephalopathy agent strain control glycosylation of PrP. J. Gen. Virol. 80:1865-1872. [DOI] [PubMed] [Google Scholar]
- 36.Stack, M., M. J. Chaplin, and J. Clark. 2002. Differentiation of prion protein glycoforms from naturally occurring sheep scrapie, sheep-passaged scrapie strains (CH1641 and SSBP1), bovine spongiform encephalopathy (BSE) cases and Romney and Cheviot breed sheep experimentally inoculated with BSE using two monoclonal antibodies. Acta Neuropathol. 104:279-286. [DOI] [PubMed] [Google Scholar]
- 37.Stack, M., M. Jeffrey, S. Gubbins, S. Grimmer, L. Gonzalez, S. Martin, M. Chaplin, P. Webb, M. Simmons, Y. Spencer, P. Bellerby, J. Hope, J. Wilesmith, and D. Matthews. 2006. Monitoring for bovine spongiform encephalopathy in sheep in Great Britain, 1998-2004. J. Gen. Virol. 87:2099-2107. [DOI] [PubMed] [Google Scholar]
- 38.Sweeney, T., T. Kuczuis, M. McElroy, M. G. Parada, and M. H. Groschup. 2000. Molecular analysis of Irish sheep scrapie cases. J. Gen. Virol. 81:1621-1627. [DOI] [PubMed] [Google Scholar]
- 39.Tagliavini, F., F. Prelli, L. Verga, G. Giaccone, R. Sarma, P. Gorevic, B. Ghetti, F. Passerini, E. Ghibaudi, G. Forloni, M. Salmona, O. Bugiani, and B. Frangione. 1993. Synthetic peptides homologous to prion protein residues 106-147 form amyloid-like fibrils in vitro. Proc. Natl. Acad. Sci. USA 90:9678-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vincent, B., E. Paitel, P. Saftig, Y. Frobert, D. Hartmann, B. de Strooper, J. Grassi, E. Lopez-Perez, and F. Checler. 2001. The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol-esters-regulated normal cleavage of the cellular prion protein. J. Biol. Chem. 276:37743-37746. [DOI] [PubMed] [Google Scholar]
- 41.Wadsworth, J. D., A. F. Hill, S. Joiner, G. S. Jackson, A. R. Clarke, and J. Collinge. 1999. Strain-specific prion-protein conformation determined by metal ions. Nat. Cell Biol. 1:55-59. [DOI] [PubMed] [Google Scholar]
- 42.Wood, J. L., I. McGill, S. H. Done, and R. Bradley. 1997. Neuropathology of scrapie: a study of the distribution of brain lesions in 222 cases of natural scrapie in sheep. Vet. Rec. 140:167-174. [DOI] [PubMed] [Google Scholar]
- 43.Yadavalli, R., R. P. Guttmann, T. Seward, A. P. Centers, R. A. Williamson, and G. C. Telling. 2004. Calpain-dependent endoproteolytic cleavage of PrPSc modulates scrapie prion propagation. J. Biol. Chem. 279:21948-21956. [DOI] [PubMed] [Google Scholar]